Abstract
A growing number of web-based databases and tools for protein research are being developed. There is now a widespread need for visualization tools to present the three-dimensional (3D) structure of proteins in web browsers. Here, we introduce our 3D modeling program—Web3DMol—a web application focusing on protein structure visualization in modern web browsers. Users submit a PDB identification code or select a PDB archive from their local disk, and Web3DMol will display and allow interactive manipulation of the 3D structure. Featured functions, such as sequence plot, fragment segmentation, measure tool and meta-information display, are offered for users to gain a better understanding of protein structure. Easy-to-use APIs are available for developers to reuse and extend Web3DMol. Web3DMol can be freely accessed at http://web3dmol.duapp.com/, and the source code is distributed under the MIT license.
INTRODUCTION
Protein structure visualization is an essential task in bioinformatics and structural biology. Owing to the rendering efficiency bottleneck of three-dimensional (3D) computer graphics, traditional protein structure viewers are often limited to the native desktop environment (1–3). PyMOL (http://www.pymol.org/) is a commonly used and representative software for molecular visualization, owing to its high rendering efficiency and fine graphical quality. However, beginners, especially users in medical or biological fields, must devote time and energy to gain familiarity with the command-line interaction of PyMOL, and it is inconvenient for desktop software to present their graphical results through the Internet.
Unlike desktop software, web applications are more flexible in fetching, handling and sharing online resources. However, until recently, web browsers have not able to render 3D graphics without the support from additional plugins. Jmol (http://sourceforge.net/projects/jmol/) (4) and OpenAstexViewer (http://www.openastexviewer.net/) (5) are web-based molecule viewers implemented as Java Applets. Their performance is good, but a Java runtime plugin must be preinstalled into every web browser accessing Jmol or OpenAstexViewer, and a series of security options must be found and checked off before the programs can finally run. Given the complexity and insecurity of Java Applets, JSmol (http://sourceforge.net/projects/jsmol/) (6) was created as a Jmol interpreter. JSmol directly executes without the support of plugins, but its performance is not yet adequate—obvious lagging can be observed, particularly with large and complex 3D visualizations.
With the popularity of HTML5, WebGL (http://www.khronos.org/webgl/) has been widely accepted and supported by web browser manufacturers. As an embedded implementation of OpenGL (http://www.opengl.org/), WebGL has built-in JavaScript APIs to render computer graphics by using graphics processing unit (GPU) acceleration. These features make WebGL an ideal substitute for Java Applet or Flash for 3D presentation (7). GLmol (http://www.glmol.com/), PV (http://github.com/biasmv/pv), NGL (http://github.com/arose/ngl) (8) and 3DMol.js (http://3dmol.csb.pitt.edu/) (9) are representative web-based molecule viewers using WebGL. GLmol and NGL are based on a packaged framework called Three.js (http://threejs.org/), while PV and 3DMol.js directly use low-level APIs. These tools offer basic solutions to present molecules in web browsers. GLmol is reused in several applications, such as iview (http://istar.cse.cuhk.edu.hk/iview/) (10) and ChemDoodle (http://web.chemdoodle.com/) (11); PV is the default visualization engine of SWISS-MODEL (http://www.swissmodel.expasy.org/) (12); and both NGL and PV have been adopted by the RCSB Protein Data Bank (http://www.rcsb.org/) (13) as online viewers. However, these applications have some insufficiencies: the performance of GLmol is not stable during rendering of large and complex molecules; the representation and functionality of PV and 3DMol.js is not diverse in comparison with the alternatives; and NGL is not flexible enough to present a molecule in an integrated way, that is to say, different parts of a protein cannot be presented as different modes.
To date, the web-based viewers described above and other similar viewers focused solely on the visualization of 3D structures, while some meta-information recorded in PDB archives, such as primary sequence, source of organism and crystallographic information, is entirely ignored.
Here, we present Web3DMol, a novel web-based protein structure visualization tool that provides interactive 3D presentation of protein structures and allows users without any programing experience to construct desirable graphical results with minimal time and effort. Compared with alternative web-based molecule viewers, Web3DMol has unique features. For example, the primary and secondary structures of the visualized proteins are illustrated as a plot in order to reveal the correspondence between the linear sequence and 3D structure. Different parts of a molecule can be presented as different modes by using the fragment segmentation. Distances and angles among atoms in a molecule can be measured through simple mouse interactions. Related meta-information recorded in the PDB archive is also listed for further exploration. The only requirement to use Web3DMol is a web browser that supports HTML5, no matter on Windows, Mac OS or Linux with graphical interface (Supplementary Table S1).
MATERIALS AND METHODS
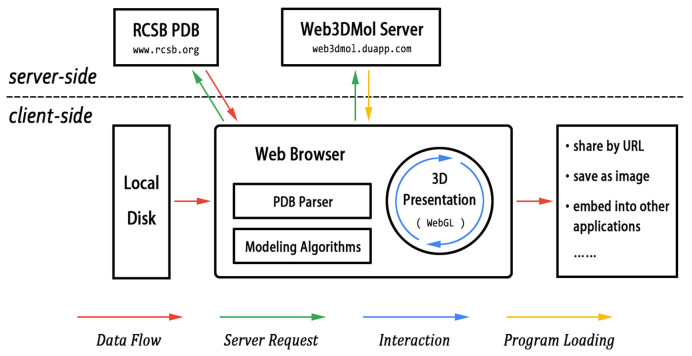
The technology architecture of Web3DMol is shown in Figure 1. An easy way to access Web3DMol is by visiting http://web3dmol.duapp.com/ in a modern web browser. Notably, although Web3DMol can be accessed through a URL, the program is implemented with JavaScript and completely executes on client-side. Thus, Web3DMol can also be run by downloading the source code to a local computer and then opening the HTML file in a web browser.
Figure 1.
The architecture of Web3DMol. The web browser plays a key role in Web3DMol, in which server requests are initiated, PDB archives are parsed, modeling algorithms execute and graphical results are presented. The red arrows indicate that PDB data never flows from the client-side to the server-side.
The inputs of Web3DMol are PDB archives, which can be either loaded automatically from the RCSB PDB by submitting a four-digit identification code or loaded manually by users from their local disks. The PDB archive is then parsed line by line to extract useful information such as the 3D coordinates of atoms, residue sequences and secondary structures. Next, the modeling algorithms compute the vertices and triangles, which are used to compose the final 3D presentations. Users have an interactive visualization of the 3D structure that they can manipulate with simple mouse operations, and the graphical results can also be shared with others through URLs, saved as images or embedded into other web applications.
The web browser plays a key role in Web3DMol, in which PDB archives are parsed, modeling algorithms execute and graphical results are presented. As for the 3D graphical engine, Web3DMol uses WebGL, rather than Java 3D (in Java Applet) or Stage3D (in Flash), to obtain hardware acceleration as well as avoid the requirement of plugin pre-installation.
PRESENTATION
Diverse representation, color and label modes are supported to construct complex 3D graphics (Figure 2). To give users enough choices to control the presentation of every residue, we incorporated an important concept—fragment, which dramatically enhances the application's flexibility. A fragment is a segment of a chain in a molecule, and it has independent representation, color and label mode from the global configuration. Users can freely add and remove fragments to manipulate the details of the 3D structure and make the resulting graphics more appealing (Figure 2).
Figure 2.
The structure of a potassium channel (PDB ID: 1BL8) (14). This molecule consists of four chains, which are shown in different representation and color modes. Chain A is presented as stick mode colored by element; Chain B is presented as spheres colored by hydrophobicity; Chain C is presented as tubes colored by chain spectrum; Chain D is presented as cartoon mode colored by secondary structure. A region of chain B is labeled with B-Factor values. This integrated graphical result is constructed with several fragments.
Users can modify most of the default configuration values, including the size of geometries, color schemes, light effects, fog effects, surface material, labeling font, animation speeds and some auxiliary options. User-modified configurations can be saved in local storage, and when Web3DMol is restarted in the same web browser, the local configurations override the default ones.
FUNCTIONALITY
Correspondence
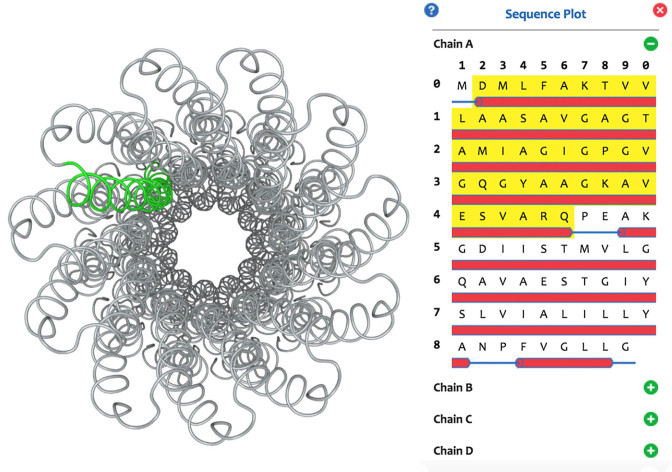
The primary and secondary structures of a protein molecule are illustrated as a sequence plot (Figure 3). The residue sequence is listed in order, and the α-helices, β-sheets and random coils are shown as small red cylinders, green arrows and blue strips, respectively. Single residues or secondary structures can be recognized automatically when the mouse hovers over the plot, and the recognized segment can be highlighted or hidden through left or right clicking. Pressing and holding the left button of the mouse adds the recognized segment as a fragment. By using the sequence plot, users can intuitively inspect the mapping from the linear sequence to 3D structure.
Figure 3.
The structure of a Na+-ATPase (PDB ID: 1YCE) (15) and its sequence plot. Correspondence between the linear sequence and 3D structure is illustrated by highlighting a region of the molecule.
Moreover, a segment can be picked up from the 3D structure and mapped to the linear sequence. Right clicking a region in the graphics, the information about residue, secondary structure and chain at the clicked site is listed in a pop-up dialog. Users can choose to highlight or hide these segments or simply add them as fragments.
Measurement
Geometric measurements are useful in structural biology and computational chemistry. Web3DMol supports several types of measurement, including distance, vector angle, dihedral angle and area among atoms, which can be carried out through simple mouse interactions. User can click ‘Add Measurement’ to start a measuring procedure, then clicking the 3D structure to assign the guide points order by A, B, C and D according to the illustration. The calculation results are depicted on the graphical canvas.
Meta-information
PDB archives record not only the 3D coordinate data but also some related information about the molecules, such as the molecular classification in biology, the structural resolution and the experimental details. Archives are written in plain text with a particular format, so it is possible, though tedious and error-prone for users to find the buried meta-information by reading the source files. Web3DMol retrieves and lists this meta-information for potential further exploration. For example, if a researcher is interested in a protein shown in Web3DMol, the original online page of the related paper can be accessed through the PubMed or the DOI links listed at the right side of Web3DMol.
Sharing by URL
It is inconvenient for desktop software to share graphical results through the Internet in an interactive way. The only feasible method is saving the results as static images, and then posting the images to web pages. Users have no interaction with this kind of presentation of protein structure. Web-based tools have the inherent ability to avoid incompatibility between the desktop and web environments, because these programs are originally run in web browsers. Web3DMol's graphical results can be easily shared with other users or presented on other web servers through the automatically generated sharing-URL. This URL copies the graphical results with complete interactive behaviors, such as rotating, zooming and translating. If an online database uses Web3DMol rather than static image to present protein structure in their web pages, users will be able to experience more intuitive observation and understanding of the molecule's 3D structure.
EXTENSIONS
There are two options for extending Web3DMol. For HTML developers, the <iframe> tag can be used to directly embed Web3DMol into web pages (Supplementary Figure S1A). The HTML code is very easy to write because the only requirement is a URL, which is set as the ‘src’ attribute of the <iframe> tag. The URL can be constructed manually with the configuration keys and values or simply by copying the sharing-URL instead. Another option is for JavaScript developers, which is somewhat more difficult but has much more flexibility because the visualization details can be controlled and customized. In this case, the Web3DMol library must initially be included by using a <script> tag; then, the initial function must be invoked to start the rendering process, and other functions are invoked to customize the graphical results (Supplementary Figure S1B). Well-formed JavaScript APIs are available, covering the entire procedure from the initialization to the output.
LIMITATIONS
3D modeling and rendering are both resource-intensive calculations, and as a form of interpreted language, JavaScript is not good at high-efficiency calculations. Therefore, when the size of a molecule becomes very large, Web3DMol sacrifices some graphical quality to maintain the efficiency. Another restriction is from web browser manufacturers. For example, V8, the JavaScript engine in Google Chrome, has a threshold for maximum heap memory usage, so that when the number of atoms is too large, Chrome will crash. In fact, for very large molecules, RCSB PDB does not offer common PDB archives to download. Instead, structural data are recorded in a more complicated format such as mmCIF (16). Under these conditions, we recommend use of desktop software, for example PyMOL, whose modeling and rendering algorithms are written in C++ (a compiled language).
JavaScript was originally designed for web page interaction, and its function libraries are not abundant. Therefore, it is difficult for Web3DMol to handle PDB archives if they are in zipped format or to record a small movie while users are manipulating the 3D structure.
The support for WebGL on mobile devices is uneven. Web3DMol runs well on some of the latest cellphones, but not very smoothly on most mobile devices at present (Supplementary Table S1). The memory capacity, the performance of microchips and the touch-based interactions limit the user experience of Web3DMol on mobile devices.
DISCUSSION
Web3DMol provides a solution to visualize protein structures in web browsers. Users with little computational expertise can easily create 3D models with simple mouse interactions. It saves users the time and energy spent on installing of software or plugins and gaining familiarity with command-lines. For web-based databases and tools, Web3DMol is helpful for visualizing protein structure data without a need to parse PDB archives and construct geometries from the beginning. Compared with existing web-based molecule viewers, Web3DMol has unique functions, including sequence plot, fragment segmentation, measure tool and meta-information display. We expect that Web3DMol will be a valuable tool for both researchers and developers who are interested in protein structure research. More usage instructions are available at the Help Page (http://web3dmol.duapp.com/help.html).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Xu Zhang in M.Q.Z.'s laboratory for sharing his ideas and enthusiasm to make this work possible. Furthermore, we show high esteem to Warren Lyford DeLano, the author of PyMOL.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
State Key Research Development Program of China [2016YFC1200303]; National Natural Science Foundation of China [31361163004, 31671383]. Funding for open access charge: National Natural Science Foundation of China [31361163004, 31671383].
Conflict of interest statement. None declared.
REFERENCES
- 1. O’Donoghue S.I., Goodsell D.S., Frangakis A.S., Jossinet F., Laskowski R.A., Nilges M., Saibil H.R., Schafferhans A., Wade R.C., Westhof E. et al. . Visualization of macromolecular structures. Nat. Methods. 2010; 7:S42–S55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pirhadi S., Sunseri J., Koes D.R.. Open source molecular modeling. J. Mol. Graph. Model. 2016; 69:127–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mura C., McCrimmon C.M., Vertrees J., Sawaya M.R.. An introduction to biomolecular graphics. PLoS Comput. Biol. 2010; 6:e1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanson R.M. Jmol—a paradigm shift in crystallographic visualization Journal of Applied Crystallography. J. Appl. Crystallogr. 2010; 43:1250–1260. [Google Scholar]
- 5. Hartshorn M.J. AstexViewer: a visualisation aid for structure-based drug design. J. Comput. Aided Mol. Des. 2002; 16:871–881. [DOI] [PubMed] [Google Scholar]
- 6. Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L.. JSmol and the next-generation web-based representation of 3D molecular structure as applied to Proteopedia Israel Journal of Chemistry. Israel J. Chem. 2013; 53:207–216. [Google Scholar]
- 7. Chavent M., Levy B., Krone M., Bidmon K., Nomine J.P., Ertl T., Baaden M.. GPU-powered tools boost molecular visualization. Brief. Bioinform. 2011; 12:689–701. [DOI] [PubMed] [Google Scholar]
- 8. Rose A.S., Hildebrand P.W.. NGL viewer: a web application for molecular visualization. Nucleic Acids Res. 2015; 43:W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rego N., Koes D.. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015; 31:1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H., Leung K.S., Nakane T., Wong M.H.. iview: an interactive WebGL visualizer for protein-ligand complex. BMC Bioinformatics. 2014; 15:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burger M.C. ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics. J. Cheminform. 2015; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. et al. . SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rose P.W., Prlic A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. et al. . The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R.. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998; 280:69–77. [DOI] [PubMed] [Google Scholar]
- 15. Meier T., Polzer P., Diederichs K., Welte W., Dimroth P.. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005; 308:659–662. [DOI] [PubMed] [Google Scholar]
- 16. Westbrook J.D., Bourne P.E.. STAR/mmCIF: an ontology for macromolecular structure. Bioinformatics. 2000; 16:159–168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.