Abstract
Background.
Despite a multiplicity of clinical trials testing immune checkpoint inhibitors, the frequency of expression of potential predictive biomarkers is unknown in glioma.
Methods.
In this study, we profiled the frequency of shared biomarker phenotypes. To clarify the relationships among tumor mutational load (TML), mismatch repair (MMR), and immune checkpoint expression, we profiled patients with glioma (n = 327), including glioblastoma (GBM) (n = 198), whose samples had been submitted for analysis from 2009 to 2016. The calculation algorithm for TML included nonsynonymous mutation counts per tumor, with germline mutations filtered out. Immunohistochemical analysis and next-generation sequencing were used to determine tumor-infiltrating lymphocyte expression positive for programmed cell death protein 1 (PD-1), PD ligand 1 (PD-L1) expression on tumor cells, MMR (MLH1, MSH2, MSH6, and PMS2) protein expression and mutations, and DNA polymerase epsilon (POLE) mutations.
Results.
High TML was only found in 3.5% of GBM patients (7 of 198) and was associated with the absence of protein expression of mutL homolog 1 (MLH1) (P = .0345), mutS homolog 2 (MSH2) (P = .0099), MSH6 (P = .0022), and postmeiotic segregation increased 2 (PMS2) (P = .0345) and the presence of DNA MMR mutations. High and moderate TML GBMs did not have an enriched influx of CD8+ T cells, PD-1+ T cells, or tumor-expressed PD-L1. IDH1 mutant gliomas were not enriched for high TML, PD-1+ T cells, or PD-L1 expression.
Conclusions.
To clarify the relationships among TML, MMR, and immune checkpoint expression, we profiled the frequency of shared biomarker phenotypes. On the basis of a variety of potential biomarkers of response to immune checkpoints, only small subsets of glioma patients are likely to benefit from monotherapy immune checkpoint inhibition.
Keywords: glioblastoma, glioma, immune checkpoint, mismatch repair, mutational load
Importance of the study
With over a dozen open clinical trials, immune checkpoint inhibitor clinical trials have generated unprecedented enthusiasm. A variety of response biomarkers, such as PD-1/PD-L1 expression, tumor mutational load or burden, and DNA MMR defects, have all been thought to be associated with response. In this analysis, we show that these biomarkers are expressed infrequently in GBM, and without substantial overlap. A comprehensive biomarker analysis will be required to identify the enrichment criteria for advanced-stage clinical trials of these agents.
An unprecedented number of clinical trials are under way that are exploring the clinical utility of immune checkpoint blockade in glioblastoma (GBM), spurred by the recent FDA approvals of immune checkpoint inhibitors in other advanced cancers.1–6 When immune checkpoint inhibitors were initially investigated in clinical trials, tumor expression of programmed cell death protein 1 (PD-1) and PD ligand 1 (PD-L1) was presumed to be associated with clinical response.7 As the field of biomarkers evolved, focus shifted to assessments of tumor mutational load (TML), which is associated with the abundance of neoantigens8,9 and increased immunogenicity.10 The techniques and cut points for defining TML in glioma have not been harmonized; thus, other easier-to-measure indices have been proposed, such as determining mutations in the exonuclease domain of polymerase epsilon (POLE), which leads to hypermutations and neoantigen load.11 In metastatic urothelial carcinoma, hypermutation of more than 12 mutations per megabase was a significant indicator of response to immune checkpoint inhibition.12 Similarly in colorectal cancer, tumors with a mutational load of ≥17 mutations per megabase may be hypermutated (unpublished data), which may be a potential indicator of immunotherapy response. However, it is likely that the cut points for TML response to immune checkpoint therapeutics will vary among cancer types.9,13,14 Several early-stage clinical trials of immune checkpoint inhibitors have demonstrated that defective mismatch repair (MMR) is associated with clinical responses to immune checkpoint inhibitors.13,15,16 Treatment with nivolumab (a PD-1 immune checkpoint inhibitor) resulted in significant clinical and radiographic responses in a recent study of 2 siblings diagnosed with recurrent multifocal biallelic MMR-deficiency GBM; both had significantly higher neoantigen loads and hypermutant profiles than did those with sporadic cancers.16
Even though gliomas have been shown to carry an average TML which is lower than cancer types in which immune checkpoint inhibitors are highly active,17 GBMs with DNA repair defects may demonstrate a “hypermutator” phenotype18 that could render them sensitive to immune checkpoint blockade. A hypermutator phenotype has been described in GBM specimens with MSH6 mutations.19 Hypermutated samples typically harbor mutations in at least one of the MMR genes MLH1, MSH2, MSH6, and PMS2.20 Several studies have highlighted the higher rate of acquired MMR deficiency during the treatment and recurrence of GBM.19,21,22 For example, exome sequencing of matched initial and recurrent GBM from individual centers and The Cancer Genome Atlas (TCGA) demonstrated that subsets of recurrent tumors harbor somatic mutations in MMR genes. Specifically, approximately 26% of recurrent tumors acquired mutations in MSH6 and demonstrated increased mutational rates. All of these patients received alkylating agents (most commonly temozolomide) as part of their initial treatment, and the resulting mutation pattern is indicative of alkylator-induced mutations in the setting of MMR defects.19,21,22 Taken together, these observations suggest that a small number of newly diagnosed and a larger proportion of recurrent GBM tumors have inherent or acquired MMR defects or the hypermutator phenotype. Depending on the tumor, these MMR defects and the hypermutator phenotype may be present at diagnosis, emerge during initial treatment with radiotherapy and temozolomide, develop at recurrence, and subsequently imply sensitivity to immune checkpoint inhibitor therapy.
It is currently unknown whether PD-1, PD-L1, TML, MMR defects, or POLE mutations are predictive of GBM patient responses to immune checkpoints; this can only be elucidated in the context of a well-designed clinical trial. It is unknown whether all of these biomarkers will need to be assessed to identify a biomarker of response or whether some of them co-associate, thereby reducing the hypersegmentation of the population and simplifying the analysis. In this study, we profiled the frequency of shared biomarker phenotypes in glioma tissue samples. We also hypothesized that TML would be associated with MMR defects and POLE mutations, tumor T-cell influx, and PD-1/PD-L1 expression as an evolutionary mechanism to help tumors with a high antigen burden to remain undetected by the immune system.
Materials and Methods
All cases were from patients with gliomas and had been submitted worldwide to Caris Life Sciences for genomic analysis between 2009 and 2016. The initial histological diagnosis, based on the World Health Organization (WHO) 2007 classification, was confirmed. This study included 327 consecutive glioma patients with TML data. Immunohistochemistry (IHC) data for PD-1 on tumor-infiltrating lymphocytes (TILs) and for PD-L1 on tumor cells were available for 152 and 295 gliomas, respectively. In addition, IHC and next-generation sequencing (NGS) were used to determine the expression and mutational status of the MMR system (MLH1, MSH2, MSH6, and PMS2) and POLE.
Immunohistochemistry Analysis
An IHC analysis was performed on the entire section of formalin-fixed, paraffin-embedded tumor samples using automated staining techniques. Dilutions and conditions were performed on the basis of package insert instructions; they were optimized and validated and met the standards and requirements of the Clinical Laboratory Improvement Amendments/College of American Pathologists and the International Organization for Standardization. IHC results were evaluated independently by 6 board-certified pathologists. The primary antibody used against PD-L1 was SP142 (Spring Biosciences).12 Antibody specificity and lot-to-lot reproducibility were assessed by western blot analysis on tumor cell lysate. The chromogenic reporter 3,3′-diaminobenzidine was used to allow colorimetric visualization of the antibody, yielding a brown stain that could be analyzed with a light microscope. For the SP142 clone, the Rabbit LINKER visualization system (Dako) was used. The staining was regarded as positive if its intensity on the membranes of the tumor cells was >1+ (on a semiquantitative scale of 0–3: 0 for no staining, 1+ for weak cytoplasmic staining, 2+ for moderate membranous staining, and 3+ for strong membranous staining), and the percentage of positively stained cells was >5%. The primary antibody used for PD-1 was MRQ-22 (Ventana), and staining was scored as positive if the number of PD-1+ TILs was >1 cell per high-power field.23 PD-1 TIL density was evaluated using a hotspot approach. The whole tumor sample was reviewed at low power (4x objective), and the areas of the highest density of TILs in direct contact with malignant tumor cells were enumerated, at a 400x visual field (40x objective×10x ocular). MMR protein expression was also tested by IHC using antibody clones from Ventana (MLH1: M1; MSH2: G219-1129; MSH6: 44; and PMS2: EPR3947). The complete absence of protein expression (0+ in 100% of cells) was determined as a loss of MMR. The primary antibody used for CD8 was SP57 (Ventana), and the number of CD8+ TILs per high-power field was recorded. After the single field with the highest density of CD8+ cells was identified on a low power 4x scan, CD8+ cells were enumerated in 10 consecutive high-power fields. During the validation process for each IHC, any interpathologist variability was addressed in a scope session with all the pathologists, led by the medical director. In addition, weekly training sessions were held, using randomly selected samples, to help all pathologists reach the same scores.
Next-Generation Sequencing
We performed NGS on genomic DNA isolated from formalin-fixed, paraffin-embedded tumor tissue using the Illumina NextSEQ platform.22 An Agilent custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets. All variants were detected with >99% confidence. TML was calculated by counting all nonsynonymous missense mutations that had not previously been reported as germline alterations. High TML was defined as more than 20 mutations per 1.4 megabase (Mb); moderate TML was defined as 11–20 mutations per 1.4 Mb; and low TML was defined as 10 or fewer mutations per 1.4 Mb.
Statistical Methods
Fisher’s exact test was used to examine the association between the biomarkers of IHC/NGS and binary TML. The Cochran Armitage test24 was used when TML was evaluated in 3 categories (high, medium, and low). The Wilcoxon rank sum test was used to compare raw TMLs. Spearman’s rank correlation test was used to examine the overall association between TML and tumor grade. Because the counts of CD8+ cells were right censored at 200, the log-rank test was used to examine their association with MMR. To adjust for multiple testing and limit the risk of false-positive results, we used the Benjamini–Hochberg procedure to correct the P-values resulting from the Fisher’s exact tests for associations with mutations.25 All reported P-values are 2 sided and corrected for multiple comparisons using the Benjamini–Hochberg procedure or false discovery rate (FDR) correction. P-values <.05 were declared statistically significant. All the analyses were implemented using the statistical software R v3.3.1, with the packages coin v1.0-24 and survival v2.39-4.
Ethics Statement
Human subjects are de-identified prior to analysis and this research is exempt under the Code of Federal Regulations 45 CFR 46.101(b)(4) from 45 CFR part 46 requirements.
Results
Characteristics of the Analyzed Patient Cohort
This study included 327 consecutive glioma patients with TML data obtained globally. The study cohort included grade IV GBM (n = 198), grade IV gliosarcoma (n = 10), high-grade not otherwise specified (NOS) (n = 16), grade III astrocytoma, grade III oligoastrocytoma, grade III oligodendroglioma (n = 34), grade II astrocytoma, grade II oligoastrocytoma, grade II oligodendroglioma (n = 45), grade I astrocytoma (n = 5), and 19 other types (glioma NOS, ependymoma of various grades, ganglioglioma, oligoastrocytoma NOS, etc.). As expected, there was enrichment of GBM cases in males, and grade was associated with age.
High-Grade Gliomas Demonstrate High but Infrequent TML
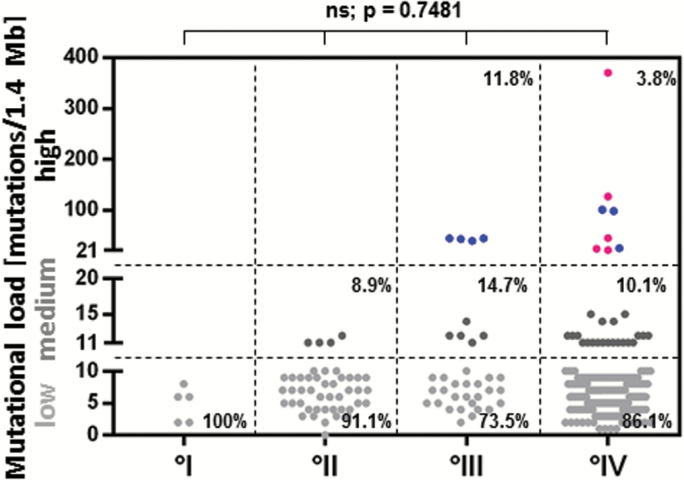
The range of glioma TML was 0–370 mutations/1.4 Mb. Most gliomas (85.3% [279 of 327]) had low TML, defined as 10 mutations/1.4 Mb or less. Approximately 10.1% (33 of 327) possessed moderate TML (11–20 mutations/1.4 Mb). Only 4.6% (15 of 327) had high TML, defined as more than 20 mutations/1.4 Mb. Three of these had no definitive grade and therefore are not shown in Fig. 1A. Among the high TML-expressing gliomas, 40% (6 of 15) were newly diagnosed and 60% (9 of 15) were recurrent. These cases included grade III glioma (n = 4), grade IV GBM and gliosarcoma (n = 8), and high-grade glioma NOS (n = 3). Of the high-TML GBMs, 57% (4 of 7) were newly diagnosed, and the remainder were recurrent. Only high-grade gliomas demonstrated high TML (Fig. 1). Specifically, 3.5% of GBMs (7 of 198) had high TML and 10% of GBMs (20 of 198) had moderately elevated TML. Including the gliosarcoma cases, 3.8% of grade IV cases (8 of 208) had high TML and 10% (21 of 208) had moderately elevated TML. Spearman’s rank correlation was −0.0178 (P = .7481), indicating that TML was independent of glioma grade. No differences were found between TML and age or sex.
Fig. 1.
TML is associated with WHO tumor grade. High TML was defined as more than 20 mutations per 1.4 Mb, moderate TML as 11–20 mutations per 1.4 Mb, and low TML as 10 or fewer mutations per 1.4 Mb. Of the high TML-expressing gliomas, 40% (6/15) were newly diagnosed and 60% (9 of 15) were recurrent. Within the high-TML GBM subset, 57.1% (4 of 7) were newly diagnosed and the remaining were recurrent. °I = grade I glioma, °II = grade II glioma, °III = grade III glioma, °IV = grade IV glioma. Red = newly diagnosed glioma. Blue = recurrent glioma. ns = not significant.
MMR Protein Expression Loss Corresponds to High TML in GBM
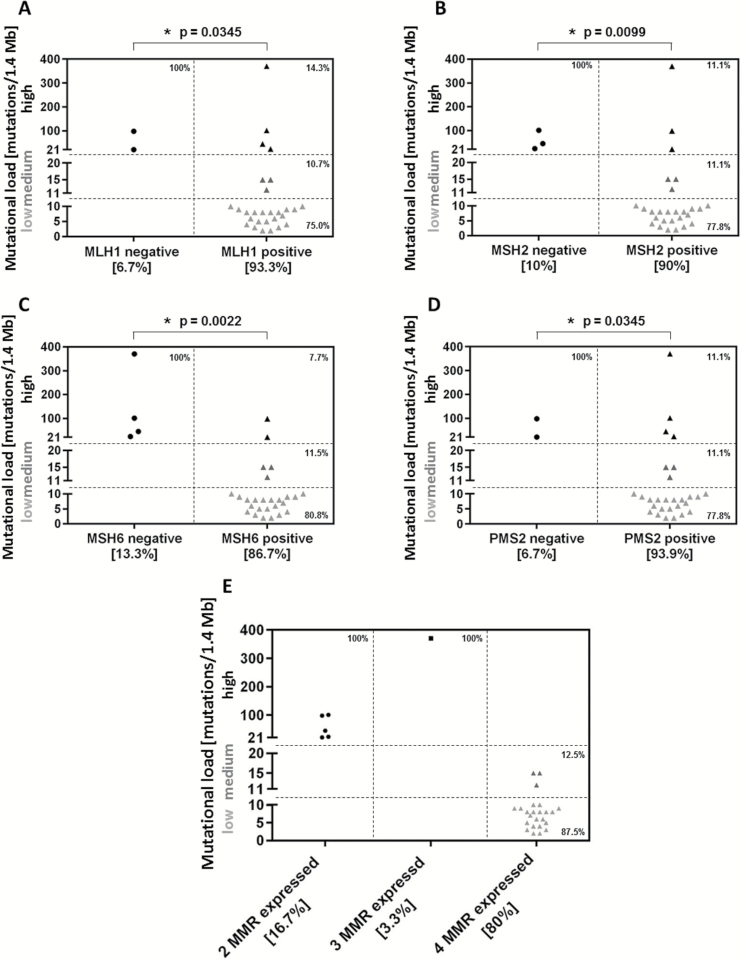
To ascertain whether MMR protein expression is commonly associated with TML, we conducted a subanalysis in the GBM specimens using MMR protein expression data (n = 30). MMR protein expression was evident in the majority of GBM cases, with only 6.7% (2 of 30) demonstrating a complete loss of mutL homolog 1 (MLH1), 10% a loss of mutS homolog 2 (MSH2) (3 of 30), 13.3% (4 of 30) a loss of MSH6, and 6.7% (2 of 30) a loss of postmeiotic segregation increased 2 (PMS2) (Table 1, Fig. 2A–D). A high TML was associated with loss of IHC expression of MLH1 (P = .0345), MSH2 (P = .0099), MSH6 (P = .0022), and PMS2 (P = .0345). Of the 30 GBM cases that were stained for MMR protein expression, 6 showed a high TML; these 6 tumors lost the expression of at least one of the 4 analyzed MMR enzymes (Fig. 2E). In some instances, the loss of expression of one MMR protein was associated with the loss of another, such as MLH1 with PMS2 (P = .0001) and MSH2 with MSH6 (P < .0001), including within molecularly defined GBM (ie, isocitrate dehydrogenase 1 [IDH1] wild-type only: P = .0023 and P = .001, respectively).
Table 1.
Pathological composition and characteristics of the analyzed mutational load patient cohort
| Pathology | All Patients | GBM | Gliosarcoma | High Grade NOS | Grade III | Grade II | Grade I | Other |
|---|---|---|---|---|---|---|---|---|
| WHO grade | I + II + III + IV | IV | IV | III/IV | III | II | I | ? |
| Number of patients (%) | 327 | 198 | 10 | 16 | 34 | 45 | 5 | 19 |
| Parameters | ||||||||
| Sex, no. (%) | ||||||||
| Male | 179 (54.7) | 110 (55.6) | 6 (60.0) | 8 (50.0) | 19 (55.9) | 23 (51.1) | 3 (60.0) | 10 (52.6) |
| Female | 148 (45.3) | 88 (44.4) | 4 (40.0) | 8 (50.0) | 15 (44.1) | 22 (48.9) | 2 (40.0) | 9 (47.4) |
| Age, mean y (range) | 52.1 (0-85) | 56.6 (4-85) | 52.9 (35-63) | 53.9 (28-81) | 46.7 (21-73) | 42.4 (18-78) | 19.4 (0-56) | 44.3 (15-74) |
| <50 y | 121 (37.0) | 44 (22.2) | 3 (30.0) | 6 (37.5) | 18 (52.9) | 34 (75.6) | 4 (80.0) | 13 (68.4) |
| ≥50 y | 206 (63.0) | 154 (77.8) | 7 (70.0) | 10 (62.5) | 16 (47.1) | 11 (24.4) | 1 (20.0) | 6 (31.6) |
| IDH1 mutation | ||||||||
| Wild type | 248 | 182 | 10 | 12 | 10 | 16 | 5 | 13 |
| Mutated | 79 | 16 | 0 | 4 | 24 | 29 | 0 | 6 |
| Mutational load | ||||||||
| <10 mutations/1.4 Mb (low) | 279 | 171 | 8 | 13 | 25 | 41 | 5 | 16 |
| 11-20 mutations/1.4 Mb (moderate) | 33 | 20 | 1 | 1 | 5 | 4 | 0 | 2 |
| >20 mutations/1.4 Mb (high) | 15 | 7 | 1 | 2 | 4 | 0 | 0 | 1 |
| Mean TML ± SD | 10.2 (±26.2) | 9.8 (±27.7) | 18.8 (±38.2) | 11.1 (±17.5) | 11.4 (±11.9) | 6.7 (± 2.8) | 4.8 (±2.7) | 17.7 (±51.2) |
| PD-1/PD-L1 expression | ||||||||
| PD-1 on TILs, no. / total no. (%) | ||||||||
| Positive | 59/152 (38.8) | 43/94 (45.7) | 3/4 (75.0) | 3/8 (37.5) | 14/16 (87.5) | 1/16 (6.2) | 1/2 (50.0) | 6/12 (50.0) |
| PD-L1 on tumor cells, no. / total no. (%) | ||||||||
| Positive | 24/295 (8.1) | 19/189 (10.1) | 3/10 (30.0) | 1/16 (6.2) | 2/16 (12.5) | 0/41 (0) | 0/5 (0.0) | 1/18 (5.5) |
| MMR protein expression | ||||||||
| MLH1, no. / total no. | ||||||||
| Expression | 33/35 | 28/30 | 0/1 | 2/2 | 0/1 | 3/3 | 1/1 | 3/3 |
| MSH2, no. / total no. | ||||||||
| Expression | 33/35 | 27/30 | 0/1 | 2/2 | 0/1 | 3/3 | 1/1 | 3/3 |
| MSH6, no. / total no. | ||||||||
| Expression | 31/34 | 26/30 | 0/1 | 2/2 | 0/1 | 2/2 | 1/1 | 3/3 |
| PMS2, no. / total no. | ||||||||
| Expression | 32/35 | 28/30 | 0/1 | 2/2 | 0/1 | 3/3 | 1/1 | 2/3 |
Fig. 2.
MMR protein expression corresponds to TML in GBM. MMR protein expression was determined by IHC in GBM cases (n = 30). Only 6.7% (2 of 30) demonstrated loss of MLH1 (A), 10% (3 of 30) loss of MSH2 (B), 13.3% (4 of 30) loss of MSH6 (C), and 6.7% (2 of 30) loss of PMS2 (D). When these GBM cases were analyzed for TML, 6 showed high TML. Of the GBMs with high TML, at least 1 MMR enzyme was found to have defective expression (E). High TML was defined as more than 20 mutations per 1.4 Mb, moderate TML as 11–20 mutations per 1.4 Mb, and low TML as 10 or fewer mutations per 1.4 Mb. ns = not significant.
MMR Mutations Are Associated with High TML but Are Rare in Glioma
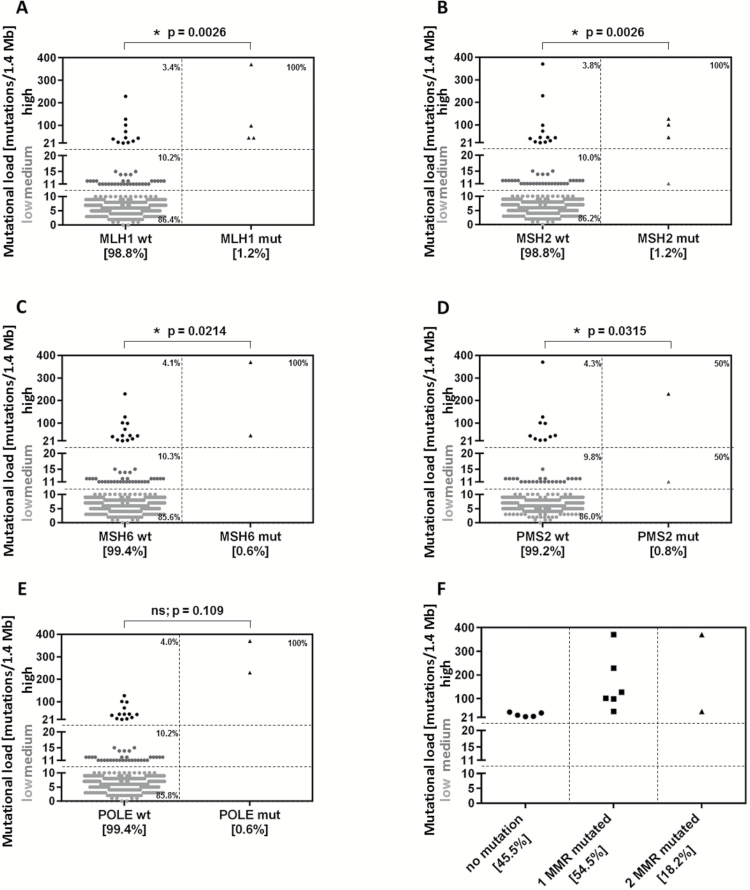
An association was found between TML and MLH1, MSH2, MSH6, and PMS2 mutations in gliomas (P = .0026, .0026, .0214, and .0315, respectively) (Fig. 3A–D), indicating that a high TML is associated with MMR mutations in gliomas. Per statistical methods, the P-value was corrected for multiple comparisons using a stringent FDR correction (Benjamini–Hochberg procedure) to reduce the probability of false positive results. Of the glioma samples analyzed for all 4 mutations in MMR, we found 11 cases with a high TML; 54.5% (6 of 11) of these cases exhibited at least one mutated MMR gene, 18% (2 of 11) carried 2 mutations, and the remainder had no MMR mutation (Fig. 3F). An association was also found between TML and MLH1, MSH2, and MSH6 mutations specifically in GBM (P = .0011, .0011, and .0011, respectively). We found no mutations of PMS2 among the GBM patients. In many instances, the gliomas with loss of MMR protein expression also demonstrated MMR mutations (MLH1 = 4 cases with loss of protein expression, 3 of them mutated; MSH2 = 5 cases with loss of protein expression, 3 of them mutated; and MSH6 = 6 cases with loss of protein expression, 2 of them mutated), but not all had loss of MMR protein expression, since epigenetic alterations were not accounted for.
Fig. 3.
MMR mutations do not correspond to TML in glioma. Mutations in MMR and POLE were analyzed by NGS. An association was found between TML and the occurrence of (A) MLH1 mutation, (B) MSH2 mutation, (C) MSH6 mutation, (D) PMS2 mutations, and (E) POLE mutations (P = .109; P = .0144 before FDR adjustment). (F) Of the samples analyzed for mutations in the 4 MMR enzymes, 11 showed high TML; 54.5% (6 of 11) were mutated in at least 1 MMR, and 18% (2 of 11) were mutated in at least 2 MMR. ns = not significant.
We then conducted a secondary analysis to ascertain whether a single genetic mutation from the 592 expanded NGS panel could be used as a surrogate for TML. We analyzed GBM cases with high TML (>20 mutations/1.4 Mb, n = 7) and found that MLH1 (P = .0069, Fisher’s exact test for a 2×2 table), MSH2 (P = .0069), MSH6 (P = .0069), ATM (P = .0069), and PIK3CA (P = .0262) mutations were statistically significantly associated with a high TML; this significance remained after adjustment when TML was assessed as high, moderate, or low using the Cochran Armitage test.
POLE Mutations in Glioma
The highest TMLs of 327 tumors (229 and 370 mutations/1.4 Mb) were found in 2 tumors (1 GBM and 1 glioma NOS) that exhibited POLE mutations via NGS (Fig. 3E); however, this association was not statistically significant after FDR correction, likely due to the small incidence of unevenly distributed POLE mutations found in only the 2 highest TML patients (P = .109; P = .0144 before FDR adjustment), but it implied strong clinical importance for future validation.
TML Is Not Associated with CD8+ T-Cell Influx or PD-1 and PD-L1 Expression
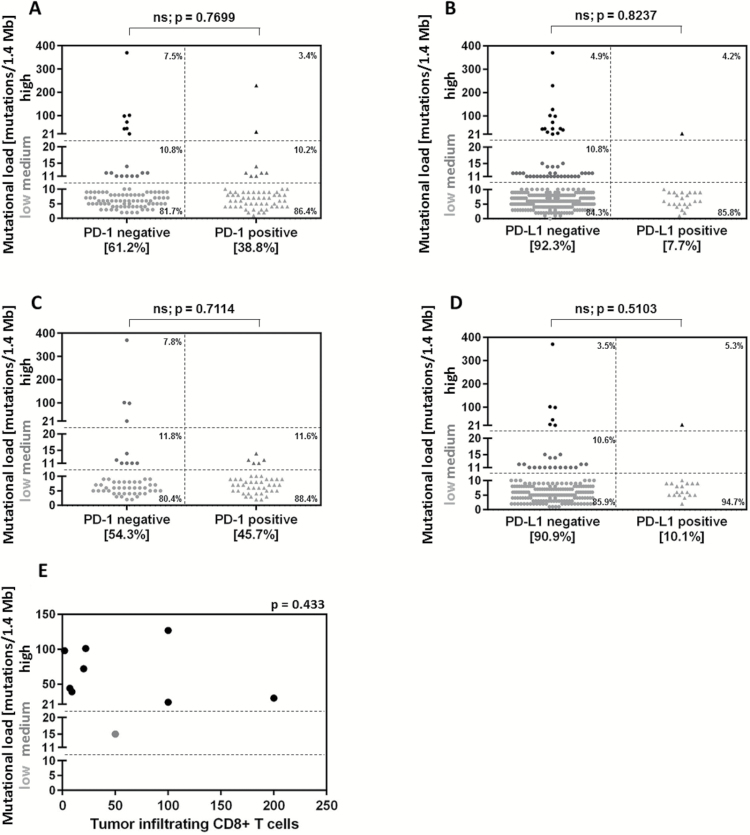
PD-1 staining results were available for 152 cases, and PD-L1 staining results were available for 310 cases. Fifty-nine of 152 (38.8%) cases were PD-1 positive and 24 of 310 (7.7%) cases were PD-L1 positive. No association was found between TML and PD-1/PD-L1 expression (P = .7699 and 0.8237, respectively) (Fig. 4A, B). Of the GBM patients, 43 of 94 patients (45.7%) were PD-1 positive and 19 of 189 patients (10.1%) were PD-L1 positive, findings that are consistent with a prior report.26 When these GBMs were analyzed for TML, there was no association with PD-1 (P = .7114) or PD-L1 (P = .5103) (Fig. 4C, D). We then performed a secondary analysis of the gliomas enriched for higher TML, quantified the tumor-infiltrating CD8+ T cells (n = 9 total; IDH1 wild-type gliosarcoma [n = 1], IDH1 mutated high-grade glioma NOS [n = 1], IDH1 wild-type high-grade glioma NOS [n = 1], IDH1 mutated grade II oligodendroglioma [n = 1], IDH1 wild-type grade III anaplastic astrocytoma [n = 1], IDH1 wild-type GBM [n = 1], and IDH1 mutated GBM [n = 3]), and found no association (Fig. 4E) (P = .433). Therefore, in gliomas with high or moderate TML, there was no significant correlation with CD8+ T-cell influx. Furthermore, there was no significant relationship found between CD8+ T-cell counts and PD-1/PD-L1 expression (data not shown). The loss of the MSH2/MSH6 pair (P = .0255) was more likely to be associated with high CD8+ TIL counts in tumors.
Fig. 4.
TML is not associated with PD-1 and PD-L1 expression or CD8+ T cell influx. Of all analyzed samples, 38.8% (59 of 152) were PD-1 positive (A) and 7.7 % (24 of 310) were PD-L1 positive (B). Of the GBM patients, 45.7% (43 of 94) were PD-1 positive (C) and 10.1% (19 of 189) were PD-L1 positive (D). No association was found between TML and PD-1/PD-L1 expression. PD-1 and PD-L1 expression levels were determined using IHC staining. (E) CD8+ T-cell influx was determined by IHC in a subset of gliomas with high and moderate TML (n = 9). No association with TML was found.
Subgroup Analysis of IDH1 Wild-type GBM
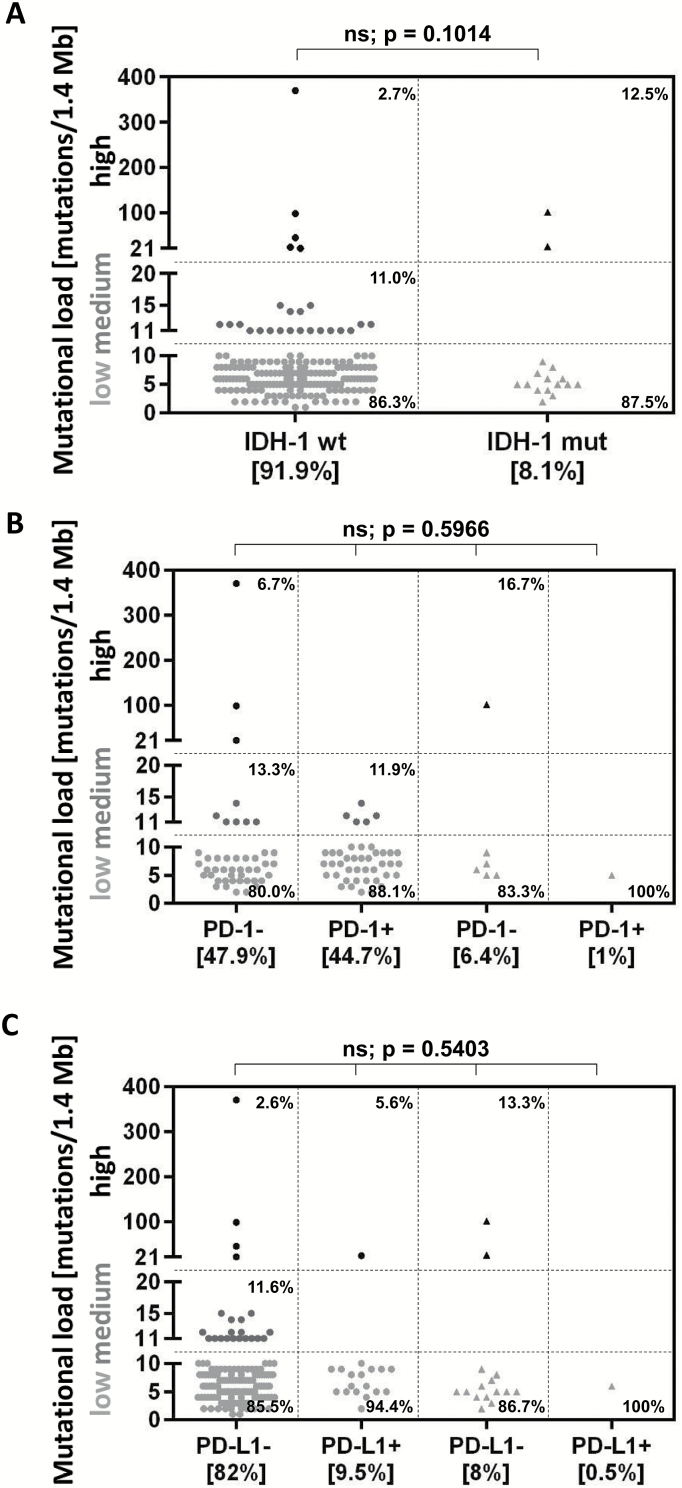
To ascertain whether there were any significant differences in the immunobiological characteristics of IDH1 wild-type (n = 182) and mutated (n = 16) GBM (Table 1), we compared TMLs and found that 5 of 182 (2.7%) IDH1 wild-type GBM patients had high TML versus 2 of 16 (12.5%) IDH1 mutated GBM patients (P = .1014) (Fig. 5A). However, PD-1+ T cells were almost exclusively identified in IDH1 wild-type GBMs, and this was not associated with TML (P = .5966). None of the high-TML IDH1 wild-type GBMs demonstrated PD-1+ T cell influx (Fig. 5B). Furthermore, PD-L1 expression was predominantly detected in IDH1 wild-type tumors and was not associated with high TML (P = .5403) (Fig. 5C).
Fig. 5.
Subgroup analysis of IDH1 wild-type GBM. TML was compared between IDH1 wild-type (n = 182) and mutated (n = 16) GBM; 2.7% (5 of 182) of IDH1 wild-type cases and 12.5% (2 of 16) of mutated cases showed high TML (P = .1014) (A). Both PD-1 (B) and PD-L1 expression (C) were predominantly expressed in IDH1 wild-type cases and were not associated with high TML.
Discussion
Currently, there are an unprecedented number of clinical trials evaluating the effects of immune checkpoint inhibitors in GBM. Preliminary clinical trial experience indicates that only a select subset of these patients may experience a response.27–31 Several candidate biomarkers that may enrich for patient populations that respond to immune checkpoint inhibitors have been proposed in other oncological indications. However, it is unclear whether these are operational in GBM and if so, to what extent. It is possible that several of these candidate biomarkers co-associate; thus, a correlation assessment to clinical response could be streamlined and prioritized for these trials.
In this comprehensive biomarker analysis from patients worldwide, although we found an association between high TML and loss of MMR protein expression, we found no statistically significant association among TML, influx of cytotoxic CD8+ T cells, and immune checkpoint expression. These results strongly indicate that all of these biomarkers will need to be interrogated in the context of the aforementioned clinical trials to identify those that are associated with response. A high TML usually results in more tumor antigens and neoantigens, which leads to increased immunogenicity and presumably increased CD8 antitumor reactivity. However, in gliomas with high or moderate TML, there was no significant association with CD8+ T-cell influx. This may be secondary to the convention of sampling immune cells from within the tumor as opposed to the infiltrating invasive edge where they may predominate, or it may be due to immunosuppressive mechanisms that are operational in GBM, including those that induce T-cell apoptosis and that are not related to immune checkpoints. Moreover, we evaluated the CD8+ T-cell count in only 9 tumors with mostly high and moderate TML, which is a limitation.
In this study, TML was not significantly associated with age or tumor grade. In a recent study of the glioma dataset of TCGA, tumor grade was associated with TML, as grade IV gliomas had significantly higher mutational burden (average number of nonsilent mutations = 57) than grade III (32.4) or grade II gliomas (20.6).32 Older age was also associated with a higher TML in the entire TCGA dataset and among histologically and molecularly defined glioma subtypes (n = 542). In our study, TML was higher overall in grade III and grade IV gliomas than in lower-grade histological types. Moreover, the 2 patients with the highest TML (370 and 229 mutations/1.4 Mb) were quite young (7 and 15 years of age, respectively); however, it must be noted that these patients also had POLE mutations, which may confound the results. In our study, the majority of cases were GBM (n = 198), and only a few were grade III (n = 23) or grade II (n = 19); interestingly, although there were more GBM cases in our dataset, the proportions of high TML were similar in both grade III gliomas and GBM.
Similar to the findings in studies in other cancers,13,33–35 the loss of DNA MMR enzymes was associated with a high TML. This was especially the case when the expression was lost in more than one enzyme. This association was not as evident when high TML was associated with mutations within these enzymes, likely secondary to the mutations’ inability to always alter functional activity or expression. Certain genetic mutations, such as MLH1, MSH2, MSH6, PIK3CA, and ATM, were associated with an increased TML in GBM but were not absolute indicators of a high TML. Mutations in both the MMR gene MLH132 and the kinase PIK3CA32,37 have previously been shown to be negative prognosticators for survival in GBM patients. When mutated, PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha) stimulates a second messenger signaling pathway to promote cell proliferation and cancer formation in various cancer types including GBM.32,37–40PIK3CA mutations may play an indirect role in high TML, due to increased risk of genetic aberration with increased cellular proliferation. The DNA damage kinase ataxia telangiectasia mutated (ATM) is a tumor suppressor that responds to DNA double-strand breaks, and when mutated, it leads to GBM formation and progression.41 Thus, it is not surprising that mutations in ATM could result in increased TML.
We found POLE genetic mutations in 2 of the glioma specimens (1 GBM and 1 glioma NOS). Mutations within the exonuclease domain of the POLE gene affect the DNA polymerase proofreading function, and such mutations are associated with an ultramutated phenotype in colon and endometrial cancers.11,42 A 2015 study published in Neuro-Oncology identified a unique molecular subgroup consisting of 4 high-grade glioma cases with ultramutated POLE mutations (3 GBMs and 1 anaplastic astrocytoma) out of a total of 55 cases from the Yale and TCGA databases. These 4 patients were statistically significantly younger (35.5 vs 58 y, P = .005) at the time of diagnosis and had longer progression-free survival (26.93 vs 6.93 mo, P = .03) than did the other 51 POLE wild-type primary high-grade glioma patients.43 Similarly, in our study, the 2 patients with POLE mutations were young (7 and 15 y) (no survival data are available for these patients).
Another point to consider is which mutations are ideal or appropriate targets for the immune system. In a recent Science paper, McGranahan et al demonstrate that lung tumors with a low neoantigen subclonal fraction and high mutational load had durable clinical benefit to pembrolizumab, a PD-1 blockade therapy. However, chemotherapy-induced subclonal neoantigens (leading to increased TML) were found in poor responders to immune checkpoint blockade.44 In our study among the high TML-expressing GBMs, 57% were newly diagnosed and 43% were recurrent. The recurrent GBMs with high TML may not theoretically respond to immune checkpoint blockade, given that radiation and chemotherapy may induce de novo subclonal mutations that do not elicit an effective antitumor immune response. Indeed, trunk mutations (evaluated in our study), which may have been present during the initial events of the cancer, may be superior immunological targets, but additional functional studies would be needed to ascertain this definitively. Unfortunately, and a limitation of our study, clinical information including survival for this dataset is not available and therefore it is not possible to ascertain the association of mutational load with treatment regimens and outcomes.
It is highly unlikely that clinical responders to immune checkpoints will be identified by a single biomarker, especially given the complex biology of gliomas and the immunological cascade that must be triggered in an anti-tumor immune response. In addition, it is likely that these latest biomarkers will be superseded by those that account for the functional complexity of an effective immune-tumor interaction. In conclusion, our data indicate that on the basis of several potential biomarkers of response to immune checkpoints, only a small subset of glioma patients are likely to benefit from immune checkpoint inhibition.
Funding
This research was funded by the Provost Retention Fund and National Institutes of Health grants CA 120813, P50 CA127001, and P30 CA016672.
Conflict of interest statement. A.B.H. serves on the Caris Life Sciences Scientific Advisory Board and is a stockholder in the company. J.X. and D.S. are employees of Caris Life Sciences.
Acknowledgments
The authors thank Audria Patrick and Ann Sutton BA for their administrative and editorial support.
References
- 1. Weber JS, D’Angelo SP, Minor D, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16(4):375–384. [DOI] [PubMed] [Google Scholar]
- 2. Motzer RJ, Rini BI, McDermott DF, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33(13):1430–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Westin JR, Chu F, Zhang M, et al. Safety and activity of PD1 blockade by pidilizumab in combination with rituximab in patients with relapsed follicular lymphoma: a single group, open-label, phase 2 trial. Lancet Oncol. 2014;15(1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369(2):134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rizvi NA, Mazières J, Planchard D, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16(3):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schumacher TN, Kesmir C, van Buuren MM. Biomarkers in cancer immunotherapy. Cancer Cell. 2015;27(1):12–14. [DOI] [PubMed] [Google Scholar]
- 9. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Champiat S, Ferté C, Lebel-Binay S, Eggermont A, Soria JC. Exomics and immunogenics: bridging mutational load and immune checkpoints efficacy. Oncoimmunology. 2014;3(1):e27817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howitt BE, Shukla SA, Sholl LM, et al. Association of polymerase e-Mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1(9):1319–1323. [DOI] [PubMed] [Google Scholar]
- 12. Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le DT, Uram JN, Wang H, et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 17. Colli LM, Machiela MJ, Myers TA, Jessop L, Yu K, Chanock SJ. Burden of nonsynonymous mutations among TCGA cancers and candidate immune checkpoint inhibitor responses. Cancer Res. 2016;76(13):3767–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee V, Murphy A, Le DT, Diaz LA., Jr Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016;21(10):1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13(7):2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. The Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216): 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66(8):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15(14):4622–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gatalica Z, Snyder C, Maney T, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2965–2970. [DOI] [PubMed] [Google Scholar]
- 24. Agresti A. Categorical Data Analysis, New York: John Wiley & Sons; 2012. [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc. 1995;57(1): 289–300. [Google Scholar]
- 26. Garber ST, Hashimoto Y, Weathers SP, et al. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro Oncol. 2016;18(10):1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown NF, Carter T, Shaw HM, et al. Sequential immune checkpoint inhibition with concurrent bevacizumab for relapsed glioblastoma: a single centre experience. ASCO Meeting Abstracts. 2016;34(15_suppl): e13514. [Google Scholar]
- 28. Schaff L, Donovan L, Lassman AB, et al. IMCT-18PD-1 inhibitors for recurent high grade glioma (HGG). Neuro-Oncology. 2015;17(suppl 5): v111. [Google Scholar]
- 29. Reardon DA, De Groot JF, Colman H, et al. Safety of pembrolizumab in combination with bevacizumab in recurrent glioblastoma (rGBM). ASCO Meeting Abstracts. 2016; 34(15_suppl): 2010. [Google Scholar]
- 30. Reardon DA, Sampson JH, Sahebjam S, et al. Safety and activity of nivolumab (nivo) monotherapy and nivo in combination with ipilimumab (ipi) in recurrent glioblastoma (GBM): updated results from checkmate-143. ASCO Meeting Abstracts. 2016; 34(15_suppl): 2014. [Google Scholar]
- 31. Sampson J, Omuro A, Vlahovic G, et al. IMCT-03safety and activity of nivolumab monotherapy and nivolumab in combination with ipilimumab in recurrent glioblastoma: updated results from checkmate-143. Neuro-Oncology. 2015; 17(suppl 5): v107. [Google Scholar]
- 32. Draaisma K, Wijnenga MM, Weenink B, et al. PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients. Acta Neuropathol Commun. 2015;3:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stadler ZK, Battaglin F, Middha S, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34(18):2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tetzlaff MT, Singh RR, Seviour EG, et al. Next-generation sequencing identifies high frequency of mutations in potentially clinically actionable genes in sebaceous carcinoma. J Pathol. 2016;240(1):84–95. [DOI] [PubMed] [Google Scholar]
- 35. Shlien A, Campbell BB, de Borja R, et al. ; Biallelic Mismatch Repair Deficiency Consortium. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. [DOI] [PubMed] [Google Scholar]
- 36. Stark AM, Doukas A, Hugo HH, Mehdorn HM. The expression of mismatch repair proteins MLH1, MSH2 and MSH6 correlates with the Ki67 proliferation index and survival in patients with recurrent glioblastoma. Neurol Res. 2010;32(8):816–820. [DOI] [PubMed] [Google Scholar]
- 37. Liang H, Wang R, Jin Y, Li J, Zhang S. MiR-422a acts as a tumor suppressor in glioblastoma by targeting PIK3CA. Am J Cancer Res. 2016;6(8):1695–1707. [PMC free article] [PubMed] [Google Scholar]
- 38. Ogino S, Lochhead P, Giovannucci E, Meyerhardt JA, Fuchs CS, Chan AT. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene. 2014;33(23):2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64(21):7678–7681. [DOI] [PubMed] [Google Scholar]
- 40. Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. [DOI] [PubMed] [Google Scholar]
- 41. Blake SM, Stricker SH, Halavach H, et al. Inactivation of the ATMIN/ATM pathway protects against glioblastoma formation. Elife. 2016; 5(e08711): 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahn SM, Ansari AA, Kim J, et al. The somatic POLE P286R mutation defines a unique subclass of colorectal cancer featuring hypermutation, representing a potential genomic biomarker for immunotherapy. Oncotarget. 2016;7(42):68638–68649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Erson-Omay EZ, Çağlayan AO, Schultz N, et al. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol. 2015;17(10):1356–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]