Abstract
Similarity between the DNA substrates and products of integrase-mediated site-specific recombination reactions results in a single recombinase enzyme being able to catalyze both the integration and excision reactions. The control of directionality in these reactions is achieved through a class of small accessory factors that favor one reaction while interfering with the other. These proteins, which we will refer to collectively as recombination directionality factors (RDFs), play architectural roles in reactions catalyzed by their cognate recombinases and have been identified in conjunction with both tyrosine and serine integrases. Previously identified RDFs are typically small, basic and have diverse amino acid sequences. A subset of RDFs, the cox genes, also function as transcriptional regulators. We present here a compilation of all the known RDF proteins as well as those identified through database mining that we predict to be involved in conferring recombination directionality. Analysis of this group of proteins shows that they can be grouped into distinct sub-groups based on their sequence similarities and that they are likely to have arisen from several independent evolutionary lineages. This compilation will prove useful in recognizing new proteins that confer directionality upon site-specific recombination reactions encoded by plasmids, transposons, phages and prophages.
INTRODUCTION
Recombination directionality factors (RDFs) are a diverse group of proteins involved in controlling the directionality of integrase-mediated site-specific recombination reactions. Typically, RDFs are small DNA-binding proteins acting as accessory factors to influence the choice of substrates that are recombined by their cognate recombinase. While the majority of the RDFs that have been described are components of phage-encoded site-specific recombination systems [e.g. Lambda (1)], RDF proteins are also associated with a variety of other recombination systems including those encoded by plasmids [e.g. pSAM2 (2)] and transposons [e.g. Tn916 (3)].
The best studied RDF is that encoded by phage Lambda, Xis. The phage-encoded integrase, a member of the tyrosine family of site-specific recombinases, catalyzes both integration and excision reactions. The integration reaction utilizes the phage attP and bacterial attB DNA sites as substrates and generates recombinant junctions, attL and attR, as products. The excision reaction involves recombination between attL and attR to generate attP and attB as products. Both of these reactions require the integration host factor (IHF) in addition to integrase (4,5). Lambda Xis is required for the excision reaction, but inhibits integrative recombination (6).
Lambda Xis determines the directionality of recombination by influencing the formation of specific protein–DNA architectures. The Lambda integrase is composed of three domains, two of which confer different DNA binding specificities; both DNA binding valences can be occupied simultaneously leading to both intramolecular and intermolecular integrase-mediated bridges. The formation of these bridges is facilitated by IHF, which binds to specific sites in the DNA substrates and introduces DNA bends (7). Lambda Xis also binds to specific recognition sequences and introduces sharp DNA bends, which in the attR substrate promotes the formation of protein–DNA structures that can undergo excisive recombination, but which in attP prevents the formation of the architectures needed for integration (8,9). There is also some additional evidence that the C-terminal end of Lambda Xis interacts with the integrase protein. However, this interaction is not absolutely required and seems primarily to be involved in stabilizing binding of integrase to the DNA (10).
Although more than 100 phage-encoded integrases have been described (11), not all of these belong to the tyrosine family of recombinases (12,13). More than two dozen large serine recombinases have been described of which nearly half function in phage integration and excision. These recombinase proteins contain a 140 residue N-terminal domain with strong similarity to the catalytic domain of transposon resolvases and DNA invertases which utilize a serine near the N-terminus as the catalytic residue, but they also have a C-terminal domain that is much larger than that of the typical resolvase or invertase. We will thus refer to this class of integrase as the serine-integrases (Int-S) to distinguish them from the tyrosine-integrases (Int-Y). However, little is known about how the directionality of these systems is regulated. In the best-studied Int-S example, encoded by Streptomyces phage φc31, the recombination sites are small, there is no evidence of a host integration factor and no RDF has been identified (14). In phage TP901 (15) and in the heterocyst development system in Anabaena (16), RDF proteins have been identified although their mechanism of action is not understood.
In the case of the Lambda recombination system, the only known role of the Xis protein is in controlling the directionality of recombination. However, this is not true for the RDFs encoded by phages HP1 and P2 where they also act as transcriptional regulators (17,18). These are referred to as Cox proteins, and they can both negatively and positively regulate transcription initiation. Initial characterization of the HP1 excision reaction showed that the Cox protein binds specifically to attP DNA and forms several specific DNA–protein complexes in the presence of attP and Cox (17). In spite of the fact that the Cox proteins are also transcriptional regulators it seems likely that they regulate recombination directionality in a similar way as described for the Lambda integration system.
While a number of RDFs have been identified through experimental approaches, others have proven difficult to find using comparative sequence methods, mainly because the proteins are small, typically containing <100 amino acids, and contain few, if any, highly conserved residues. Moreover, while many RDFs are basically charged, there are several examples of others that are acidic. Because of the great diversity of these RDF proteins, there previously has been no systematic effort to catalog, classify or explore the possible evolutionary relationships among them.
In this study, we present our attempt to identify all of the likely RDFs for which sequence information has been determined, including those that have not been identified previously. A variety of database search techniques were used to identify sequences that have similarity to known RDFs, generating a list of 63 known or putative RDFs which can be further sub-classified into at least seven smaller groups on the basis of sequence similarity. This compilation of proteins has been analyzed for homologous groups and different chemical characteristics. Many of the identified RDFs fall into specific groups for which some have discernible differences in amino acid composition.
Since the group of proteins influencing the directionality of site-specific recombination reactions are highly diverse, are sometimes involved in additional processes, and are likely to have originated more than once during the course of evolution, we have chosen to describe them as RDFs. While xis and cox are adequate as gene names, the term RDF avoids the use of the potentially misleading terms ‘excise’ and ‘excisionase’.
RESULTS AND DISCUSSION
The identification of RDFs has previously proven difficult due to their small size and sequence diversity. In order to compile as complete a list of RDFs as possible we used a two-step approach. First, we used text-based searches to identify all of the previously annotated RDFs, noting which of these were accompanied by experimental support for their function. Secondly, we conducted a broad database search of both protein and nucleotide records using each of the previously identified RDFs as query sequences.
Determination of annotated excisionases
To collate all of the currently annotated RDFs, we first identified all existing GenBank records containing the text terms excisionase, excisase or xis. From this search a total of 204 records were found, of which 55 were duplicate records with the same accession number, and 62 of the remaining 149 records were duplicate entries of the same sequence with different accession numbers. After these were removed, the list contained records representing 87 unique sequences. Closer examination of these revealed that 39 were included only because the records contained cross-references to RDFs (24 to cognate integrases and 15 to other proteins) and these were also removed to leave a list of 48 putative RDFs.
There are compelling reasons to think that several of the protein entries appearing in this list do not in fact function as RDFs. For example, in mycobacteriophages L5 and D29, gene 34 was initially annotated as a putative excisionase since it encodes a small protein and is closely linked to the integrase gene (19,20). However, another reading frame, 34.1, was subsequently considered as a more likely candidate for encoding Xis since it is more highly conserved between these two phages (21). It is now known that neither 34 nor 34.1 of these phages encodes the RDF and there is good experimental evidence that this activity is provided by gene 36 (22). However, the incorrect annotations resulted in the errant assignment of orf53 from pREAT701 (23) which was annotated as a putative excisionase based on its similarities to D29 34.1. Each of these entries was removed from the list. In three other cases, in plasmid pME2200 and phages phiAR29 and A2, a gene was assigned as a putative excisionase on the basis that it is adjacent to an integrase gene, but without any additional supporting evidence. Since none of these genes has sequence features shared by other RDFs (see below) they were also removed from this list, although we obviously cannot exclude the possibility that they do provide RDF activity. We also removed gene SCE39.01c since it is incomplete and represents part of the longer gene SCE29.20c. Of the 38 remaining RDFs, all have either been shown experimentally to be required for excisive recombination, or are reasonably close relatives of those that have.
It is possible that other RDFs were missed in the text search simply due to variations in nomenclature. For example, the P2 cox gene—which acts as a transcriptional regulator as well as controlling directionality in recombination—was only included because of a linked protein record which identified it as an excisionase. There are three other annotated cox genes (encoded by phages HP1, K139 and WPhi) that were missed in the initial list, but were identified in subsequent sequence searches (see below). Finally, there are two serine integrases, encoded by phage TP901 (15) and Anabaena (XisF) (16) that require an RDF for excisive recombination. There is good experimental evidence for the RDF encoded by phage TP901 as well as the involvement of two proteins, XisH and XisI, in XisF-mediated recombination. The addition of these three proteins raises the number of unique RDFs that we have identified through text searches of existing database entries to 41.
Searches for unidentified excisionases
The compilation of RDFs was expanded by using sequence similarity based methods to search GenBank’s non-redundant (nr) database of proteins. A variety of search algorithms were used including PROBE (24), BLAST (25) and PSI-BLAST (26) (all three algorithms available from NCBI) and each of the 41 annotated RDFs were used as query sequences. At the beginning of the search we recognized that the small size and sequence diversity of this group of proteins may present serious difficulties in identifying RDFs based on sequence characteristics alone. This was particularly evident with the PROBE program, which utilizes iterative rounds of BLAST searches in conjunction with model building to search for distantly related proteins. While this is an effective method for identifying protein families (24), it was not helpful in identifying other RDFs since we frequently found that the resulting list of similar proteins contained none of the original sequences (presumably due to the small size of the query sequences). We therefore focused on using manual iterations of BLAST, and PSI-BLAST, in which proteins that were obviously not RDFs, typically larger proteins containing small segments of similarity, could be excluded.
Since these sequence-based searches generated lists containing many sequences, of which we suspected that only a subset are likely to function as RDFs, we used a secondary criterion to screen the results. Each of the proteins found with a moderate BLAST score (E < 10) was examined for the presence of a nearby integrase gene since RDF and recombinase genes are frequently closely linked. However, for putative RDFs within a bacterial genome (most likely as part of a prophage) we looked as far as 50 kb in either direction for an associated integrase, since this is a reasonable distance that an RDF and recombinase gene could be separated by in an integrated phage genome if these genes flank the attachment site. Using these two criteria, we identified a total of 16 previously unidentified RDFs.
An additional three putative RDFs were identified that did not appear to be accompanied by a recombinase gene. These are encoded by phages DR1455, phi-R67 and TM1. However, these all appeared with good BLAST scores (E < 10–4) and these three were thus included in the RDF compilation. Two other closely related proteins (B2168_C1_172 and SCE68.26c) also did not appear to have a cognate recombinase although their BLAST scores are also relatively poor (E = 10–3). Both of these protein sequences came from incomplete genome projects (of Mycobacteria leprae and Streptomyces coelicolor respectively) so that a thorough examination of the flanking sequences was not possible. However, we noted that in the case of B2168_C1_172, there was a good match (E = 10–22, identified through a TBLASTN search) to a segment of an integrase-like pseudogene located ∼41 kb away. Both of these putative RDFs were added to the list.
Since RDF genes are small, and some even overlap adjacent integrase genes, it is easy for them to escape annotation during analysis of the nucleotide sequence. We therefore collected the GenBank nucleotide records containing ‘bacteriophage’ or ‘integrase’ as key words using ENTREZ. These sequences were translated in six frames (using translate.pl written for this work; J.A.Lewis, unpublished) and formatted as a separate database (using FORMATDB from NCBI) that could be more readily searched using the BLAST program. By using each of the identified RDFs as query sequences, we found two new putative RDFs, one of which was previously described as a pseudogene in prophage DLP12 (27). Since only part of the gene could be identified this was not included in the list. The second candidate (Pspu, encoded by Pseudomonas putida, Table 1) overlaps an adjacent integrase gene and has strong similarity to the Lambda family of RDFs. This was added to the list to generate a final compilation of 63 known or putative RDF proteins. The complete list of RDFs is shown in Table 1.
Table 1. The 63 RDFs analyzed in this study.
| Familya |
Nameb |
Locationc |
Host |
Evidd |
Refs |
RDF gi # |
Rec typee |
Rec gi # |
| L5 | B2168_C1_172 | Prophage | Mycobacteria leprae | P | (34) | 467073 | ND | ND |
| L5 | DR1455 | Prophage | Deinococcus radiodurans | P | (35) | 6459215 | ND | ND |
| L5 | L5 | Phage | mycobacteria | E | (22) | 15892 | Tyr | 465416 |
| L5 | Rv2657c | Prophage | Mycobacteria tuberculosis | P | (32,36) | 1550698 | Tyr | 1550700 |
| L5 | SCE68.26c | Prophage | Streptomyces coelicolor | P | (37) | 5123673 | ND | ND |
| L5 | D29 | Phage | mycobacteria | P | (21,38) | 2358239 | Tyr | 3172283 |
| P22 | APSE-1 | Phage | Acyrthosiphon pisum | P | (39) | 6118035 | Tyr | 6118033 |
| P22 | P22 | Phage | Escherichia coli | E | (40) | 75990 | Tyr | 76009 |
| P22 | SfV | Phage | Shigella flexneri | P | (41) | 2465478 | Tyr | 2465477 |
| P22 | SfX | Phage | S.flexneri | P | (42) | 4099029 | Tyr | 4099030 |
| pSAM2 | pNL1 | Plasmid | Sphingomonas aromaticivorans | P | (43) | 3378297 | Tyr | 3378303 |
| pSAM2 | pSAM2 | Plasmid | Streptomyces ambofaciens | E | (44,45) | 3043524 | Tyr | 3043525 |
| pSAM2 | pSE101 | Plasmid | Saccharopolyspora erythraea | P | (46) | 1076058 | Tyr | 541467 |
| pSAM2 | pSE211 | Plasmid | S.erythraea | E | (47) | 152673 | Tyr | 152674 |
| pSAM2 | Rv2310 | Prophage | M.tuberculosis | P | (36) | 3261643 | Tyr | 1524291 |
| pSAM2 | Rv3750c | Prophage | M.tuberculosis | P | (36) | 2960174 | Tyr | 2960175 |
| pSAM2 | SCE29.20c | Prophage | S.coelicolor | P | (37) | 4490998 | Tyr | 4490997 |
| pSAM2 | TM1 | Prophage | Arthrobacter sp. TM1 | P | (48) | 8517283 | ND | ND |
| SLP1 | P4 | Phage | E.coli | P | (49) | 140147 | Tyr | 15176 |
| SLP1 | phi-R73 | Phage | E.coli | P | (50) | 93825 | Tyr | 93827 |
| SLP1 | SLP1 | Plasmid | S.coelicolor | E | (51) | 312936 | Tyr | 312937 |
| SLP1 | yp43 | Prophage | Yersinia pestis | P | (52) | 4106643 | Tyr | 4106629 |
| HP1 | HP1 | Phage | Haemophilus influenzae | E | (53) | 459180 | Tyr | 459175 |
| HP1 | K139 | Phage | Vibrio cholerae | P | (54) | 4530499 | Tyr | 4530503 |
| HP1 | phi-R67 | Prophage | E.coli | P | (55,56) | 141342 | ND | ND |
| HP1 | S2 | Phage | H.influenzae | P | (57) | 1679810 | Tyr | 1679808 |
| P2 | P2 | Phage | E.coli | E | (58,59) | 76820 | Tyr | 6136261 |
| P2 | WPhi | Phage | E.coli | P | (60) | 5824357 | Tyr | 5824355 |
| L54a | L54a | Phage | staphylococcus | E | (28) | 76013 | Tyr | 76011 |
| L54a | pXO1 | Plasmid | Bacillus anthracis | P | (61) | 4894317 | Tyr | 4894320 |
| L54a | T12 | Phage | Streptococcus pyogenes | P | (62) | 1877428 | Tyr | 1877429 |
| Tn916 | Tn1545 | Transposon | Streptococcus pneumoniae | E | (63) | 75987 | Tyr | 76007 |
| Tn916 | Tn1549 | Transposon | Enterococcus faecalis | P | (64) | 8100683 | Tyr | 8100684 |
| Tn916 | Tn5382 | Transposon | Enterococcus faecium | P | (65) | 3243184 | Tyr | 3243185 |
| Tn916 | Tn916 | Transposon | E.faecalis | E | (66,67) | 532534 | Tyr | 532535 |
| Lambda | 434 | Phage | E.coli | P | (68) | 801887 | Tyr | 215353 |
| Lambda | e14 | Prophage | E.coli | P | (69) | 7466710 | Tyr | 3024035 |
| Lambda | H19J | Phage | E.coli | P | (70) | 4490351 | Tyr | 4490352 |
| Lambda | HK022 | Phage | E.coli | E | (71) | 15761 | Tyr | 15760 |
| Lambda | HK97 | Phage | E.coli | P | (72) | 6901614 | Tyr | 6901614 |
| Lambda | Lambda | Phage | E.coli | E | (1,73) | 215134 | Tyr | 215133 |
| Lambda | Pspu | Prophage | Pseudomonas putida | P | (74) | 4520377f | Tyr | 4520380 |
| Lambda | P21 | Phage | E.coli | E | (75,76) | 215449 | Tyr | 215450 |
| Tn5276 | ICESt1 | Transposon | Streptococcus thermophilus | P | (77) | 6782410 | Tyr | 6782411 |
| Tn5276 | Tn5252 | Transposon | S.pneumoniae | P | (78) | 1361379 | Tyr | 1361380 |
| Tn5276 | Tn5276 | Transposon | L.lactis | E | (3) | 1075727 | Tyr | 1075733 |
| misc | 11 | Phage | staphylococcus | E | (79) | 455128 | Tyr | 166159 |
| misc | 16–3 | Phage | Rhizobium meliloti | E | (80) | 5824336 | Tyr | 5824335 |
| misc | 186 | Phage | E.coli | E | (81) | 3337276 | Tyr | 3337277 |
| misc | D3 | Phage | Pseudomonas aeruginosa | P | (82) | 9635627 | Tyr | 9635596 |
| misc | Gifsy-1 | Phage | Salmonella typhimurium | P | (83) | 3294479 | Tyr | 3294478 |
| misc | mv4 | Phage | Lactobacillus plantarum | P | (84) | 684926 | Tyr | 684925 |
| misc | Mx8 | Phage | Myxococcus xanthus | P | (29) | 2105132 | Tyr | 2149006 |
| misc | phi-80 | Phage | E.coli | E | (40) | 75989 | Tyr | 75992 |
| misc | phig1e | Phage | lactobacillus | P | (85) | 1926325 | Tyr | 1926326 |
| misc | SgiI | Prophage | Salmonella enterica DT104 | P | (86) | 9944850 | Tyr | 9944851 |
| misc | Tn4555 | Transposon | Bacteroides fragilis | P | (87) | 5453491 | Tyr | 5453489 |
| misc | VT2-Sa (933W) | Phage | E.coli | P | (88,89) | 5881594 | Tyr | 5881593 |
| misc | ydaQ | Prophage | E.coli | P | (69) | 1787608 | Tyr | 1787607 |
| misc | Rv1584c | Prophage | M.tuberculosis | P | (36) | 7476829 | Ser | 7476830 |
| misc | TP901-1 | Phage | L.lactis | E | (15) | 2924238 | Ser | 6808404 |
| misc | xisH | Chromosomal | anabaena | E | (16) | 1613875 | Ser | 1075645 |
| misc | xisI | Chromosomal | anabaena | E | (16) | 1613876 | Ser | 1075645 |
aThe family groupings based on sequence similarity.
bThe name used for the protein in this paper. For phages, plasmid and transposons locations, the name of the element is used to name the protein. For prophage and chromosomal locations, the gene name is used directly.
cLocation of the gene as found in database searches.
dType of evidence indicating functionality as an RDF: E, experimental; P, putative.
eThe type of recombinase as classified by its catalytic residue (Tyr or Ser).
fThe xis gene has not been annotated as an open reading frame. It is in this nucleotide record (4773–4531).
The main concern in establishing this list of putative RDFs was that we include as many candidate RDFs as possible while excluding all of those that do not function as RDFs. While we believe that in general this was achieved, we also identified proteins that matched some but not all of the criteria, and for which excision function cannot be ruled out. These borderline protein sequences can be viewed at the web site http://www.pitt.edu/~gfh/rdf.html.
Recombination directionality factor classification
Throughout the course of the extensive database searching described above it became evident that the RDFs and putative RDFs do not belong to a single group of closely related proteins. Attempts to align them all using multiple sequence alignment programs were problematic and we could not clearly identify any residues that were highly conserved in all or most members of the group. We therefore attempted to place them into family groups that were more closely related to each other than they were to other putative RDFs. An indication of appropriate groupings was first obtained by examining phenogram representations using guide trees generated by ClustalX (available from BioWeb at http://www.web.tiscalinet.it/biologia) analysis (Fig. 1). While many different trees can be generated, depending on the input order of the sequences and other features of the heuristics (only one example is represented in Fig. 1), those sequences lying close together in the phenogram typically remain together, suggesting that the list might be further sub-divided into smaller groups. This is supported by further PSI-BLAST analyses, where searches identify predominantly those proteins that are near one another in the phenograms.
Figure 1.
Phenogram of RDFs. A tree based on degrees of similarities between RDFs was calculated with CLUSTALX (using the default parameters from http://web.tiscalinet.it/biologia/) and rendered with the DrawGram program (from the PHYLIP package at http://evolution.genetics.washington.edu). The vertical bars indicate groups of RDFs that stay together during multiple cycles of tree generation. The groups are named (as shown on the right) according to a member for which there is experimental evidence of RDF activity.
From these analyses, we propose that 46 of the protein sequences can be assembled into 10 distinct groups or families. Since each of these families contains at least one RDF for which there is experimental support for its function, we named the families after such a member (i.e. P22, L5, pSAM2, SLP1, Tn916, L54a, P2, HP1, Lambda and Tn5276 families). Seventeen sequences did not assemble into families and are listed as miscellaneous. For most of the 10 families the membership is fairly obvious and family members are identified in early rounds of reiterative PSI-BLAST searches; in at least four of the families (Tn5275, Lambda, Tn916 and L54a) the PSI-BLAST searches converged without inclusion of any other putative RDFs. In contrast, members of the L5, pSAM2 and SLP1 subgroups repeatedly identified matches to each other, and these were thus joined into a larger family that we will refer to as the L5–SAM–SLP1 family. Members of the P22 family also appeared in these PSI-BLAST searches, but because of several insertions that are common to the P22 family and absent from the L5–SAM–SLP1 family, we will consider the P22 group as a separate family. The P2 and HP1 groups also appeared to be sufficiently similar to warrant inclusion into a single family (P2–HP1 family). This grouping is also supported by the observation that at least one member of both groups has been shown experimentally to function as cox genes in transcriptional regulation as well as recombination.
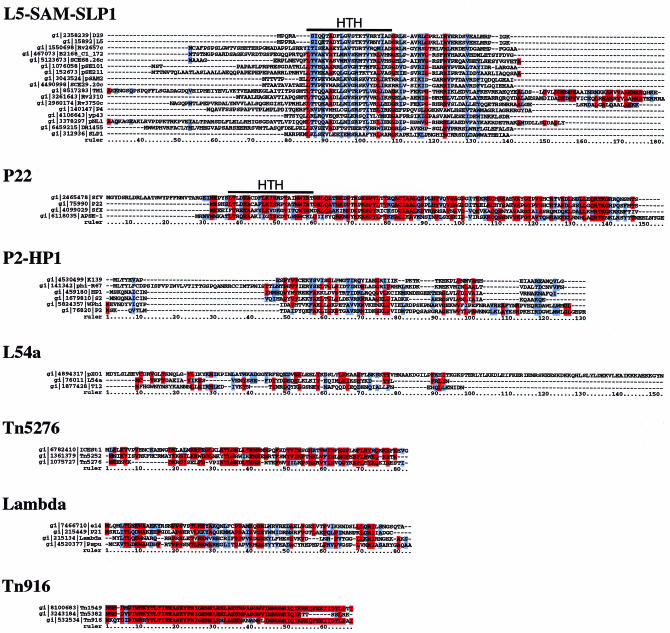
Amino acid sequence alignment illustrates the relationships among the RDF family members (Fig. 2). The largest group (with 17 members) is the L5–SAM–SLP1 family. The members of this group vary considerably in distance from the initiating amino acid to the closely related core segment of ∼50 residues which they all share. While this may indicate poor conservation of these parts of the proteins, it could also result from errant assignment of the translation initiation codons. There are no amino acids that are absolutely conserved among all the members, but there are many positions where the chemical character of the amino acids is shared (Fig. 2). The P2–HP1 family (which has six members) has a similar character with several well conserved residues but not that are common to all members.
Figure 2.
Sequence alignments of RDFs. RDFs within individual families are shown using alignments derived from CLUSTALX analysis (using the default parameters as in Fig. 1). Amino acid residues that are identical in 65% of the sequences are highlighted in red and residues that are similar among at least 75% of sequences are shown in blue. Similarity groupings were based on positive scoring substitutions as determined by the BLOSUM 85 substitution matrix (33). The location of a putative helix–turn–helix DNA binding motif is shown above the L5–SAM–SLP1 and P22 families. In cases where RDFs from different sources are of identical sequence, only one was used in the alignment and the view of each alignment is limited to a 150 residue segment containing the related sequences.
The remaining five groups each have fewer members, and it is perhaps not surprising that there are a number of absolutely conserved residues. In the P22 family, there is a segment of ∼110 residues, of which 21 are present in all four members. The Tn916 family is also a tight group (although with only three members; the Tn1545 Xis was not included since it is identical to that of Tn916, and their cognate integrases are also extremely similar), with a common segment of ∼70 residues of which 42 are present in all members. The Lambda, Tn5276 and L54a groups are rather more diverse than these. The Lambda group contains eight family members although the Xis encoded by phages HK97, HK022, 434 and H19J were excluded from the alignment because they are identical to the Lambda Xis. The L54a family is clearly the most diverse of these groups although the pXO1, L54a and T12 RDFs do appear to be more similar to each other than other RDFs, and they share properties regarding protein charge that are unique to this family (see below).
Protein charge
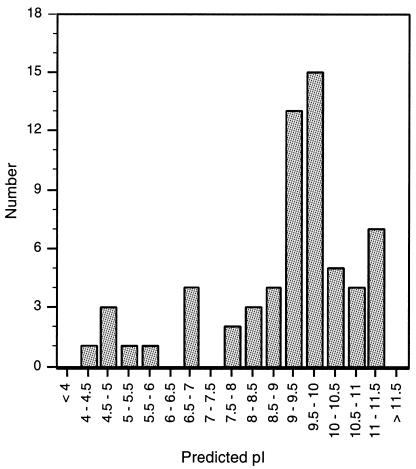
It has been reported previously that phage RDFs are typically basic, with pIs in the range of 9–10 (22,28). However, analysis of the pI values of the RDFs shown in Table 1 shows that while about half of the proteins fall into this category, there is actually a much broader range of pI values represented (Fig. 3). Interestingly, 10 RDFs have a pI <7, and these are listed in Table 2. This includes all three of the L54a family members as well as two (Mv4 and phig1e) that are in the miscellaneous group, but which appear to function with integrases that, along with those associated with the L54a RDF family, are in the LC3 group of recombinases (11) (Table 3). It therefore seems plausible that at least these five may have a shared ancestry.
Figure 3.
Distribution of isoelectric focusing point (pI) values among RDFs. The predicted pI was calculated for each RDF using Compute pI/MW (http://www.expasy.ch). The number of RDFs with pIs within 0.5 pH intervals was determined and plotted. The majority of the proteins are basic, with only 10 of the 63 RDFs having a pI < 7. Five of the seven proteins (Lambda, HK97, HK022, 434, H19J) in the 11–11.5 range have identical sequences.
Table 2. Acidic RDFs.
| Name |
pI |
RDF family |
Integrase familya |
| Mv4 | 6.51 | misc | LC3 |
| phig1e | 4.57 | misc | LC3 |
| L54a | 4.53 | L54a | LC3 |
| T12 | 5.15 | L54a | LC3 |
| pXO1 | 6.87 | L54a | misc |
| 11 | 4.13 | misc | phi11 |
| WPhi | 6.58 | P2 | P2 |
| orfA | 6.93 | pSAM2 | N/A |
| xisH | 4.78 | misc | serine |
| xisI | 5.88 | misc | serine |
aIntegrase family was based on Esposito’s classifications (11).
Table 3. RDF versus integrase classifications.
| Namea |
RDF family |
Integrase familyb |
| L5 | L5 | FRAT1 |
| Rv2657c | L5 | FRAT1 |
| D29 | L5 | FRAT1 |
| APSE-1 | P22 | P22 |
| P22 | P22 | P22 |
| SfV | P22 | P22 |
| SfX | P22 | P22 |
| pNL1 | pSAM2 | misc |
| pSAM2 | pSAM2 | misc |
| pSE101 | pSAM2 | pSE |
| pSE211 | pSAM2 | pSE |
| SCE29.20c | pSAM2 | pSE |
| P4 | SLP1 | P4 |
| phi-R73 | SLP1 | P4 |
| SLP1 | SLP1 | P4 |
| yp43 | SLP1 | P4 |
| Tn1545 | Tn916 | Tn916 |
| Tn1549 | Tn916 | Tn916 |
| Tn5382 | Tn916 | Tn916 |
| Tn916 | Tn916 | Tn916 |
| ICESt1 | Tn5276 | LC3 |
| Tn5252 | Tn5276 | LC3 |
| Tn5276 | Tn5276 | LC3 |
| L54a | L54a | LC3 |
| pXO1 | L54a | misc |
| T12 | L54a | LC3 |
| HP1 | HP1 | P2 |
| K139 | HP1 | P2 |
| S2 | HP1 | P2 |
| P2 | P2 | P2 |
| Wphi | P2 | P2 |
| 434 | Lambda | Lambda |
| e14 | Lambda | Lambda |
| H19J | Lambda | Lambda |
| HK022 | Lambda | Lambda |
| HK97 | Lambda | Lambda |
| Lambda | Lambda | Lambda |
| Pspu | Lambda | Lambda |
| P21 | Lambda | Lambda |
| 11 | misc | phi11 |
| 16-3 | misc | misc |
| 186 | misc | P2 |
| D3 | misc | P22 |
| Gifsy-1 | misc | Phi-80 |
| mv4 | misc | LC3 |
| Mx8 | misc | misc |
| phi-80 | misc | Phi-80 |
| phig1e | misc | LC3 |
| SgiI | misc | misc |
| Tn4555 | misc | misc |
| VT2-Sa (933W) | misc | phiCTX |
| ydaQ | misc | p4 |
Other RDF properties
Previous studies suggested that some RDFs have a putative helix–turn–helix DNA binding motif that is responsible for DNA binding (22,29). We thus examined all of the RDFs for the presence of this motif. Using the HTHpred program [written for this work using the method of Dodd and Egan (30)], 23 of the putative RDFs have a probability of >25% of containing this motif, and three others (SCE29.20c, SCE68.26c and TM1) had scores of 2.2 or greater, only just missing the 2.5 cut-off value representing a probability of 25% (30); these are therefore also reasonable candidates for having this motif (Fig. 2). The majority of the predicted DNA binding domains (19 of the 26), are in proteins that are in the L5, P22, pSAM2 and SLP1 families. Thus, of the 22 total members of these families, only three (pSE101, pSE211, SLP1) were not predicted to contain a helix–turn–helix motif by this analysis. None of the Lambda group of RDFs was predicted to contain a helix–turn–helix motif (31).
Evolutionary considerations
The compilation and grouping of these known and putative RDFs reveals at least three important insights into their evolution. First, while the members of some groups (e.g. the Lambda group) are encoded by phages that infect related hosts, this is not true for all families. For example, in the L5 sub-family, there are members encoded by mycobacteriophages as well as an RDF encoded by a putative prophage of Deinococcus radiodurans. In the pSAM2 sub family there are RDFs encoded by both plasmids and prophages within hosts as diverse as Sphingomonas aromaticivorans, Arthrobacter sp., Streptomyces ambofaciens, Saccharopolyspora erythraea, Mycobacterium tuberculosis and S.coelicolor. Assuming that the members of each family or sub-family do indeed arise from common ancestry, this suggests that the RDFs have disseminated broadly throughout the phage and plasmid population by extensive lateral exchange, which appears to be a common theme in phage evolution (32).
The second observation is that the RDFs appear to co-evolve with their cognate integrases. A comparison of independently generated families for the RDFs (this work) and phage integrases (11) reveals considerable congruence (Table 3). For 52 of the RDFs in Table 1, cognate full-length tyrosine integrase sequences have been identified, 40 of which were previously classified (11) and another 12 which have only recently been identified (D.Esposito, Invitrogen Corporation, personal communication). Most of the RDFs that can be grouped into families form groups which correspond to the family groupings of their cognate recombinases. For example, the four members of the Tn916 family of RDFs form the same group as those of their cognate integrases (Table 3). The simple interpretation of this pattern is that although the RDF genes may indulge in widespread lateral movement throughout the phage population that they tend to do so in partnership with their cognate integrase.
A significant departure of this general pattern is seen with the L54a and Tn5276 families of RDFs. In this case, the cognate integrases (ICESt1, Tn5252 and Tn5276 in the Tn5276 family, and L54a, pXO1 and T12 in the L54a family) all form a single family of LC3 integrases (the pXO1 integrase has been classified as a miscellaneous member). The lack of congruence cannot be simply explained as an artifact of the sequence analysis of the RDFs, since the L54a and Tn5276 RDF families are not only evidently different in their primary sequences (Fig. 2) but are substantially different in their overall protein charge (Fig. 3). It therefore seems likely that, at least in this case, the integrase and RDFs have followed distinct evolutionary paths.
The third consideration is whether all of these RDFs are derived from a single common ancestor or whether it is plausible that they have multiple origins. We suggest that there are likely to have been at least four separate origins for the following reasons. First, the L54a group not only shares little or no sequence similarity with the other RDF families but they are quite acidic with respect to their charge, in marked contrast with the other RDF families. It thus seems likely that these arose independently from other RDFs. Secondly, the P2–HP1 cox genes differ from other RDFs in that they are the only ones that also act as transcriptional regulators. We therefore suggest that these may also have arisen independently. Thirdly, the L5, SAM, SLP1 and P22 families all are predicted to contain helix–turn–helix DNA binding motifs, which are not observed in the Tn916, Tn5276 or Lambda families. Taken together with the extent of sequence divergence, it seems plausible that these could also have arisen from an independent origin. Finally, while the Tn916, Tn5276 and Lambda families are distinctly different from each other with regard to their primary structures, they are all small, basic proteins and their evolutionary relationships to each other are less clear. Finally, we recognize that speculating on the origins of these proteins must be cautious, since their small sizes and roles as architectural rather than catalytic molecules may enable more frequent acquisition of function through convergent evolution than would be expected for larger enzymes.
Further considerations
In light of the great diversity of RDFs described here and the likelihood of multiple origins, what proportion of RDFs has been identified? We note that more than 120 Int-Y have been described (11), most of which are likely to utilize an accessory protein to regulate directionality. Since the total number of RDFs described here is only ∼50% of this number, it seems that there are many RDFs for which sequence information already exists, whose functions have not yet been identified. Since we have not found these by the exhaustive database analyses described above, their identification will have to await experimental dissection of other recombination systems.
The architectural role of Xis in Lambda recombination is well established and it seems probable that other RDFs associated with tyrosine recombinases will act similarly. However, the mechanism of directionality regulation has not been clearly established in any of the integration systems that use a large serine recombinase. The only such system that has been investigated biochemically is that encoded by Streptomyces phage φc31, although no φc31 RDF has been identified. However, the φc31 attP site is substantially smaller than those of the Int-Y that have been studied, suggesting a quite different recombinational process that does not require the formation of complex higher order protein–DNA structures (13,14). Nevertheless, there are at least four RDFs that are associated with an Int-S, all of which are classified as miscellaneous. One putative RDF (Rv1584c) associated with an Int-S (Rv1586c) has weak sequence similarity to the L5 family of RDFs, of which Rv2657c is a member. Rv2657c, which is associated with an Int-Y (Rv2659c), and Rv1584c are both found in prophage-like elements (φRv2 and φRv1 in M.tuberculosis) that have a colinear arrangement of these genes, adding further support to the identification of Rv1584c as a putative RDF. There is experimental evidence supporting the function of the TP901 RDF as well as Anabaena XisH and XisI (15,16). However, the role that the RDF proteins play in control of directionality in the Int-S systems remains to be elucidated.
Finally, the compilation and classification of RDF and putative RDFs should be helpful in the future annotation of plasmid, phage and prophage sequences. As new RDFs are identified and added to this list we expect to see the formation of additional families of RDF proteins and to gain further insights into their evolution and function.
Web site
Further details on the RDFs, putative RDFs, and borderline protein sequences are available on the World Wide Web at http://www.pitt.edu/~gfh/rdf.html. Additional information on the cognate integrases is available at the tyrosine recombinase web site at http://members.home.net/domespo/trhome.html.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dominic Esposito for his help in classifying new integrases and his comments on the manuscript. We also like to thank Aisha Mitchell for technical support. This work was supported by NIH grants GM59968 and AI45683.
References
- 1.Nash H.A. (1981) Integration and excision of bacteriophage Lambda: the mechanism of conservation site specific recombination. Annu. Rev. Genet., 15, 143–167. [DOI] [PubMed] [Google Scholar]
- 2.Martin C., Mazodier,P., Mediola,M.V., Gicquel,B., Smokvina,T., Thompson,C.J. and Davies,J. (1991) Site-specific integration of the Streptomyces plasmid pSAM2 in Mycobacterium smegmatis. Mol. Microbiol., 5, 2499–2502. [DOI] [PubMed] [Google Scholar]
- 3.Rauch P.J. and de Vos,W.M. (1994) Identification and characterization of genes involved in excision of the Lactococcus lactis conjugative transposon Tn5276. J. Bacteriol., 176, 2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nash H.A. and Robertson,C.A. (1981) Purification and properties of the Escherichia coli protein factor required for Lambda integrative recombination. J. Biol. Chem., 256, 9246–9253. [PubMed] [Google Scholar]
- 5.Bushman W., Thompson,J.F., Vargas,L. and Landy,A. (1985) Control of directionality in Lambda site specific recombination. Science, 230, 906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abremski K. and Gottesman,S. (1982) Purification of the bacteriophage Lambda xis gene product required for Lambda excisive recombination. J. Biol. Chem., 257, 9658–9662. [PubMed] [Google Scholar]
- 7.Robertson C.A. and Nash,H.A. (1988) Bending of the bacteriophage Lambda attachment site by Escherichia coli integration host factor. J. Biol. Chem., 263, 3554–3557. [PubMed] [Google Scholar]
- 8.Yin S., Bushman,W. and Landy,A. (1985) Interaction of the Lambda site-specific recombination protein Xis with attachment site DNA. Proc. Natl Acad. Sci. USA, 82, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moitoso de Vargas L. and Landy,A. (1991) A switch in the formation of alternative DNA loops modulates Lambda site-specific recombination. Proc. Natl Acad. Sci. USA, 88, 588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z., Gumport,R.I. and Gardner,J.F. (1998) Defining the structural and functional roles of the carboxyl region of the bacteriophage Lambda excisionase (Xis) protein. J. Mol. Biol., 281, 651–661. [DOI] [PubMed] [Google Scholar]
- 11.Esposito D. and Scocca,J.J. (1997) The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res., 25, 3605–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christiansen B., Brondsted,L., Vogensen,F.K. and Hammer,K. (1996) A resolvase-like protein is required for the site-specific integration of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol., 178, 5164–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorpe H.M. and Smith,M.C. (1998) In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl Acad. Sci. USA, 95, 5505–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorpe H.M., Wilson,S.E. and Smith,M.C. (2000) Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol. Microbiol., 38, 232–241. [DOI] [PubMed] [Google Scholar]
- 15.Breuner A., Brondsted,L. and Hammer,K. (1999) Novel organization of genes involved in prophage excision identified in the temperate lactococcal bacteriophage TP901-1. J. Bacteriol., 181, 7291–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramaswamy K.S., Carrasco,C.D., Fatma,T. and Golden,J.W. (1997) Cell-type specificity of the Anabaena fdxN-element rearrangement requires xisH and xisI. Mol. Microbiol., 23, 1241–1249. [DOI] [PubMed] [Google Scholar]
- 17.Esposito D., Wilson,J.C. and Scocca,J.J. (1997) Reciprocal regulation of the early promoter region of bacteriophage HP1 by the Cox and Cl proteins. Virology, 234, 267–276. [DOI] [PubMed] [Google Scholar]
- 18.Saha S., Haggard-Ljungquist,E. and Nordstrom,K. (1987) The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J., 6, 3191–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatfull G.F. and Sarkis,G.J. (1993) DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol., 7, 395–405. [DOI] [PubMed] [Google Scholar]
- 20.Ribeiro G., Viveiros,M., David,H.L. and Costa,J.V. (1997) Mycobacteriophage D29 contains an integration system similar to that of the temperate mycobacteriophage L5. Microbiology, 143, 2701–2708. [DOI] [PubMed] [Google Scholar]
- 21.Ford M.E., Sarkis,G.J., Belanger,A.E., Hendrix,R.W. and Hatfull,G.F. (1998) Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol., 279, 143–164. [DOI] [PubMed] [Google Scholar]
- 22.Lewis J.A. and Hatfull,G.F. (2000) Identification and characterization of mycobacteriophage L5 excisionase. Mol. Microbiol., 35, 350–360. [DOI] [PubMed] [Google Scholar]
- 23.Takai S., Hines,S.A., Sekizaki,T., Nicholson,V.M., Alperin,D.A., Osaki,M., Takamatsu,D., Nakamura,M., Suzuki,K., Ogino,N. et al. (2000) DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun., 68, 6840–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuwald A.F., Liu,J.S., Lipman,D.J. and Lawrence,C.E. (1997) Extracting protein alignment models from the sequence database. Nucleic Acids Res., 25, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsey D.F., Mullin,D.A. and Walker,J.R. (1989) Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J. Bacteriol., 171, 6197–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Z.H. and Lee,C.Y. (1989) Nucleotide sequence and genetic characterization of staphylococcal bacteriophage L54a int and xis genes. J. Bacteriol., 171, 4146–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmi D., Magrini,V., Hartzell,P.L. and Youderian,P. (1998) Genetic determinants of immunity and integration of temperate Myxococcus xanthus phage Mx8. J. Bacteriol., 180, 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodd I.B. and Egan,J.B. (1990) Improved detection of helix–turn–helix DNA-binding motifs in protein sequences. Nucleic Acids Res., 18, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho E.H., Alcaraz,R.,Jr, Gumport,R.I. and Gardner,J.F. (2000) Characterization of bacteriophage Lambda excisionase mutants defective in DNA binding. J. Bacteriol., 182, 5807–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrix R.W., Smith,M.C., Burns,R.N., Ford,M.E. and Hatfull,G.F. (1999) Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl Acad. Sci. USA, 96, 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robison K. (1994) GenBank accession no. AAA17257.1.
- 35.White O., Eisen,J.A., Heidelberg,J.F., Hickey,E.K., Peterson,J.D., Dodson,R.J., Haft,D.H., Gwinn,M.L., Nelson,W.C., Richardson,D.L. et al. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole S.T., Brosch,R., Parkhill,J., Garnier,T., Churcher,C., Harris,D., Gordon,S.V., Eiglmeier,K., Gas,S., Barry,C.E.,III et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 37.Murphy L., Harris,D., James,K.D., Parkhill,J., Barrell,B.G. and Rajandream,M.A. (1999) EMBL accession no. T36276.
- 38.Peña C.E., Stoner,J. and Hatfull,G.F. (1998) Mycobacteriophage D29 integrase-mediated recombination: specificity of mycobacteriophage integration. Gene, 225, 143–151. [DOI] [PubMed] [Google Scholar]
- 39.van der Wilk F., Dullemans,A.M., Verbeek,M. and van den Heuvel,J.F. (1999) Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology, 262, 104–113. [DOI] [PubMed] [Google Scholar]
- 40.Leong J.M., Nunes-Duby,S.E., Oser,A.B., Lesser,C.F., Youderian,P., Susskind,M.M. and Landy,A. (1986) Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J. Mol. Biol., 189, 603–616. [DOI] [PubMed] [Google Scholar]
- 41.Huan P.T., Bastin,D.A., Whittle,B.L., Lindberg,A.A. and Verma,N.K. (1997) Molecular characterization of the genes involved in O-antigen modification, attachment, integration and excision in Shigella flexneri bacteriophage SfV. Gene, 195, 217–227. [DOI] [PubMed] [Google Scholar]
- 42.Guan S. and Verma.,N.K. (1996) GenBank accession no. AAD10294.1.
- 43.Romine M.F., Stillwell,L.C., Wong,K.K., Thurston,S.J., Sisk,E.C., Sensen,C., Gaasterland,T., Fredrickson,J.K. and Saffer,J.D. (1999) Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol., 181, 1585–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boccard F., Smokvina,T., Pernodet,J.L., Friedmann,A. and Guerineau,M. (1989) The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J., 8, 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raynal A., Tuphile,K., Gerbaud,C., Luther,T., Guerineau,M. and Pernodet,J.L. (1998) Structure of the chromosomal insertion site for pSAM2: functional analysis in Escherichia coli. Mol. Microbiol., 28, 333–342. [DOI] [PubMed] [Google Scholar]
- 46.Brown D.P., Idler,K.B., Backer,D.M., Donadio,S. and Katz,L. (1994) Characterization of the genes and attachment sites for site-specific integration of plasmid pSE101 in Saccharopolyspora erythraea and Streptomyces lividans. Mol. Gen. Genet., 242, 185–193. [DOI] [PubMed] [Google Scholar]
- 47.Katz L., Brown,D.P. and Donadio,S. (1991) Site-specific recombination in Escherichia coli between the att sites of plasmid pSE211 from Saccharopolyspora erythraea.Mol. Gen. Genet., 227, 155–159. [DOI] [PubMed] [Google Scholar]
- 48.Gartemann K.-H., Fiedler,J., Schmitz,A., Zellermann,E.-M. and Eichenlaub,R. (1999) GenBank accession no. AAF76245.1.
- 49.Halling C., Calendar,R., Christie,G.E., Dale,E.C., Deho,G., Finkel,S., Flensburg,J., Ghisotti,D., Kahn,M.L., Lane,K.B. et al. (1990) DNA sequence of satellite bacteriophage P4. Nucleic Acids Res., 18, 1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J., Inouye,M. and Inouye,S. (1991) Association of a retroelement with a P4-like cryptic prophage (retronphage phi R73) integrated into the selenocystyl tRNA gene of Escherichia coli. J. Bacteriol., 173, 4171–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brasch M.A. and Cohen,S.N. (1993) Excisive recombination of the SLP1 element in Streptomyces lividans is mediated by Int and enhanced by Xis. J. Bacteriol., 175, 3075–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchrieser C., Rusniok,C., Frangeul,L., Couve,E., Billault,A., Kunst,F., Carniel,E. and Glaser,P. (1999) The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun., 67, 4851–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esposito D. and Scocca,J.J. (1994) Identification of an HP1 phage protein required for site-specific excision. Mol. Microbiol., 13, 685–695. [DOI] [PubMed] [Google Scholar]
- 54.Nesper J., Blass,J., Fountoulakis,M. and Reidl,J. (1999) Characterization of the major control region of Vibrio cholerae bacteriophage K139: immunity, exclusion and integration. J. Bacteriol., 181, 2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu M.Y., Inouye,M. and Inouye,S. (1990) Retron for the 67-base multicopy single-stranded DNA from Escherichia coli: a potential transposable element encoding both reverse transcriptase and Dam methylase functions. Proc. Natl Acad. Sci. USA, 87, 9454–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dodd I.B. and Egan,J.B. (1996) The Escherichia coli retrons Ec67 and Ec86 replace DNA between the cos site and a transcription terminator of a 186-related prophage. Virology, 219, 115–124. [DOI] [PubMed] [Google Scholar]
- 57.Skowronek K. and Baranowski,S. (1997) The relationship between HP1 and S2 bacteriophages of Haemophilus influenzae. Gene, 196, 139–144. [DOI] [PubMed] [Google Scholar]
- 58.Yu A. and Haggard-Ljungquist,E. (1993) The Cox protein is a modulator of directionality in bacteriophage P2 site-specific recombination. J. Bacteriol., 175, 7848–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eriksson J.M. and Haggard-Ljungquist,E. (2000) The multifunctional bacteriophage P2 cox protein requires oligomerization for biological activity. J. Bacteriol., 182, 6714–6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu T. and Haggard-Ljungquist,E. (1999) The transcriptional switch of bacteriophage WPhi, a P2-related but heteroimmune coliphage. J. Virol., 73, 9816–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okinaka R.T., Cloud,K., Hampton,O., Hoffmaster,A.R., Hill,K.K., Keim,P., Koehler,T.M., Lamke,G., Kumano,S., Mahillon,J. et al. (1999) Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol., 181, 6509–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McShan W.M., Tang,Y.F. and Ferretti,J.J. (1997) Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol. Microbiol., 23, 719–728. [DOI] [PubMed] [Google Scholar]
- 63.Poyart-Salmeron C., Trieu-Cuot,P., Carlier,C. and Courvalin,P. (1989) Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J., 8, 2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garnier F., Taourit,S., Glaser,P., Courvalin,P. and Galimand,M. (2000) Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology, 146, 1481–1489. [DOI] [PubMed] [Google Scholar]
- 65.Carias L.L., Rudin,S.D., Donskey,C.J. and Rice,L.B. (1998) Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol., 180, 4426–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flannagan S.E., Zitzow,L.A., Su,Y.A. and Clewell,D.B. (1994) Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid, 32, 350–354. [DOI] [PubMed] [Google Scholar]
- 67.Marra D. and Scott,J.R. (1999) Regulation of excision of the conjugative transposon Tn916. Mol. Microbiol., 31, 609–621. [DOI] [PubMed] [Google Scholar]
- 68.Baker J., Limberger,R., Schneider,S.J. and Campbell,A. (1991) Recombination and modular exchange in the genesis of new lambdoid phages. New Biol., 3, 297–308. [PubMed] [Google Scholar]
- 69.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt H. and Karch,H. (2000) Chromosomal insertion sites of Shiga toxin 1-converting bacteriophage H19J. GenBank accession no. CAB38714.1.
- 71.Yagil E., Dolev,S., Oberto,J., Kislev,N., Ramaiah,N. and Weisberg,R.A. (1989) Determinants of site-specific recombination in the lambdoid coliphage HK022. An evolutionary change in specificity. J. Mol. Biol., 207, 695–717. [DOI] [PubMed] [Google Scholar]
- 72.Juhala R.J., Ford,M.E., Duda,R.L., Youlton,A., Hatfull,G.F. and Hendrix,R.W. (2000) Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol., 299, 27–51. [DOI] [PubMed] [Google Scholar]
- 73.Ball C.A. and Johnson,R.C. (1991) Efficient excision of phage Lambda from the Escherichia coli chromosome requires the Fis protein. J. Bacteriol., 173, 4027–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nashimoto H. (1999) NCBI GenBank accession no. BAA75916.1 GI:4520380.
- 75.Schneider S.J. (1992) PhD dissertation, Stanford University, Stanford, CA.
- 76.Wang H., Yang,C.H., Lee,G., Chang,F., Wilson,H., del Campillo-Campbell,A. and Campbell,A. (1997) Integration specificities of two lambdoid phages (21 and e14) that insert at the same attB site. J. Bacteriol., 179, 5705–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burrus V., Roussel,Y., Decaris,B. and Guedon,G. (2000) Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol., 66, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kilic A.O., Vijayakumar,M.N. and al-Khaldi,S.F. (1994) Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J. Bacteriol., 176, 5145–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ye Z.H., Buranen,S.L. and Lee,C.Y. (1990) Sequence analysis and comparison of int and xis genes from staphylococcal bacteriophages L54a and phi 11. J. Bacteriol., 172, 2568–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Semsey S., Papp,I., Buzas,Z., Patthy,A., Orosz,L. and Papp,P.P. (1999) Identification of site-specific recombination genes int and xis of the Rhizobium temperate phage 16-3. J. Bacteriol., 181, 4185–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dodd I.B., Reed,M.R. and Egan,J.B. (1993) The Cro-like Apl repressor of coliphage 186 is required for prophage excision and binds near the phage attachment site. Mol. Microbiol., 10, 1139–1150. [DOI] [PubMed] [Google Scholar]
- 82.Kropinski A.M. (2000) Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol., 182, 6066–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Figueroa-Bossi N. and Bossi,L. (1999) Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol., 33, 167–176. [DOI] [PubMed] [Google Scholar]
- 84.Dupont L., Boizet-Bonhoure,B., Coddeville,M., Auvray,F. and Ritzenthaler,P. (1995) Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol., 177, 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kodaira K.I., Oki,M., Kakikawa,M., Watanabe,N., Hirakawa,M., Yamada,K. and Taketo,A. (1997) Genome structure of the Lactobacillus temperate phage phi g1e: the whole genome sequence and the putative promoter/repressor system. Gene, 187, 45–53. [DOI] [PubMed] [Google Scholar]
- 86.Boyd D.A., Peters,G.A., Ng,L. and Mulvey,M.R. (2000) Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica typhymurium DT104. FEMS Microbiol. Lett., 189, 285–291. [DOI] [PubMed] [Google Scholar]
- 87.Tribble G.D., Parker,A.C. and Smith,C.J. (1999) Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid, 42, 1–12. [DOI] [PubMed] [Google Scholar]
- 88.Miyamoto H., Nakai,W., Yajima,N., Fujibayashi,A., Higuchi,T., Sato,K. and Matsushiro,A. (1999) Sequence analysis of Stx2-converting phage VT2-Sa shows a great divergence in early regulation and replication regions. DNA Res., 6, 235–240. [DOI] [PubMed] [Google Scholar]
- 89.Plunkett G.,III, Rose,D.J., Durfee,T.J. and Blattner,F.R. (1999) Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol., 181, 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]