Abstract
Along protein sequences, co-evolution analysis identifies residue pairs demonstrating either a specific co-adaptation, where changes in one of the residues are compensated by changes in the other during evolution or a less specific external force that affects the evolutionary rates of both residues in a similar magnitude. In both cases, independently of the underlying cause, co-evolutionary signatures within or between proteins serve as markers of physical interactions and/or functional relationships. Depending on the type of protein under study, the set of available homologous sequences may greatly differ in size and amino acid variability. BIS2Analyzer, openly accessible at http://www.lcqb.upmc.fr/BIS2Analyzer/, is a web server providing the online analysis of co-evolving amino-acid pairs in protein alignments, especially designed for vertebrate and viral protein families, which typically display a small number of highly similar sequences. It is based on BIS2, a re-implemented fast version of the co-evolution analysis tool Blocks in Sequences (BIS). BIS2Analyzer provides a rich and interactive graphical interface to ease biological interpretation of the results.
INTRODUCTION
In recent years, a particular focus has been drawn to the study of co-evolving residues within a protein and among proteins. Co-evolving residues in a protein structure, possibly a complex, correspond to groups of residues whose mutations have arisen simultaneously during the evolution of different species and this is due to several possible reasons involving the 3D shape of the protein: functional interactions, conformational changes and folding. Several studies addressed the problem of extracting signals of co-evolution between residues. All these methods provide sets of co-evolved residues that are usually physically close in the 3D structure (1–9) and form connected networks covering roughly a third of the entire structure. Co-evolved residues have been demonstrated, for a few protein complexes (for which experimental data are available), to play a crucial role in allosteric mechanisms (1,3,10), to maintain short paths in network communication and to mediate signaling (11,12). Methods such as Direct Coupling Analysis (DCA) (5), EVcouplings (4) and PSICOV (7) are applicable to protein families displaying a large number of evolutionarily related sequences and sufficient divergence, these characteristics constituting the bottleneck of today co-evolution analysis methods (13). The requirement on the large number of sequences has been dropped in recently developed methods (14) but the divergence of the sequences remains a mandatory constraint.
For many proteins, characteristic of vertebrate or viral species, the statistics that current co-evolution methods require (to estimate the ‘background noise’ and the relevance of the co-evolution signals) are not applicable because of the reduced number of sequences, either coming from species or from populations and their conservation. Hence, alternative paradigms should be followed. To overcome these difficulties, we developed a fast algorithm for the co-evolution analysis of relatively small sets of sequences (where ‘small’ means <50 sequences) displaying high similarity, called BIS2 (15). BIS2 is a computationally efficient version of Blocks In Sequences (BIS) (16). BIS2 is a combinatorial method that could successfully handle highly conserved proteins, such as the amyloid β peptide, playing an important role in Alzheimer's disease, and families of very few sequences, such as the adenosine triphosphatase (ATPase) protein families, characterized by conserved motifs. These studies also highlighted that co-evolving protein fragments and not only residues, are indicators of important information explaining: folding intermediates, peptide assembly, key mutations with known roles in genetic diseases and distinguished subfamily-dependent motifs. They could capture, with high precision, experimentally verified hotspots residues (15,16).
BIS2 high performance (15) allows, today, to open the way to co-evolution studies of protein–protein interaction networks in viral genomes at the genotype level. With BIS2, a complete co-evolution analysis of the small Hepatitis C virus (HCV) genome of 10 proteins and the reconstruction of the associated interaction network at the residue/domain resolution was possible (15).
Web servers for co-evolution analyzes have been proposed for DCA (http://dca.rice.edu/) and EVcouplings (http://evfold.org/evfold-web/) among others (17–24); in particular, we notice CAPS (http://caps.tcd.ie/; (25)) and ContactMap (http://raptorx.uchicago.edu/ContactMap; (14)), designed to work with very small sets of sequences. Such services allow the user to provide a MSA, run the analysis, and download the results. Usually, it is possible to map co-evolved residues on a protein structure, or to get a contact map highlighting the ability of the method to correctly predict 3D contacts from sequence information. To the best of our knowledge, previously published web servers for protein co-evolution analysis do not allow the user to customize output visualization, even though some of them provide highly detailed information.
Below, we describe BIS2Analyzer, a web server that combines an easy-to-use interface for BIS2 co-evolution analysis and a rich and interactive output visualization. BIS2Analyzer was conceived to identify specific mutagenesis sites, find evidence for protein-protein interactions and/or conformational changes, find specific residues for guiding folding or docking, design experimental cross-linking.
BIS2Analyzer WORKFLOW
BIS2Analyzer takes as input a multiple sequence alignment (MSA) of homologous protein sequences and, optionally, a phylogenetic tree built on the alignment. It is run with a number of parameter values, which are automatically set, but can be fully customized by the user. The output consists of a set of clusters of co-evolving residues, possibly associated to different parameters (dimension, block mode, alphabet—see below). Statistical significance and similarity scores are provided for each prediction. BIS2Analyzer graphical interface design offers a framework helping the user to easily analyze the results and to reason on potential experimental hypotheses built from identified correlations. Co-evolution clusters are displayed on the MSA and can be mapped to a reference sequence of choice or on the protein structure, when available. Inter-protein co-evolution analysis is specifically addressed, with the possibility to visualize two structures for interactive inspection of binding modes between the proteins. Visualization of clusters of co-evolving residues can be enabled/disabled interactively. The user can also access and download all html files displaying the results of BIS2 analysis. Textual files containing full output information are provided for ease of further analyzes.
THE BIS2 ALGORITHM
The description of BIS2 algorithm, used in BIS2Analyzer, appeared in (15,16). BIS2 is a combinatorial method structured in three main steps. First, it detects co-evolving residues among each pair of alignment positions and associates a co-evolution score to the pairs. To do this, each position of the alignment, called a hit, is considered as a starting point for a search of all other positions in the alignment that present a similar distribution of amino acids as the hit. The co-evolution score, defined in the interval [0, 1] (0 stands for absence and 1 for perfect signal of co-evolution), describes the amino acid distribution in a pair of positions of the sequence alignment. Second, BIS2 constructs a co-evolution score matrix, where entries in the matrix correspond to co-evolution scores for pairs of positions. Third, BIS2 clusters the co-evolution matrix with CLusters AGgregation (CLAG) (26) and identifies groups of positions displaying similar co-evolution patterns with all other positions in the alignment.
Note that BIS2 has been designed for alignments with relatively high conservation levels (16) and it can be parameterized accordingly. In particular, it deals with very few conserved sequences and allows the analysis of sequences in genotypic viral populations and conserved vertebrate protein families. The list of BIS2 parameters is as follows:
Dimension parameter
Given a position in the MSA, exceptions are distinct amino acid types occurring only once in the position. The dimension of a position is the number of its exceptions. Given a maximum dimension D, BIS2 is run for each dimension d ≤ D. This has the effect of discarding all positions with more than D exceptions from the analysis.
Blocks analysis
BIS2 can be run for identifying co-evolving blocks, that is protein fragments (16), instead of just residues. Each hit is extended to a block by considering the maximum number of positions around the hit that preserve the same distribution.
Alphabet reduction
Amino acid variability within the same physico-chemical class can be neglected by reducing the alphabet. Namely, BIS2 can be run on a MSA where each amino acid is replaced by a letter corresponding to the physico-chemical class it belongs to (see below). This feature is useful when analyzing datasets displaying moderate residue variability.
EXAMPLES OF BIS2Analyzer PREDICTIONS
BIS2 is specifically designed to find co-evolution signals on conserved sequences or conserved motifs. In (16), we have applied the method to several protein families characterized by a limited number of sequences and relatively high API (Average Pairwise Identity). We have also shown that the method can successfully detect signals between conserved motifs lying in rather diverged sequences. This was done on the ATPase protein family Upf1, comprising 18 sequences with API 0.58 and 677 positions to be analyzed. Co-evolved residue pairs were detected among a number of known conserved ATPase motifs and the prediction was realized with high specificity (0.99) and accuracy (0.92). BIS2 was also applied to the 10 proteins comprising the genome of HCV (15), opening the way to co-evolution analysis in viral populations.
Here, we illustrate the usefulness and accuracy of BIS2Analyzer on other proteins (see Table 1 for their characteristics), coming from either species or viral populations. We highlight predictions of direct correlations between pairs of residues or of networks of residues, within and between proteins. These residues need not be in physical proximity within crystallographic structures, but may come close to each other upon conformational changes taking place along the life of the protein. Therefore, BIS2 aims at finding signals that are possibly different than direct contacts, in contrast to many existing co-evolution analysis methods, especially designed to predict 3D contacts. Finally, we suggest a new computational strategy to analyze large datasets of diverged sequences with BIS2Analyzer.
Table 1. BIS2Analyzer computational time on different protein families.
| # Seqs | AL (aa) | API (%) | Time* | |
|---|---|---|---|---|
| Amyloid β peptide | 80 | 43 | 87 | 13’ |
| B domain/protein A | 452 | 62 | 82 | 2΄49΄ |
| c-KIT | 81 | 976 | 67 | 1h47΄30΄ |
| HCV NS5A | 40 | 451 | 94 | 3΄37″ |
| HCV NS3-NS5B | 27 | 1222 | 92 | 37΄40″ |
| Morbillivirus protein N | 144 | 387 | 73 | 21΄39″ |
# Seqs = number of sequences; AL = Alignment Length; API = Average Pairwise Identity; * execution realized on an Intel(R) Xeon(R) CPU E5-2440 0 @ 2.40GHz.
In the first two examples below, BIS2Analyzer was compared to several web servers and programs: EVcouplings (with option PLM, pseudo-likelihood maximization approach (21,27,28)), DCA and PSICOV, all of them producing a list of predicted pairs of co-evolving positions ranked according to best confidence values. Given that the reliability of statistical methods strongly depends on the number of input sequences, we ran the above tools on two MSAs, in addition to the ones described in Table 1, consisting of larger sets of sequences. For each method, we considered the top 50 predicted pairs. Other two comparisons were realized with CAPS and ContactMap.
Fragments of residues in contact within a protein
Amyloid β is a peptide playing a crucial role in Alzheimer. There is experimental evidence that six regions (32 aa in whole) of the protein sequence play a role in the disease. BIS2Analyzer finds 5 co-evolution clusters, for a total of 30 aa, and 26 of these residues overlap the 6 regions with known function (16). On their 50 top scored pairs, EVcouplings, DCA and PSICOV do not provide successful results compared to BIS2Analyzer, as reported in Table 2. Similarly, low performance is reported for CAPS and ContactMap.
Table 2. Performances of various co-evolution analysis methods.
| Amyloid β peptide | B domain/protein A | |||
|---|---|---|---|---|
| C/G/P (TP) | Pr (TP)(R) | C/G/P (TP) | Pr (TP) | |
| BIS2 | 5 (4) | 30 (26)(6) | 5 (2) | 28 (10) |
| CAPS | 3 (2) | 14 (6)(1) | 6 (0) | 22 (1) |
| ContactMap* | 50 (13) | 28 (14)(1) | 50 (16) | 25 (11) |
| DCA | 50 (8) | 29 (14)(3) | 50 (1) | 30 (4) |
| DCAa | 50 (26) | 37 (23)(5) | 50 (2) | 48 (11) |
| DCAb | 50 (13) | 29 (15)(3) | 50 (1) | 42 (10) |
| EVcouplings | 50 (2) | 28 (13)(1) | 50 (0) | 32 (3) |
| EVcouplingsa | 50 (2) | 29 (19)(1) | 50 (0) | 26 (1) |
| EVcouplingsb | 50 (2) | 27 (16)(1) | 50 (3) | 46 (9) |
| PSICOV | 50 (11) | 33 (20)(3) | 50 (1) | 24 (4) |
| PSICOVa | 50 (20) | 30 (26)(3) | 50 (4) | 45 (8) |
| PSICOVb | - | - | 50 (4) | 44 (12) |
C/G/P = predicted Cluster/Group/Pairs (outputs: C for BIS2, G for CAPS and P for all other methods) depending on the method; Pr = predicted residues (that is, the total number of different residues in C/G/P); TP = True Positives; R= experimental functional regions that are, at least partially, predicted (at least one pair of residues lies within the same C/G/P).
* ContactMap built a MSA of 73 sequences and 27% API for amyloid β peptide and a dataset of 56 sequences and 40% API for B domain/protein A. All other methods are run on the MSA described in Table 1, unless specified differently.
a MSA for amyloid β peptide: 919 sequences (NCBI PF03494 entry), 90% API; B domain/protein A: 11116 sequences (NCBI PF02216 entry), 74% API.
b MSA for amyloid β peptide: 273 sequences (Uniprot PF03494 entry), 87% API (PSICOV output is empty on this sequence set); B domain/protein A: 919 sequences (Uniprot PF02216 entry), 87% API.
Hotspot residues within a protein
BIS2Analyzer applied to B domain of protein A (16) identifies 28 co-evolving residues organized in 5 clusters and finds co-evolution among 10 hotspots over 13 known to be important for the folding of the protein. Among the 50 top scored pairs, EVcouplings, PSICOV and DCA do not perform well as shown in Table 2. However, notice that on the MSA described in Table 1, DCA detects 26 contacts in the 3D structure out of 50 predictions. CAPS performance is very low, while ContactMap identifies 11 hotspots within a relatively low number of predicted residues.
Finding correlations in unfolded structures
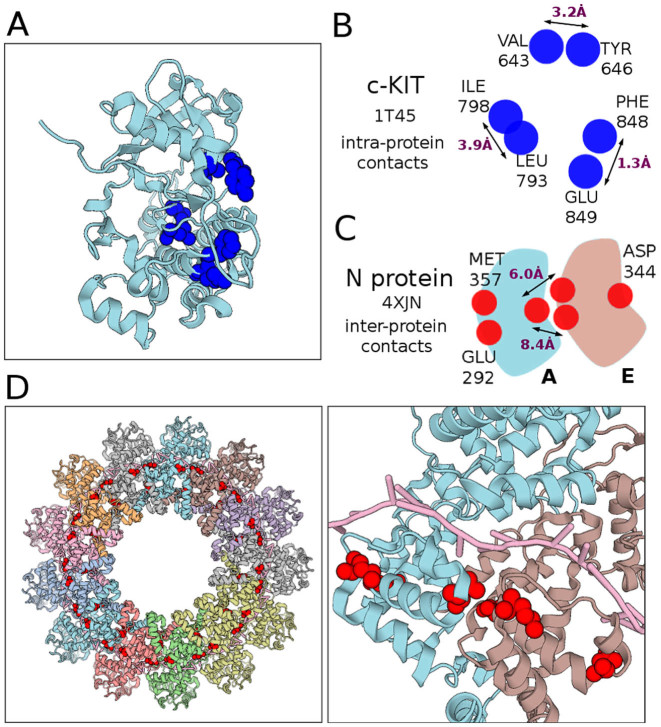
c-KIT is a receptor tyrosine kinase of type III implicated in signaling pathways crucial for cell growth, differentiation and survival (29–31). The Juxta Membrane Region (JMR) is folded in the c-KIT inactive form while it becomes unfolded in the active form. BIS2Analyzer highlights a cluster of three co-evolving residues, lying in JMR, that are in physical contact in the inactive form (Figure 1C, left). BIS2Analyzer visualization of both structures helps to reason on the structural role of the residues in disordered regions.
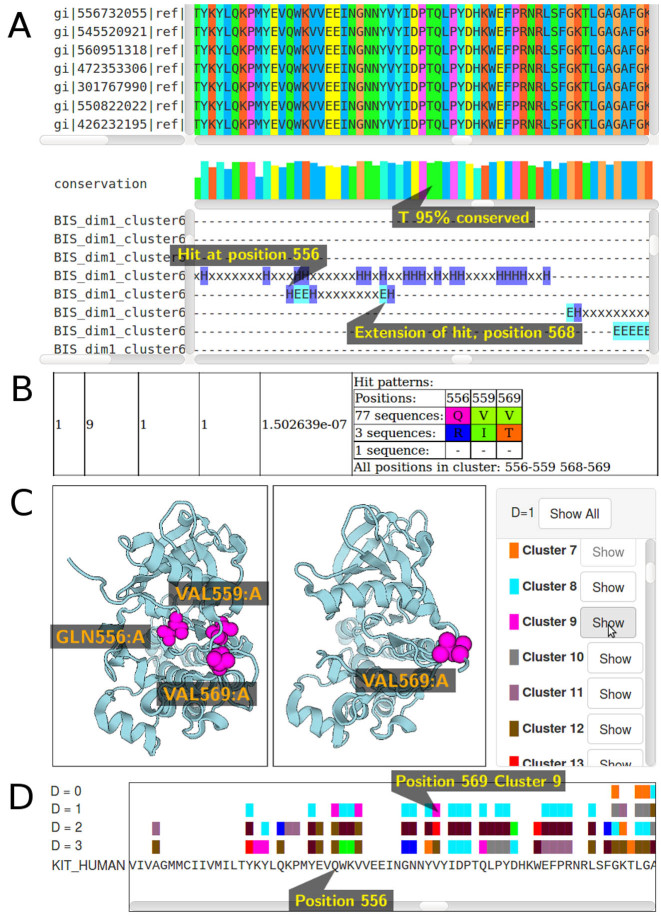
Figure 1.
Visualization of c-KIT tyrosine kinase analysis on Bis2Analyzer. (A) Part of the sequence alignment where cluster 9 is localized. (B) Description of the three hits comprising cluster 9. (C) Display of cluster 9 on c-KIT inactive form (left, 1T45) and on its active form (right, 1PKG). Note that the active form has an unfolded N-terminal that has been partially removed in the crystal (right). (D) Plot of cluster 9 (green dots) on a multiple sequence alignmen (MSA) sequence.
Finding long distance correlations
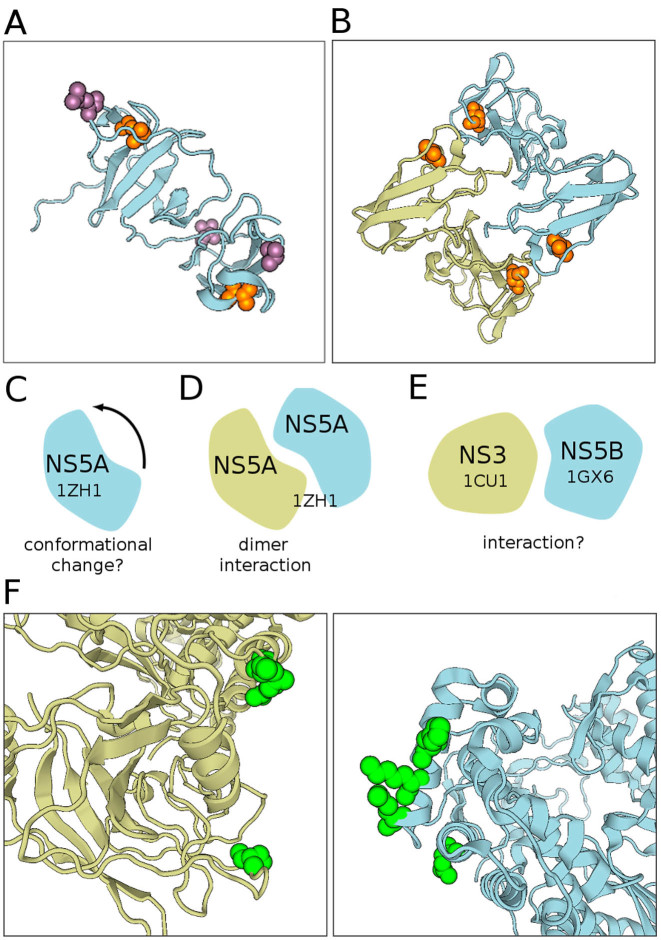
BIS2Analyzer analysis of HCV genotype 1b-MD sequences of the zinc-binding phosphoprotein NS5A highlighted two clusters of co-evolving residues (orange and violet in Figure 2A) localized in the same two regions of the protein. The co-localization of the residues allows to propose biologically interesting hypotheses explaining the correlations. We can hypothesize a conformational change of the protein and a potential functional role of the co-evolved residue pairs in the possible allosteric movement (Figure 2C). Note that a third independent pair of co-evolving residues localized in the same regions was found in genotype 2b sequences (see Figure 9 in (15)) adding confidence in the hypothesis. Also, note that the pair of orange residues are located one in front of the other at the interface of the D1 domain of NS5A (Figure 2B), suggesting a structural role of these residues in the dimeric contact (32,33) (Figure 2D).
Figure 2.
(A) Visualization of two clusters (orange and violet) obtained by BIS2Analyzer on Hepatitis C Virus (HCV) protein NS5A (1ZH1). These co-evolving residues are localized in the same regions of the protein, suggesting a conformational change (see schema in (C)). (B) The orange co-evolving residues in (A), localized far in the monomer structure of NS5A, are found in close proximity in the dimer (1ZH1, see schema in (D)). (F) Co-evolving residues (green; P-value < 1.2e−5) located on the two HCV protein structures NS3 (1CU1, light blue) and NS5B (1GX6, beige) illustrate BIS2Analyzer possibility to visualize inter-protein co-evolved residues (see schema in (E)) and inspect potential interactions.
Predicting contacts between proteins
Co-evolution analysis was performed on the RNA polymerase protein NS5B and the serine protease NS3 by concatenating the sequences of the two HCV proteins, for a set of genomes belonging to genotype 4 (15). By inspecting co-evolution between proteins, we could hypothesize the existence of inter-protein contacts between NS5B and NS3 (Figure 2E). Residue mapping on the appropriate portion of the MSA is done automatically. BIS2Analyzer supports mapping on two different structures, a feature that is particularly useful when partnership is known, but the interaction mode is not. In this manner, the user can inspect the binding mode by exploring the relative positions of the co-evolving residues mapped on the structures of the two available panels (Figure 2F, left).
Predicting clusters of residues in contact within proteins
BIS2analyzer on c-KIT shows a cluster of six residues displaying the same co-evolution signal (P-value 1.2e−5; Figure 3A). They are pairwise in contact, with a <4Å distance (Figure 3B), computed as minimal distance between heavy atoms. Among them, the three pairs are at 13.5, 8.1 and 8.7Å away, respectively, and their localization on the same surface side of the protein suggests a common functional or structural role. BIS2Analyzer possibility to identify clusters of contacts instead of isolated contacts provides new opportunities for interpretation.
Figure 3.
(A) BIS2Analyzer detects six co-evolving residues (blue, P-value ≤ 1.2e−5) located at close distance on the surface of c-KIT tyrosine kinase (1T45). (B) Schema illustrating the distances among the 6 co-evolving residues in (A). (D) Three co-evolving residues (red) face opposite sites of protein N (chain A, 4XJN, right). They identify inter-protein contacts at the interface of the mononegavirales protein N assembly (4XJN, left) (see schema in (C)).
Finding distant correlations justified by a large complex assembly
BIS2Analyzer analysis of the mononegavirales protein N (Table 1) highlighted a cluster comprised of three co-evolving residues (red in Figure 3D, right) localized in two opposite faces of the protein. The co-localization of the residues is visualized in the large structure of the parainfluenza virus 5 nucleocapsid–RNA complex (4XJN), formed by 13 homodimers, where the three residues enter in contact after dimerization, as illustrated in Figure 3D, left.
Exploring large sets of divergent sequences with BIS2Analyzer
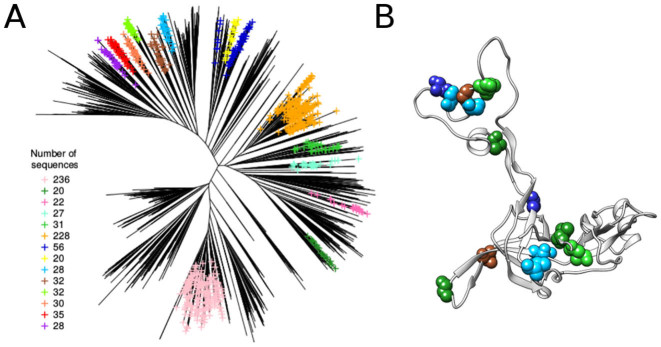
BIS2Analyzer can be used to explore large sets of divergent sequences, as the ribosomal L3 protein family. We considered 2414 sequences (alignment length 390 aa) and analyzed 14 subtrees of its distance tree (constructed with BioNJ (34)), selected to contain at least 20 sequences and displaying non-trivial clusters of co-evolving residues (Figure 4A). A cluster is considered to be trivial if (i) either it is conserved (P-value = 1), (ii) or the co-evolution pattern comprises only one amino-acid occurring more than once, (iii) or the co-evolution pattern is only due to the presence of gaps. After applying BIS2Analyzer, we retained 16 co-evolution clusters, belonging to 11 subtrees, with a P-value < 1e−3. Non-perfect co-evolution patterns (i.e. CLAG scores <1) were retained only if their significance was high (P-value < 1e−7). Among all BIS2 co-evolution clusters, 10 of them (on 8 subtrees) contain residues close in the 3D structure. The remaining 6 clusters (located on 5 subtrees) link two distant regions as illustrated in Figure 4B, one of those is the ordered extension loop, totally devoid of secondary structure. The localization of the residues suggests a possible rearrangement of the protein structure during the protein lifetime, where the pairs of residues could enter in physical contact. The native structure of L3 is not available, but there is ground to suspect that those regions come close to each other upon refolding since this is the case of 50S ribosome L4 protein, whose conformation in the complex is very similar to that of L3 (see also (35)). A guideline explaining how to realize analyzes on large sets of divergent sequences is reported in the online ‘Tutorial’ section.
Figure 4.
(A) Clustering of the phylogenetic tree constructed from a dataset of RL3 sequences (2414 sequences; subset of the UniRef90 dataset in UniProt from which too divergent sequences have been eliminated). Selected subtrees (shown in color) contain at least 20 sequences and at least one non-trivial co-evolution cluster (See text.). (B) BIS2 co-evolution clusters on the 3D structure (PDB ID: 4U26, chain BD), colored according to the subtree they belong to; the six co-evolution clusters shown above belong to five sub-trees and link the ordered extension loop with the structured region.
HOW TO RUN BIS2Analyzer
The web server provides a job submission page (‘Submit’). It requires input files formatted in a standard way and most of the parameters are automatically set, so the basic usage should be straightforward. To have a glimpse on the type of input required, sample inputs can be loaded for intra- and inter-protein co-evolution analysis. For information on how to customize the default behavior, a detailed tutorial is provided and is accessible online at the ‘Tutorial’ page. Below, we overview the usage of the web server.
Details on the input data
BIS2Analyzer accepts as input a MSA in FASTA format, either copy-pasted or uploaded as a file. Sequences must contain only upper case characters, dashes and dots. There is no restriction for sequence names. Once the job is submitted, based on a randomly generated jobID, a web link is provided allowing the user to access the data at a later time. Optionally, an e-mail address can be provided; job queuing, beginning and completion are notified. The mail reports a mnemonic jobname chosen by the user or generated otherwise.
Guidelines on input sequences
We recommend applying BIS2 either on tens of sequences, or on a few hundreds of sequences with relatively high API. In the latter case, we identify very high API (∼80% and above), where BIS2 might be run with default parameters and moderately high API (∼60 ÷ 80%), where BIS2 could be run with higher dimensions D or with the alphabet reduction option enabled; in this way, amino acid variability within the same class is neglected.
BIS2Analyzer default parameters
BIS2Analyzer generates a rooted phylogenetic tree with BIONJ (34) by default. First, it computes the distance matrix, based on Jones–Taylor–Thornton distance model (36) with Protdist, from PHYLIP version 3.696 (37); then, it uses BIONJ to build the phylogenetic tree and SeaView to re-root the tree (38).
The dimension parameter D is set to 2. By default, BIS2Analyzer enables the block mode; this means that hits are extended by conservation on neighboring positions. Alphabet reduction option is disabled.
BIS2Analyzer options
The user can provide a phylogenetic tree in NEWICK format (either copy-pasted or uploaded as a file) or set PhyML (39) in replacement of BIONJ. The tree must be rooted (SeaView (38) can be used for this purpose). The dimension ‘D’ option sets the maximum number of allowed exceptions (with maximum allowed value D ≤ 10). The ‘block’ option can be disabled to force BIS2 to report co-evolving hits only, without extending a hit into a block. By default, the ‘pc’ option reduces the amino-acid alphabet of 20 to 8 letters representing physico-chemical classes of residues, where each residue on a class is assigned the same letter. The eight physico-chemical classes are defined by default as in (38): hydrophobic (VILMFWA), negatively charged (DE), positively charged (KR), aromatic (YH), polar (NSTQ) and C, G, P are considered as special. The user can provide a custom definition of amino acid classes, by typing a string containing the 20 amino acids, with classes separated by commas (for instance: KR,AFILMVW,NQST,HY,C,DE,P,G) in the dedicated box.
Guidelines for different analyses
Co-evolution analysis within a protein complex is of paramount importance to dissect an interface or to get clues on potential interacting residues when the structural complex is not available. The procedure for such analysis conducted with BIS2Analyzer is indicated within the ‘Tutorial’ page. Also, the computational strategy for analyzing large datasets of divergent sequences is reported in the ‘Tutorial’ page.
DISPLAY OF THE RESULTS
Output
BIS2Analyzer supplies a graphical interface to inspect co-evolution clusters, resulting as BIS2 predictions.
For each dimension considered in the analysis (d ≤ D), BIS2Analyzer displays the MSA labeled with all co-evolution clusters of that dimension (see Figure 1A). At the bottom of the MSA, a histogram reports the conservation level of the most frequent character occurring at a fixed position. A graphical ruler describing each cluster helps to browse the MSA and easily identify positions belonging to the cluster (‘H’ labels a hit and ‘E’ labels a block extension; Figure 1A).
For each cluster, BIS2Analyzer allows visualization of residue types, physico-chemical properties and MSA positions (Figure 1B). Three scores are provided for each cluster: symmetric, environmental and P-value. The first two scores vary in the interval [0, 1] and are computed by the clustering algorithm CLAG (26). They express the degree of ‘similarity’ of co-evolution of positions in a cluster with respect to all other analysed positions. In particular, scores equal to 1 correspond to a cluster where all positions show an identical co-evolution pattern with all other analysed positions. High scores guarantee the confidence in a cluster and because of this, BIS2Analyzer outputs only clusters with both scores >0.5. The P-value score is computed with a Fisher test on a diagonal matrix, where the elements of the diagonal represent the co-evolution pattern satisfied by all positions in a cluster; for example, 77−3−1 in Figure 1B is a pattern representing three distinct amino-acids on three MSA positions that occur on subsets of 77, 3 and 1 sequences, respectively. The subsets are the same for the three positions and, in this case, we talk about a perfect pattern. When the pattern if not perfect, the P-value is computed on the maximum set of aligned sequences displaying a perfect pattern.
Output visualization
The user can visualize each prediction onto a sequence through the ‘Mapping to sequence’ page, or the 3D structure through the ‘Mapping to structure’ page. In addition to the clusters’ listing, the web server provides interactive ways to inspect them on one or two proteins of interest.
Mapping on a reference sequence (Figure 1D) can be done either on the MSA consensus sequence, or on any sequence present in the MSA, or on a new sequence provided by the user as a FASTA file. All co-evolution clusters are viewable on the sequence, they are labeled with different colors and can be enabled or disabled globally or one by one. If the sequence is among the ones in the MSA, the representation is done with alignment's gaps been removed. Otherwise, if a new sequence is provided, the Smith–Waterman algorithm is applied to the new sequence and the consensus sequence computed from the MSA. We adopted the same scoring scheme as for PSIBLAST, namely, we use BLOSUM62 matrix for match-mismatch scores and assign penalties of −11 and −1 to gap opening and gap extension, respectively. A match/mismatch between non-standard amino acids is scored with 1 for a match and −4 for a mismatch. Gaps at the beginning or at the end of the alignment are scored 0, so that the sequence provided can be much shorter or longer than the length of the MSA.
Mapping on a reference structure (Figure 1C) is done by providing a PDB file or PDB ID, possibly containing multiple chains. Chains can be enabled/disabled for display. A residue mapping on the MSA is done by retrieving the sequences of each chain of the PDB and by aligning each of them independently on the MSA consensus. An alignment score cut-off is set (at 0) and chains that align against the consensus of the MSA with a score lower than the cut-off are not considered. The user can enable/disable each cluster for visualization. Colors used for identifying clusters on the structure and on the sequence are consistent. Finally, the user can decide to upload up to two PDB structures for the same co-evolution analysis. This is a useful feature when either protein interactions or different foldings (e.g. disordered versus ordered regions) for the same protein are explored. The graphical interface is implemented with Protein Viewer (PV), a WebGL-based viewer for proteins and other biological macromolecules, very fast and visualizable on smartphones (http://pv.readthedocs.io/en/v1.8.1/index.html).
DISCUSSION
BIS2Analyzer conveys an automatic, though detailed and highly customizable, pipeline; it provides to the scientific community an established method for the co-evolution analysis of very few and/or highly conserved sequences. BIS2Analyzer can be used by the biologist to foster hypothesis on protein behavior and new strategies for the design of experiments.
FUNDING
Institut Universitaire de France; French Governement Funds, at UPMC, for HPC resources [‘Equip@Meso project - ANR-10-EQPX- 29-01’]; French Governement—Excellence Program ‘Investissement d'Avenir’ in Bioinformatics [‘MAPPING project-ANR-11-BINF-0003’]. Funding for open access charge: French Governement—Excellence Program ‘Investissement d'Avenir’ in Bioinformatics [‘MAPPING project-ANR-11-BINF-0003’].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lockless S., Ranganathan R.. Evolutionary conserved pathways of energetic connectivity in protein families. Science. 1999; 286:295–299. [DOI] [PubMed] [Google Scholar]
- 2. Suel G., Lockless S., Wall M., Ranganathan R.. Evolutionary conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003; 23:59–69. [DOI] [PubMed] [Google Scholar]
- 3. Baussand J., Carbone A.. A combinatorial approach to detect co-evolved amino acid networks in protein families with variable divergence. PLoS Comput. Biol. 2009; 5, doi:10.1371/journal.pcbi.1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marks D.S., Colwell L.J., Sheridan R., Hopf T.A., Pagnani A., Zecchina R., Sander C.. Protein 3D structure computed from evolutionary sequence variation. PLoS ONE. 2011; 6, doi:10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morcos F., Pagnani A., Lunt B., Bertolino A., Marks D.S., Sander C., Zecchina R., Onuchic J.N., Hwa T., Weigt M.. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hopf T.A., Colwell L.J., Sheridan R., Rost B., Sander C., Marks D.S.. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012; 149:1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones D.T., Buchan D.W.A., Cozzetto D., Pontil M.. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012; 28:184–190. [DOI] [PubMed] [Google Scholar]
- 8. Morcos F., Jana B., Hwa T., Onuchic J.N.. Coevolutionary signals across protein lineages help capture multiple protein conformations. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20533–20538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Juan D., Pazos F., Valencia A.. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013; 14:249–261. [DOI] [PubMed] [Google Scholar]
- 10. Kuriyan J. Allostery and coupled sequence variation in nuclear hormone receptors. Cell. 2004; 116:354–356. [DOI] [PubMed] [Google Scholar]
- 11. Del Sol A., Arauzo-Bravo M., Amoros D., Nussinov R.. Modular architecture of protein structures and allosteric communications: potential implications for signaling proteins and regulatory linkages. Genome Biol. 2006; 8:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Del Sol A., Fujihashi H., Amoros D., Nussinov R.. Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol Syst. Biol. 2006; 2, doi:10.1038/msb4100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hopf T., Schärfe C.P., Rodrigues J.P., Green A.G., Kohlbacher O., Sander C., Bonvin A.M., Marks D.S. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014; 3, doi:10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S., Sun S., Li Z., Zhang R., Xu J.. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 2017; 13:e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Champeimont R., Laine E., Hu S-W., Penin F., Carbone A.. Coevolution analysis of Hepatitis C virus genome to identify the structural and functional dependency network of viral proteins. Sci. Rep. 2016; 6:26401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dib L., Carbone A.. Protein fragments: functional and structural roles of their coevolution networks. PLoS One. 2012; 7, doi:10.1371/journal.pone.0048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yip K.Y., Patel P., Kim P.M., Engelman D.M., McDermott D., Gerstein M.. An integrated system for studying residue coevolution in proteins. Bioinformatics. 2008; 24:290–292. [DOI] [PubMed] [Google Scholar]
- 18. Gouveia-Oliveira R., Roque F.S., Wernersson R., Sicheritz-Ponten T., Sackett P.W., Molgaard A., Pedersen A.G.. InterMap3D: predicting and visualising co-evolving protein residues. Bioinformatics. 2009; 25:1963–1965. [DOI] [PubMed] [Google Scholar]
- 19. Ochoa D., Pazos F.. Studying the co-evolution of protein families with the Mirrortree web server. Bioinformatics. 2010; 26:1370–1371. [DOI] [PubMed] [Google Scholar]
- 20. Simonetti F.L., Teppa E., Chernomoretz A., Nielsen M., Buslje C.M.. MISTIC: mutual information server to infer coevolution. Nucleic Acids Res. 2013; 41:W8–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamisetty H., Ovchinnikov S., Baker D.. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence-and structure-rich era. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cohen O., Ashkenazy H., Karin E. L., Burstein D., Pupko T.. CoPAP: coevolution of presence-absence patterns. Nucleic Acids Res. 2013; 41:W232–W237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadreyev I.R., Ji F., Cohen E., Ruvkun G., Tabach Y.. PhyloGene server for identification and visualisation of co-evolving proteins using normalized phylogenetic profiles. Nucleic Acids Res. 2015; doi:10.1093/nar/gkv452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baker F.N., Porollo A.. CoeViz: a web-based tool for coevolution analysis of protein residues. BMC Bioinformatics. 2016; 17:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fares M.A., McNally D.. CAPS: coevolution analysis using protein sequences. Bioinformatics. 2006; 22:2821–2822. [DOI] [PubMed] [Google Scholar]
- 26. Dib L., Carbone A.. CLAG, an unsupervised non hierarchical clustering algorithm handling biological data. BMC Bioinformatics. 2012; 13:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balakrishnan S., Kamisetty H., Carbonell J.G., Lee S.I., Langmead C.J.. Learning generative models for protein fold families. Proteins. 2011; 79:1061–1078. [DOI] [PubMed] [Google Scholar]
- 28. Ekeberg M., Lövkvist C., Lan Y., Weigt M., Aurell E.. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys. Rev. E. 2013; 87:012707. [DOI] [PubMed] [Google Scholar]
- 29. Qiu F.H., Ray P., Brown K., Barker P.E., Jhanwar S., Ruddle F.H., Besmer P.. Primary structure of c-KIT, relationship with the csf-1/pdgf receptor kinase family–oncogenic activation of v-KIT involves deletion of extracellular domain and C terminus. EMBO J. 1988; 7:1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edling C.E., Hallberg B.. c-KIT – a hematopoietic cell essential receptor tyrosine kinase. Int. J. Biochem. Cell Biol. 2007; 39:1995–1998. [DOI] [PubMed] [Google Scholar]
- 31. Lemmon M.A., Schlessinger J.. Cell signaling by receptor-tyrosine kinases. Cell. 2010; 141:1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bartenschlager R., Lohmann V., Penin F.. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013; 11:482–496. [DOI] [PubMed] [Google Scholar]
- 33. Lambert S.M., Langley D.R., Garnett J.A., Angell R., Hedgethorne K., Meanwell N.A., Matthews S.J.. The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors. Protein Sci. 2014; 23:723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gascuel O. BIONJ, an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997; 14:685–695. [DOI] [PubMed] [Google Scholar]
- 35. Timsit Y., Acosta Z., Allemand F., Chiaruttini C., Springer M.. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int. J. Mol. Sci. 2009; 10:817–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones D.T., Taylor W. R., Thornton J.M.. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992; 8:275–282. [DOI] [PubMed] [Google Scholar]
- 37. DOTREE Plotree and DOTGRAM Plotgram PHYLIP-phylogeny inference package (version 3.2). Cladistics. 1989; 5:163–166. [Google Scholar]
- 38. Gouy M., Guindon S., Gascuel O.. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010; 27:221–224. [DOI] [PubMed] [Google Scholar]
- 39. Guindon S., Dufayard J-F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010; 59:307–321. [DOI] [PubMed] [Google Scholar]