Abstract
With the rise of multi-drug resistant pathogens and the decline in number of potential new antibiotics in development there is a fervent need to reinvigorate the natural products discovery pipeline. Most antibiotics are derived from secondary metabolites produced by microorganisms and plants. To avoid suicide, an antibiotic producer harbors resistance genes often found within the same biosynthetic gene cluster (BGC) responsible for manufacturing the antibiotic. Existing mining tools are excellent at detecting BGCs or resistant genes in general, but provide little help in prioritizing and identifying gene clusters for compounds active against specific and novel targets. Here we introduce the ‘Antibiotic Resistant Target Seeker’ (ARTS) available at https://arts.ziemertlab.com. ARTS allows for specific and efficient genome mining for antibiotics with interesting and novel targets. The aim of this web server is to automate the screening of large amounts of sequence data and to focus on the most promising strains that produce antibiotics with new modes of action. ARTS integrates target directed genome mining methods, antibiotic gene cluster predictions and ‘essential gene screening’ to provide an interactive page for rapid identification of known and putative targets in BGCs.
INTRODUCTION
The alarming number of antibiotic resistant pathogens and the declining rate of novel antibiotics discovered makes the search for new antibiotic compounds one of the most important tasks of this century. The majority of drugs in the clinic has been and continues to be inspired by bioactive compounds produced as secondary metabolites by living organisms (1–3). These so-called natural products have been isolated from a variety of sources including plants, fungi and bacteria. Soil dwelling bacteria of the phylum Actinobacteria have particularly been shown to be a rich source for a variety of novel compounds with diverse bioactivities (4). It is remarkable that this reservoir of discovery has only been the product of some 1% of cultivable bacteria, many of which harbor still untapped genetic potential (1).
Traditionally, drug discovery has been largely based on bioactivity-guided methods. More recently, with the advent of next generation sequencing, genome-based methods invigorate new drug discovery efforts (5). Genome mining revealed that many bacteria previously believed to not produce natural products harbor dormant potential and that even well studied sources contain orphan biosynthetic gene clusters (BGCs), with products yet to be discovered (6). Automated genome mining tools such as antiSMASH (7) and PRISM (8), as well as databases such as MIBiG (9) or the antiSMASH database (10), along with methods such as EvoMining (11), allow for effective and fast mining of the thousands of available bacterial genomes. The current largest collection of automatically mined gene clusters is the “Atlas of Biosynthetic gene Clusters”, a component of the “Integrated Microbial Genomes" Platform of the Joint Genome Institute (JGI IMG-ABC) (12). As of January 2017 this database contains more than a million gene clusters, the majority of them orphan. The biggest challenge now is to prioritize the more laborious wet lab experiments for BGCs with the most promising bioactivities.
One recently developed approach for the specific discovery of new antibacterial compounds uses resistance as a trait for selecting for antibiotic producing bacteria. Any bacterium producing an antibiotic needs to evolve a self-protection mechanism to avoid suicide (13). Based on this idea, Wright et al. designed a screen to exploit the self-protection mechanism of antibiotic producers to enrich microbial libraries for producers of selected antibiotic scaffolds (14). Resistance mechanisms vary and include the inactivation and export of antibiotics, as well as protection and modification of target proteins. In the latter case a second resistant copy of the antibiotic target gene is typically detectable in the genome, often found within the BGC of the antibiotic and often horizontally acquired with the BGC (15–17). Moore and colleagues used this concept and screened for duplicated housekeeping genes co-localized with orphan BGCs. Using this target-directed genome mining, they were able to prioritize an orphan polyketide synthase-nonribosomal peptide synthetase hybrid BGC, responsible for producing a fatty-acid synthase inhibitor, containing a fatty acid synthase resistance gene (18). This study demonstrated nicely that the search for BGCs in conjunction with duplicated housekeeping genes can not only help prioritize BGCs with antibiotic activities, but also provide insights into the mode of action of encoded natural products.
So far, this search has been done manually, checking each detected gene cluster for possible resistant housekeeping genes. Here we introduce the antibiotic resistant target seeker (ARTS), a user-friendly web tool that automatically detects known resistance, as well as possible resistant housekeeping genes in actinobacterial genomes based on three criteria: duplication, localization within a biosynthetic gene cluster and evidence of horizontal gene transfer (HGT). ARTS allows the user a fast and efficient genome-based prioritization of bacterial strains with the potential to produce antibiotics with interesting and possibly novel modes of action.
MATERIALS AND METHODS
Workflow
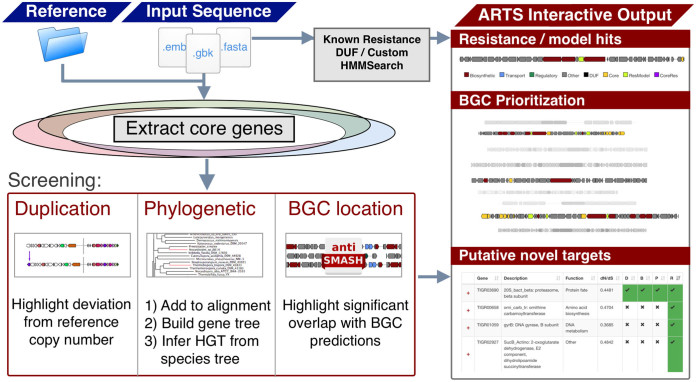
The ARTS workflow in Figure 1 illustrates automated steps for known resistance screening and novel target analysis. Users provide DNA sequences of whole genomes or gene clusters in Genbank, FASTA or EMBL format as input; alternatively an NCBI accession number or antiSMASH job id can be used to retrieve data automatically. The first steps include identifying BGCs using antiSMASH, detecting known resistance models (19–22) and detecting essential core genes (see reference set and core gene detection). During the next steps the identified core and known resistance genes are screened for their location within BGCs. Finally core genes are screened for HGT. All screening results are then summarized into an interactive output table to rapidly cross reference known and putative novel targets.
Figure 1.
Workflow of the ARTS pipeline. Input query genome sequences and reference organisms are scanned for Biosynthetic Gene Clusters (BGCs), known resistance factors and essential genes. Screening criteria for duplication, co-localization with BGCs and phylogeny are then applied and integrated into the interactive output for target directed BGC prioritization and novel target discovery.
Reference set and core gene detection
A reference set of complete genomes is used to identify essential genes, define duplication thresholds and infer HGT via species and gene tree reconciliation. The current Actinobacteria reference set is comprised of complete genomes from 189 species representing 22 different families that are available through NCBI's RefSeq (23) database (Supplementary File S1). Core genes are identified using protein families from the TIGRFAM database, where all protein models that are found in the vast majority of reference genomes are considered core genes. Unless ‘exploration mode’ is used, core genes are filtered for unlikely targets by removing regulatory, transport and biosynthetic functions associated with known BGCs. To facilitate HGT detection in later steps, alignments and gene trees are then built as detailed in the Supplementary Methods.
Screening criteria
BGC proximity
BGCs are detected using antiSMASH v3. Then core and resistance genes with intersecting locations on the same scaffold are marked as BGC proximity hits. These are presented in the visual cluster annotation as well as in the interactive summary tables.
Gene duplication
Duplication thresholds are determined comparatively using the sum of median and standard deviation for each essential gene count in the reference set. All core genes over these thresholds are then marked and recorded in the summary and duplication sections.
HGT
Phylogenetic screening is used to highlight all query organism core genes that show evidence of HGT. This is accomplished by comparing core gene trees with the species tree to highlight incongruences. This consists of first building alignments and trees for all core genes. A species tree is then inferred using ubiquitous, single copy gene trees with a coalescent tree analysis. Incongruent gene trees that are more likely explained by HGT, instead of duplications and losses through generations, is then determined as detailed in the Supplementary Methods.
Input options
Users can provide custom essential genes and resistance models for more tailored searches using the upload sections in the advanced dropdown; essential gene search thresholds can be modified in this section as well. Options to disable certain screening criteria are available to accelerate analysis and can be helpful for certain input, such as disabling phylogenetic screening for non-reference phyla genomes. By default, all criteria and trusted cutoffs are used and currently the phylogenetic reference set is selected for Actinobacteria. Non-actinobacterial genomes can be submitted for analysis however the phylogeny criteria will not be applicable. The option for ‘exploration mode’ is used for an extended search which may include more false positives but screens a larger, unfiltered, set of core genes.
Output and interactive layout
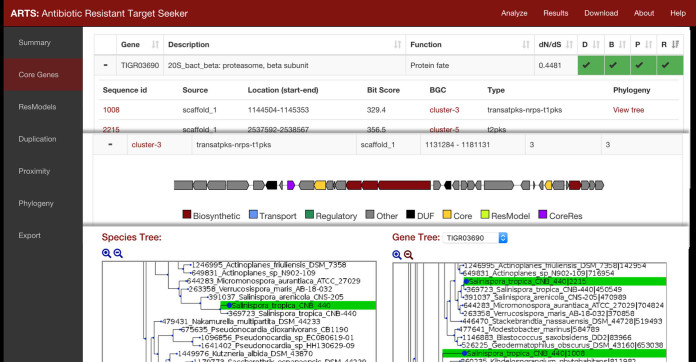
The results are presented in various sections by criteria perspectives: core genes, resistance models, gene duplications, BGC proximity and phylogeny. Dynamic tables in sections can be searched and sorted allowing for rapid prioritization of properties and their combinations (Figure 2). For example, sorting the core gene table by positive BGC hits and phylogeny will yield all core genes found in a BGC with incongruent phylogeny. Details for each item can be investigated by expanding a row to reveal links to corresponding sections. Visualizations for cluster annotations and trees highlighting the query organism are also generated for rapid confirmation of these criteria. All tables, trees and core gene alignments can be exported and saved for additional analysis from the export section. Additional metadata are provided to assess the viability of a novel target prediction which include: functional classification, average selection pressure (dN/dS) values and how widespread the essential gene is relative to reference organisms (ubiquity). This can help eliminate less promising drug target predictions, such as one that is not universally shared. Examples of all inputs and detailed tutorials on output interpretation can be found at https://arts.ziemertlab.com/help.
Figure 2.
Example output screenshot highlighting three major sections: core gene table prioritization, BGC visualization and phylogenetic incongruence confirmation.
RESULTS
Here we present ARTS, an easy to use web tool for the high-throughput screening of bacterial genomes for their potential to produce antibacterial compounds with interesting modes of action. To our knowledge, ARTS is currently the only public web server that automates an extended target-directed genome mining that includes potentially novel targets. It demonstrates significantly increased throughput compared to manual target-directed methods and allows for intuitive visualizations and rapid explorations of several resistance factors. A manual phylogenetic analysis of all core genes, with alignments and Maximum-Likelihood tree construction, took over 89 h of processing time alone on a 16-core machine. By leveraging the pre-computed reference, the analysis took 15 min using the same resources with ARTS. Aside from processing time, the interactive browser format proved to be more accessible and faster to interpret than manual methods. The ARTS web server and analysis scripts are available to the public with git repository at: https://bitbucket.org/ziemertlab/arts
Reference set and core gene analysis
To verify that core genes inferred from reference organisms are essential, comparisons to experimental examples and properties of essential genes were interrogated. A total of 664 essential gene models (Supplementary File S2) are identified using the core genome methods and 432 remain when filtered as detailed in the Supplementary Methods. Comparison to the experimentally confirmed Database of Essential Genes (24) (DEG) v13 shows 538 genes in the set match to one or more records. Functional classification reveals that the majority of genes not found in DEG are in unknown, unclassified, or energy and metabolism categories (Supplementary File S3). Additionally, all reference core gene hits, compared to all TIGRFAM hits, showed enrichment for essential functions such as: protein and amino acid synthesis, energy and metabolism, and transcription (Supplementary File S4). A variety of approaches to determine essential genes can be found in literature where ubiquity and conservation of sequence are properties frequently exploited (25–27). Nearly all of the set share these properties (Supplementary File S5) with a dN/dS mode of 0.35 and range of 0.05–1.05 illustrating strong purifying selection. While more than half of the genes are present in over 90% of genomes, many are specific to certain genera. One example is the 20S proteasome, which is essential to many actinobacterial genera but is lacking in others with only 64% global reference ubiquity. By defining core genes relative to family, this specific function is captured. All resulting trees showed valid branch support (Supplementary S6) with single copy gene trees showing monophyletic groupings.
Detection frequency and false positives
To estimate false positive rates, total hit frequencies for BGCs and complete genome sequences were evaluated as a proxy due to the unknown amount of true positives. This was done in exploratory mode to assess the upper limit of hit frequencies unless stated otherwise.
BGC clusters
We analyzed all 1409 characterized BGCs from the MIBiG database and found roughly a third (465), a quarter (348) and 12% (162) have at least one gene match to models from core gene, known resistance or both respectively. For core genes this is largely attributed to cell envelope, amino acid and protein synthesis, and energy metabolism genes. In comparison, default mode, which lacks protein families likely involved in biosynthesis, transport and regulation, showed <15% of clusters have a core gene match with a dramatic reduction in cell envelope and primary metabolism categories (Supplementary File S7). Additionally, multiple hits in the same cluster are rare and, if present, likely indicate inaccurate cluster boundaries rather than cluster participation. With respect to total genes present, <3.7 and 1.9% are marked as core gene and known resistance hits respectively.
Complete genomes
We then analyzed 200 complete Actinobacteria genomes, publicly available through NCBI, though the ARTS pipeline and detected 489 core genes per genome, on average, with only one or two genes showing three or more selection criteria (Supplementary File S8). Approximately 5% of core genes are highlighted for each criterion with the exception of phylogeny, which shows an average of 26% of all core genes highlighted. Although this initially seems high, this rate is similar to other HGT estimates of about 35% HGT for Actinobacteria (28). This shows that multiple screening criteria should be used for more confident target predictions. The occasional high counts seen in exploration mode (Supplementary File S9) can be easily and rapidly filtered by the manageable output page. Besides cross referencing with multiple criteria, single criteria hits can be narrowed by several properties such as gene prevalence, copy number statistics and function. This can dramatically filter hits and allows for expert review by the researcher. Likewise, high core hits in BGCs are easily screened using the visual annotation. Figure 3A shows added coloring to highlight multiple criteria, and hits are also seen more central in the predicted cluster. Where many core genes are captured by inaccurate cluster boundaries, as in Figure 3B, inspection can serve not only to screen possible hits but also to help define true cluster boundaries, which remains a largely unresolved challenge when dealing with bacterial BGCs.
Figure 3.
ARTS cluster visualizations from example genome Streptomyces roseochromogenes DS 12.976 where core genes are shown in yellow, known resistance in green and hits for both shown in purple. (A) Purple: positive control of resistant gyrB, Green: DNA topoisomerase IV (B) Example of cluster boundaries capturing core genes not associated with the cluster.
Positive controls
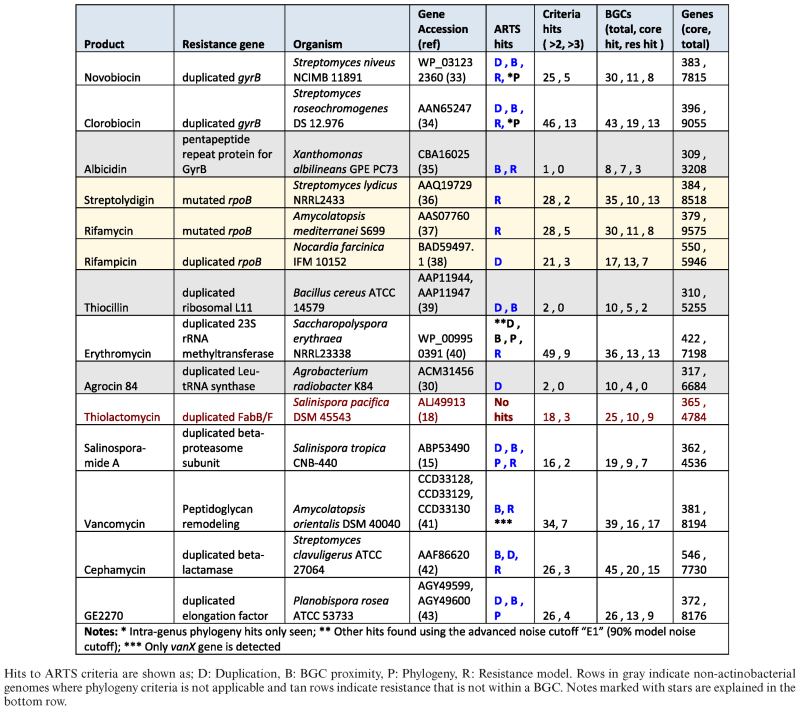
In order to test ARTS, we performed an extensive literature search and analyzed all known examples of BGCs and available bacterial genomes, that contained known examples of resistant target genes. Out of the 26 identified clusters from known examples within the MIBiG database, ARTS detected 22 clusters with resistant targets in exploration mode (Supplementary File S10). Three undetected cases were due to missing specific DNA glycosylation and CoA reductase models in both the core and known resistance set. The last missed case, the resistance conferring 23S rRNA methyltransferase, was identified in clusters for Erythromycin and Pikromycin but not Avilamycin due to its significantly shorter sequence length and low homology score. The default search mode showed that 3 positive cases for FabB/F were not detected. Both gene families are frequently used for the biosynthesis of secondary metabolites with no possibility at the moment to easily distinguish in silico if they are involved in resistance or biosynthesis. However, with expert review in exploration mode these cases were detected.
The literature search for self-resistance mechanisms that had genomes of at least draft quality available yielded 14 cases, all of which were processed with ARTS. All cases except for the resistant FabB/F showed an ARTS hit (Table 1), which is detected in exploration mode using reduced cutoffs. Two cases of rpoB resistance, single copy mutants not present in a BGC, were marked as known resistant targets with no other criteria shown. In the case of Rifampicin however, the duplicated resistance conferring rpoB was detected. The results of an ARTS analysis of Salinispora tropica CNB-440, the producer of salinosporamide A, (29), shows positive hits for all the criteria tested. The β-subunit of the proteasome is a known target of salinosporamide A and the producing cluster contains a resistant copy localized within the operon (15). The incorporated models of known resistant targets highlights the gene to indicate that both core and resistant target matches were found. From the phylogeny view the resistant copy is quickly seen as having origins from outside its species and is marked for potential HGT.
Table 1. Positive examples of genomes with known self-resistance mechanisms analyzed with ARTS default mode.
|
Distant genomes
Although ARTS is so far specialized for actinobacterial genomes, it can also be applied to organisms that are not part of the reference phylum, as demonstrated by the positive results for all three examples from Firmicutes and Proteobacteria (Table 1). One example, the Agrocin 84 producer Agrobacterium radiobacter K84, showed a duplicated resistant Leu-tRNA synthetase located on the pAgK84 plasmid that was detected but with no cluster predictions present. Further investigation showed the producing cluster is indeed part of the plasmid (30) and an extended BGC prediction using the ClusterFinder (31) algorithm yielded a putative 9kb segment which did not include this resistance gene. By highlighting potential resistance outside of predicted clusters, this example shows how ARTS can serve not only as a cluster prioritization tool but also as an orthogonal detection method to complement current methods.
DISCUSSION AND CONCLUSION
With ARTS we leveraged the recent advancements in BGC detection, phylogenetic reconstruction and HGT prediction to automate several analyses of target-directed genome mining methods into a single computationally inexpensive workflow. In addition to the standard target-directed screening of known resistance models, we have included criteria and tools for the rapid exploration of potentially novel resistance targets. While these criteria do not directly imply resistance, for example duplications due to adaptive biosynthesis, it has been shown that integration of multiple criteria can serve to highlight those involved in resistance. Two different screening modes, as well as rapid post filtering functions that are provided, allow for the fast and easy exploration of putative novel targets. This allows for the robust searching of bacterial genomes and provides utility beyond target prediction. In exploration mode for example, the detection algorithm includes genes that have been duplicated from central metabolism and have been repurposed for the biosynthesis of secondary metabolites. Indeed these gene expansions have been shown to be a valuable cluster detection technique as demonstrated with the EvoMining (11,32) method by highlighting clusters with potentially novel chemistries, such as the recent identification of arseno-organic natural products in the model actinomycetes Streptomyces coelicolor and Streptomyces lividans (11).
This iteration of ARTS is currently focused on Actinobacteria but has shown to extend to other phyla as well. We have constructed the pipeline to work easily with alternative user generated reference sets and are currently working to expand these sets for other taxa. Prioritizing putative clusters with an extra layer of antibiotic potential is the main purpose. However, examples have shown that it can also be useful in helping to define cluster boundaries. Furthermore, its use as an orthogonal cluster detection method and highlighting unknown or repurposed functions of biosynthetic enzymes is an interesting possibility. Mainly, we hope the server will provide broader access to these methods with a higher throughput of clusters and organisms screened to accelerate the discovery of competitive antibiotics.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Bradley Moore, Yvonne Mast, Dominik Pistorius, Tobias Gulder, Robin Teufel, Evi Stegmann, Paul Jensen, Alessandra Eustaquio, Chambers Hughes and their colleagues for testing ARTS and their valued feedback. The authors also thank Michelle Schorn for valued discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Center for Infectious Biology [DZIF 9.704 to N.Z.]; Novo Nordisk Foundation (to T.W., K.B.); College of Pharmacy Research Startup Funds; American Society of Pharmacognosy Research Starter Grant (to B.P.). Funding for open access charge: German Center for Infectious Biology [DZIF 9.704].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cragg G.M., Newman D.J.. Natural products: a continuing source of novel drug leads. Biochim. Biophys. Acta. 2013; 1830:3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harvey A.L. Natural products in drug discovery. Drug Discov. Today. 2008; 13:894–901. [DOI] [PubMed] [Google Scholar]
- 3. Bérdy J. Bioactive microbial metabolites. J. Antibiot. (Tokyo). 2005; 58:1–26. [DOI] [PubMed] [Google Scholar]
- 4. Nett M., Ikeda H., Moore B.S.. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009; 26:1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachmann B.O., Van Lanen S.G., Baltz R.H.. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making. J. Ind. Microbiol. Biotechnol. 2014; 41:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziemert N., Alanjary M., Weber T.. The evolution of genome mining in microbes—a review. Nat. Prod. Rep. 2016; 33:988–1005. [DOI] [PubMed] [Google Scholar]
- 7. Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Muller R., Wohlleben W. et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015; 43:W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L.H.L.H., Wyatt M.A., Magarvey N.A.. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 2015; 43:9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015; 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blin K., Medema M.H., Kottmann R., Lee S.Y., Weber T.. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017; 45:555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cruz-Morales P., Kopp J.F., Martínez-Guerrero C., Yáñez-Guerra L.A., Selem-Mojica N., Ramos-Aboites H., Feldmann J., Barona-Gómez F.. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes. Genome Biol. Evol. 2016; 8:1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hadjithomas M., Chen I.-M.A., Chu K., Ratner A., Palaniappan K., Szeto E., Huang J., Reddy T.B.K., Cimermancic P., Fischbach M.A. et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. Mbio. 2015; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wright G.D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007; 5:175–186. [DOI] [PubMed] [Google Scholar]
- 14. Thaker M.N., Waglechner N., Wright G.D.. Antibiotic resistance–mediated isolation of scaffold-specific natural product producers. Nat. Protoc. 2014; 9:1469–1479. [DOI] [PubMed] [Google Scholar]
- 15. Kale A.J., McGlinchey R.P., Lechner A., Moore B.S.. Bacterial self-resistance to the natural proteasome inhibitor salinosporamide A. ACS Chem. Biol. 2011; 6:1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freel K.C., Millán-Aguiñaga N., Jensen P.R.. Multilocus sequence typing reveals evidence of homologous recombination linked to antibiotic resistance in the genus Salinispora. Appl. Environ. Microbiol. 2013; 79:5997–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziemert N., Lechner A., Wietz M., Millán-Aguiñaga N., Chavarria K.L., Jensen P.R.. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E1130–E1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang X., Li J., Millán-Aguiñaga N., Zhang J.J., O’Neill E.C., Ugalde J.A., Jensen P.R., Mantovani S.M., Moore B.S.. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 2015; 10:2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., De Pascale G., Ejim L. et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013; 57:3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thai Q.K., Bös F., Pleiss J.. The Lactamase Engineering Database: a critical survey of TEM sequences in public databases. BMC Genomics. 2009; 10:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bush K., Jacoby G.A.. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010; 54:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibson M.K., Forsberg K.J., Dantas G.. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2014; 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo H., Lin Y., Gao F., Zhang C.T., Zhang R.. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014; 42:574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang C.H., Hsiang T., Trevors J.T.. Comparative bacterial genomics: Defining the minimal core genome. Antonie Van Leeuwenhoek. 2013; 103:385–398. [DOI] [PubMed] [Google Scholar]
- 26. Christensen H., Kuhnert P., Olsen J.E., Bisgaard M.. Comparative phylogenies of the housekeeping genes atpD, infB and rpoB and the 16S rRNA gene within the Pasteurellaceae. Int. J. Syst. Evol. Microbiol. 2004; 54:1601–1609. [DOI] [PubMed] [Google Scholar]
- 27. Alam M.T., Medema M.H., Takano E., Breitling R.. Comparative genome-scale metabolic modeling of actinomycetes: The topology of essential core metabolism. FEBS Lett. 2011; 585:2389–2394. [DOI] [PubMed] [Google Scholar]
- 28. Jeong H., Sung S., Kwon T., Seo M., Caetano-Anollés K., Choi S.H., Cho S., Nasir A., Kim H.. HGTree: Database of horizontally transferred genes determined by tree reconciliation. Nucleic Acids Res. 2016; 44:D610–D619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eustáquio A.S., McGlinchey R.P., Liu Y., Hazzard C., Beer L.L., Florova G., Alhamadsheh M.M., Lechner A., Kale A.J., Kobayashi Y. et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12295–12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ryder M.H., Slota J.E., Scarim A., Farrand S.K.. Genetic analysis of agrocin 84 production and immunity in Agrobacterium spp. J. Bacteriol. 1987; 169:4184–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cimermancic P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014; 158:412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barona-Gomez F., Cruz-Morales P., Noda-Garcia L.. What can genome-scale metabolic network reconstructions do for prokaryotic systematics. Antonie Van Leeuwenhoek. 2012; 101:35–43. [DOI] [PubMed] [Google Scholar]
- 33. Steffensky M., Mühlenweg A., Wang Z.X., Li S.M., Heide L.. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob. Agents Chemother. 2000; 44:1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmutz E., Mühlenweg A., Li S.M., Heide L.. Resistance genes of aminocoumarin producers: two type II topoisomerase genes confer resistance against coumermycin A1 and clorobiocin. Antimicrob. Agents Chemother. 2003; 47:869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hashimi S.M., Huang G., Maxwell A., Birch R.G.. DNA gyrase from the albicidin producer Xanthomonas albilineans has multiple-antibiotic-resistance and unusual enzymatic properties. Antimicrob. Agents Chemother. 2008; 52:1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sánchez-Hidalgo M., Núñez L.E., Méndez C., Salas J.A.. Involvement of the beta subunit of RNA polymerase in resistance to streptolydigin and streptovaricin in the producer organisms Streptomyces lydicus and Streptomyces spectabilis. Antimicrob. Agents Chemother. 2010; 54:1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Floss H.G., Yu T.-W.. Rifamycin-mode of action, resistance, and biosynthesis. Chem. Rev. 2005; 105:621–632. [DOI] [PubMed] [Google Scholar]
- 38. Ishikawa J., Yamashita A., Mikami Y., Hoshino Y., Kurita H., Hotta K., Shiba T., Hattori M.. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14925–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wieland Brown L.C., Acker M.G., Clardy J., Walsh C.T., Fischbach M.A.. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2549–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bibb M.J., White J., Ward J.M., Janssen G.R.. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol. Microbiol. 1994; 14:533–545. [DOI] [PubMed] [Google Scholar]
- 41. Marshall C.G., Lessard I.A.D., Park I.S., Wright G.D.. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 1998; 42:2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liras P. Biosynthesis and molecular genetics of cephamycins. Cephamycins produced by actinomycetes. Antonie Van Leeuwenhoek. 1999; 75:109–124. [DOI] [PubMed] [Google Scholar]
- 43. Sosio M., Amati G., Cappellano C., Sarubbi E., Monti F., Donadio S.. An elongation factor Tu (EF-Tu) resistant to the EF-Tu inhibitor GE2270 in the producing organism Planobispora rosea. Mol. Microbiol. 1996; 22:43–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.