Abstract
PIGSpro is a significant upgrade of the popular PIGS server for the prediction of the structure of immunoglobulins. The software has been completely rewritten in python following a similar pipeline as in the original method, but including, at various steps, relevant modifications found to improve its prediction accuracy, as demonstrated here. The steps of the pipeline include the selection of the appropriate framework for predicting the conserved regions of the molecule by homology; the target template alignment for this portion of the molecule; the selection of the main chain conformation of the hypervariable loops according to the canonical structure model, the prediction of the third loop of the heavy chain (H3) for which complete canonical structures are not available and the packing of the light and heavy chain if derived from different templates. Each of these steps has been improved including updated methods developed along the years. Last but not least, the user interface has been completely redesigned and an automatic monthly update of the underlying database has been implemented. The method is available as a web server at http://biocomputing.it/pigspro.
INTRODUCTION
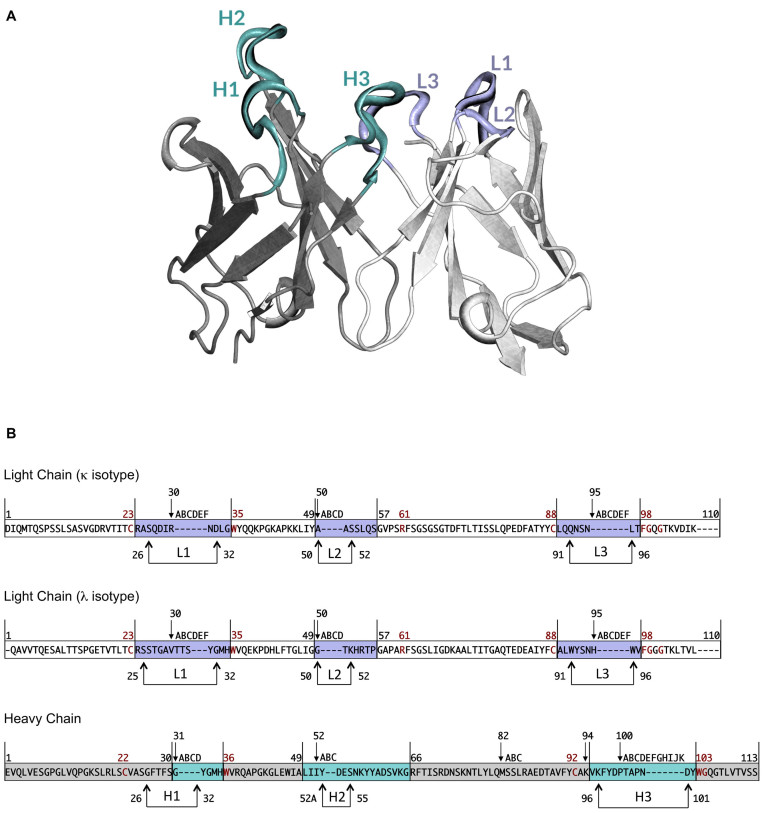
Immunoglobulins are multimeric glycoproteins secreted by B-cells and formed by two identical light and heavy chains made by structurally similar domains, two for the light and four or more for the heavy chain, respectively. These domains are very well conserved and form what is called the framework of the protein, while a few regions at the tip of the protein are very variable and constitute the antigen-binding site (1) (Figure 1A). These regions structurally correspond to loops, named L1 to L3 and H1 to H3 according to the order in which they appear in the light (L) and heavy (H) chain.
Figure 1.
(A) Variable region of an antibody molecule. Heavy and light chain framework regions are coloured in grey and white, respectively. Loops composing the antigen-binding site are coloured in pale cyan for the heavy chain and light violet for the light chain. (B) Chothia numbering scheme for VH, VK and VL. The numbers above the sequences represent the numbering of specific residues. The remaining residues are numbered consecutively. Letters correspond to insertions. Framework regions are depicted in grey for VH and in white for VK and VL. Complementarity determining regions are coloured in pale cyan for VH and in light violet for VK and VL. Arrows indicate Chothia and Lesk definition of hypervariable loops. Conserved residues are reported in dark red.
The main chain conformation of the light chain hypervariable loops and of the first two of the heavy chain have been shown to follow the canonical structure model according to which a few key residues, within or without the loops, determine their structure (2–14). The third loop of the heavy chain (H3) has a different behaviour (15–19). The region of the loop closer to the framework (named the ‘torso’(16)) follows a canonical structure model and its first four and last six residues form a bulged or non-bulged beta sheet conformation according to the identity of the residues at position 94 and 101 of the heavy chain (Figure 1B) (the numbering scheme of Chothia (8) is used throughout the paper), the remaining part has revealed to be very difficult to predict given its very high variability in both length and shape.
Given the above, the classical strategy for predicting the structure of an immunoglobulin from its amino acid sequence as implemented by us and others (20,21), follows the following steps:
The main chains of the light and heavy chain frameworks are modelled by homology using, as template, the corresponding domain from a similar immunoglobulin;
The L1-L3 and H1-H2 loops are modelled by inheriting their conformation from an immunoglobulin with the same canonical structure (i.e. bearing in the key positions the same amino acids as the target protein);
The H3 loop is modelled using loops with the highest sequence similarity to the target available or by other methods (22);
The two chains are packed together;
The side chains of the molecule are predicted using classical methods for side chain conformation prediction such as SCWRL4 (23).
The above protocol has been implemented in the original PIGS server, which has been positively evaluated in two blind assessments of the prediction accuracy of antibody modelling (24,25).
The server also included the option of selecting the two frameworks from the same immunoglobulin even though a template with a higher similarity was available for one of the two. This choice is beneficial when the difference in similarity between the two best templates for the heavy and light chain is not too high (26): selecting a less than optimal template for one chain, but coming from the same immunoglobulin used as template for the other chain, was found to minimize the error introduced by the packing of the two chains. Different protocols for antibody modelling have also been developed, using strategies different from the one here described (20,27–31).
During the past years, we continued to analyze the immunoglobulin structures taking advantage of the increased number of experimentally solved structure and have now improved the pipeline including the results of our new findings. More specifically, we integrated new strategies for: (i) selecting and aligning the framework and the loop templates based on immunoglobulin specific profiles (26); (ii) modelling the lambda light chains using an increased repertoire of canonical structures (12); (iii) packing the heavy and the light chain domains, which have been found to cluster around two dominant orientations (32); (iv) predicting the structure of the hypervariable loop H3, a rather elusive and complex problem so far, with significantly improved accuracy than that achieved by PIGS and by other methods (22). Last but importantly, our database of templates is now composed of 1691 immunoglobulins of known structures, to be compared with 312 of the original one.
We tested the performance of the new pipeline and compared it with the results obtained with PIGS. To this purpose we used the same strategy previously adopted in (26) and summarized below, as well as the same benchmark dataset. In both the older and the new version of the server, an option is provided to ‘blacklist’ an immunoglobulin of the database of known immunoglobulin structures, i.e. not to use any information about the specific blacklisted structure. This option has been used for testing the old version of the server and is used here for testing the new version. In practice, we use a leave one out procedure in which, one by one, the immunoglobulins of known structure are blacklisted and predicted without using their coordinates or the coordinates of any other immunoglobulin sharing a sequence identity equal or higher than 98% with the target (26). The produced models are then compared with the corresponding experimental structures. In summary, we were able to improve both the prediction of the overall antibody structure and of the antigen-binding site.
MATERIALS AND METHODS
There are two modes of using the server, single sequence and multiple sequence. In the first case, the user needs to input the light and heavy chain sequence of the target immunoglobulin. It can select to blacklist one of the known immunoglobulin structures, an option useful for testing the methods. A project title and an email address can be provided, but are not required.
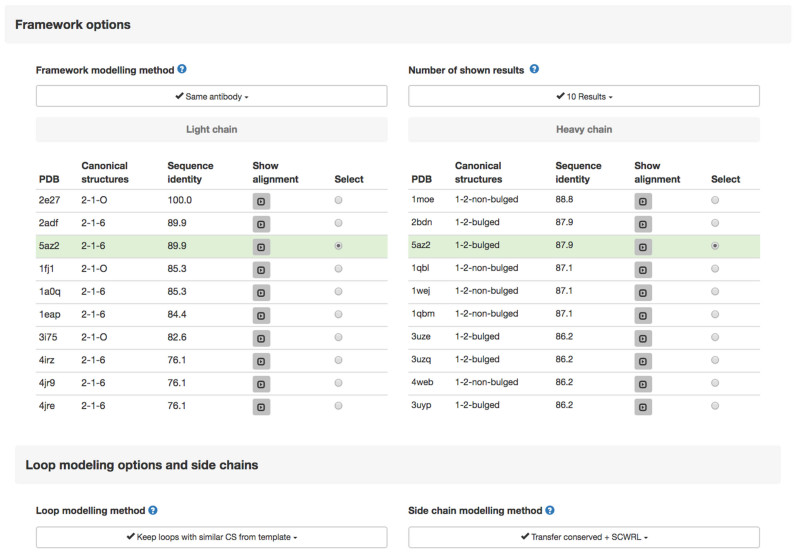
Upon selecting the Submit button, the user is presented with a new page where she/he can select a number of options (Figure 2), namely:
Figure 2.
Template selection page. Different options are provided for modelling the framework region, the loops and the side chains. Two lists of templates are displayed, for the heavy and light chain frameworks, in two separate tables. The best available templates are highlighted and automatically selected according to the ‘Framework modelling method’ and the ‘Number of shown results’. The tables report for each template, the PDB ID, the canonical structures of the loops and the target-template sequence identity. A button to visualize the target-template alignment is also provided.
Framework options
- The four available criteria are:
- Same Antibody (default) Selects the known structure that can provide a template for both the heavy and light chain, even if a different template with a higher sequence identity exists for one of the chains
- Same Canonical Structures Selects the template having loops with the same canonical structure of the target even if a different template with a higher sequence identity exists for one or both chains
- Same Antibody and Canonical Structures Selects an antibody structure that can be used as a template for both VL and VH and where the canonical structures of the loops are the same as those of the target even if a different template with a slightly better sequence identity exists for one or both chains
- Best H and L chains Selects the two chains with highest sequence identity with the corresponding chains of the target and, if needed, pack the two chains together and take the loops from a different structure
Number of shown results (default = 10) Number of results to be displayed. Templates (both in single sequence and multiple sequence modes) are chosen among these results
The page also shows the list of available templates together with the canonical structure of their loops (the definition of which can be seen by clicking an information icon) and the sequence identity between the target and each of the templates. A button allows the corresponding alignment to be displayed interactively.
The user can also select the Loop modelling method. The four options are:
Keep loops with similar Canonical Structures from template (default) If one or more of the target and template loops have the same canonical structure, keep the main chain structure of the template loops
Keep loops with similar Canonical Structures, H3LooPred for H3 If one or more of the target and template loops have the same canonical structure, keep the main chain structure for all of them excluding H3. Build the latter with the H3LooPred method (22)
Select Canonical Structures from most similar loops Take the main chain of each loop from the antibody with the same canonical structure and the highest sequence similarity
Select Canonical Structures from most similar loops, H3LooPred for H3 Take the main chain of each loop from the antibody with the same canonical structure and the highest sequence similarity. In any case build H3 with the H3LooPred method (22)
- Side chain modelling method. Criteria for the side chains modelling can be chosen among:
- Transfer Conserved + SCWRL (default). The conformation of the side chains of residues conserved between the target and the template is maintained. Non-conserved side chains are modelled using SCWRL4
- Transfer Conserved The conformation of the side chains of residues conserved between the target and the template is maintained. Only backbone atoms are included in the final model for non-conserved residues
- All SCWRL All side chains are modelled using SCWRL4
- Backbone only This will generate a ‘backbone-only’ model
In the multiple sequence mode, the user can input the sequences as a multifasta file. The file should contain both heavy and light chain sequences for each antibody. The FASTA header line should contain a name and/or a unique identifier for each antibody. Chains from the same antibody should have the same name/identifier. In this case the user selects the options at the input stage and they will be used for all the target immunoglobulins.
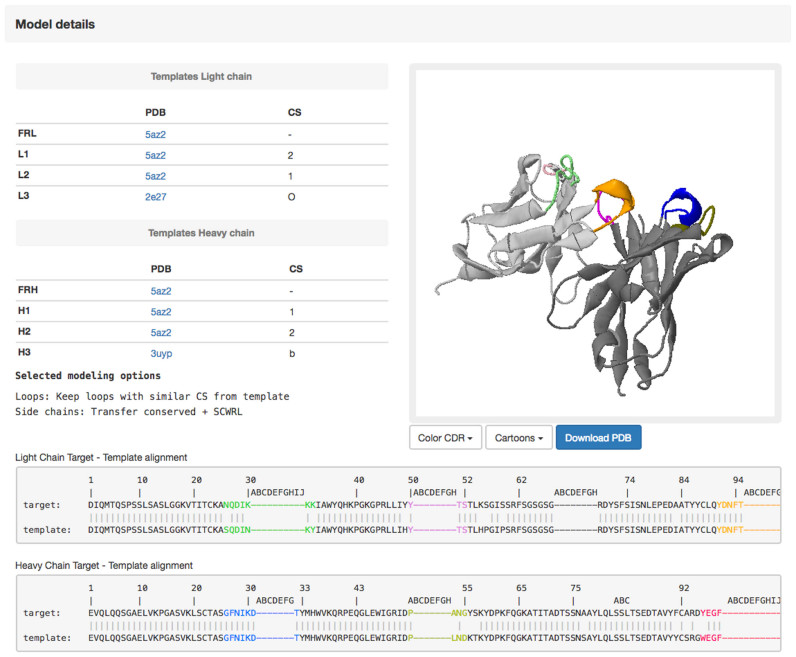
Output
The output page contains a summary of the user choices, the alignment used for building the model and the structure of the model visualized in a JSmol window (Figure 3). The final model can be downloaded as a PDB file. The REMARK records of the PDB file contain a summary of the options used (template and canonical structure for each loop).
Figure 3.
The output page. The output includes two main tables with information about the templates used to build the three-dimensional model of the target antibody. The final model can be either visualized in the jsmol window (http://www.jmol.org/) or downloaded using the ‘Download PDB’ button. The final target-template alignments for both the heavy and light chains are also shown.
RESULTS AND DISCUSSION
What is new
Prediction of immunoglobulin lambda chains
In mammals there are two types of immunoglobulin light chain, called lambda (λ) and kappa (κ). The level of expression of the two chains is different, in mouse the ratio is about 20:1 in favour of the kappa chains (33). This imbalance in the most common model animal has led to the presence of a much higher proportion of kappa chains among the immunoglobulins of known structure and, consequently, less attention has been devoted to the analysis of the conformation of the lambda type of chains. However, in human the ratio is much more balanced (34). It was found that there are recurring conformations in lambda chains, as well, and that they are not the same as those for the kappa chains. Work in our group identified several lambda-restricted canonical structures, in detail eight for L1, two for L2 and five for L3, together with the key residues determining each of them (12).
These definitions are now included in PIGSPro that is able to predict the conformation of the lambda light chains with satisfactory accuracy (Table 1). It should be mentioned that the number of available lambda structures that can be used for a comparison between the old and the new server is very limited. We could only compute the data for 15 structures and, even though these are predicted with an average Cα RMSD of 0.83 in PIGSPro compared to 0.89 for PIGS, the difference is not statistically significant.
Table 1. Cα RMSD values of the models produced by the old server (PIGS) and its updated version (PIGSPro).
| All residues | Loop residues: local | Loop residues: global | H3 residues: local | H3 residues: global | Framework residues | Lambda light chains | |
|---|---|---|---|---|---|---|---|
| PIGS | 1.36 ± 0.64 | 2.21 ± 1.48 | 2.26 ± 1.52 | 3.59 ± 2.93 | 3.67 ± 2.92 | 0.78 ± 0.28 | 0.89 ± 0.27 |
| PIGSPro | 1.16 ± 0.47 | 1.75 ± 0.95 | 1.79 ± 1.03 | 2.41 ± 2.2 | 2.45 ± 2.18 | 0.75 ± 0.24 | 0.83 ± 0.64 |
| Number of models | 252 | 252 | 252 | 252 | 252 | 252 | 15 |
The RMSD for the loop residues and H3 are computed both after superimposing their stems, i.e. the two residues before and after the loop (local) and after superposition the framework (global). Underlined values indicate a statistically significant difference (95% confidence level) with respect to the PIGS method based on an unpaired t-test.
The framework selection
In the old version of PIGS, the alignment between the target immunoglobulin sequence and that of the template(s) was based on sequence specific rules, mainly related to the position of very conserved residues (two cysteines and one tryptophan in each chain) and on the observation that insertions and deletions among immunoglobulins occur, with rare exceptions, in the loop regions (Figure 1B).
In this new version, we use a different approach that is also able to take into account unusual cases of insertions and deletions in positions other than the hypervariable loops. In particular, we built ad-hoc Hidden Markov Models (HMM) for the light (kappa and lambda) and heavy chains and use them to select the template and the alignment. As shown in previous discrimination tests, HMMs can be successfully used to distinguish members of a protein family from non-members with a high degree of accuracy (35). In our specific case, stringent criteria are used in order to ensure (i) high specificity (i.e. to distinguish immunoglobulin from other Ig-like sequences) and (ii) obtain more accurate target-template alignments, i.e. more reliable models.
The packing of the light and heavy chain framework
In (32), our group demonstrated that the immunoglobulins of known structure could be clustered according to the relative orientation of their light and heavy chains and discovered that the large majority of them can be assigned to one of two clusters. A set of residues were found to be able to discriminate between the two orientations with a classification error lower than 10%. In particular, the identity of the residue in position L44, located at the interface between the chains, permits to discriminate between the two packing modes. The specific packing of the two chains differs according to whether the residues in position L44 is or is not a Proline.
In PIGSPro, we inherit the relative orientation of the two chains by using as templates the VL–VH complex with the highest sequence similarity and superimposing the conserved residues at the interface. However, at variance with the old method, the orientation of the two chains is inherited only from complexes where the light chain of the immunoglobulin has the same residue in position L44 as the target. This, according to our previous findings (32), is expected to lead to a better prediction of the relative orientation of the chains in the model.
H3
As mentioned above, only a partial canonical structure model existed for H3. Accordingly, so far the prediction accuracy for H3 loops has been not equally satisfactory as for the other loops, an important drawback because the H3 loop is central in the binding site and therefore essential in determining the antibody–antigen interactions. We approached the problem of obtaining a better prediction for the main chain of this loop by training a Random Forest machine learning algorithm to select the closest loop among a dataset of H3 loops present in immunoglobulins of known structure. The selected putative templates are subsequently ranked according to their intramolecular interactions by comparing the predicted interactions of the modelled H3 residues with those observed in immunoglobulin of known structure (22). This strategy has proven to be sufficiently robust in identifying reliable structural templates and also provided the first evidence that the H3 environment information can be used to successfully rank large sets of conformations of the same loop (22). In summary, in terms of prediction accuracy we achieve significant improvements compared to PIGS (Table 1) and to other methods (22).
Performance improvement
We compared the performance of the new and old server on a data set of 252 structures using the leave one out procedure described above and adopted in (26). Models were only considered when the sequence identity with the corresponding template was lower than 98%.
Table 1 shows the results obtained on this dataset compared with the corresponding results for the PIGS server. As it can be seen, there is an improvement in both the prediction of the overall structure and of the antigen-binding site.
The web interface
The PIGSPro web interface has been redesigned to improve user friendliness. Responsive layouts are implemented using the Bootstrap front-end web framework, JavaScript and JQuery.
The database
PIGSPro heavily relies on the underlying database of known immunoglobulin structures. This has increased from 312 to 1691 structures and, most importantly, is now automatically updated every month. The 1691 antibodies of the database include only structures obtained from x-ray experiments with a resolution better than 3.0 Å and without any missing residue or atom.
ACKNOWLEDGEMENTS
The authors are grateful to Alessandra Rosi and Paolo Marcatili who contributed to the first version of PIGS and to Anna Chailyan who collaborated to the analysis of the lambda chain canonical structures and to the VH/VL packing.
FUNDING
EPIGEN Flagship project of the Italian Ministry of Science. Funding for open access charge: EPIGEN Flagship project of the Italian Ministry of Science and Education.
Conflict of interest statement. None declared.
REFERENCES
- 1. Narciso J.E., Uy I.D., Cabang A.B., Chavez J.F., Pablo J.L., Padilla-Concepcion G.P., Padlan E.A.. Analysis of the antibody structure based on high-resolution crystallographic studies. N. Biotechnol. 2011; 28:435–447. [DOI] [PubMed] [Google Scholar]
- 2. Chothia C., Lesk A.M.. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 1987; 196:901–917. [DOI] [PubMed] [Google Scholar]
- 3. Chothia C., Lesk A.M., Tramontano A., Levitt M., Smith-Gill S.J., Air G., Sheriff S., Padlan E.A., Davies D., Tulip W.R. et al. . Conformations of immunoglobulin hypervariable regions. Nature. 1989; 342:877–883. [DOI] [PubMed] [Google Scholar]
- 4. Tramontano A., Chothia C., Lesk A.M.. Framework residue 71 is a major determinant of the position and conformation of the second hypervariable region in the VH domains of immunoglobulins. J. Mol. Biol. 1990; 215:175–182. [DOI] [PubMed] [Google Scholar]
- 5. Chothia C., Lesk A.M., Gherardi E., Tomlinson I.M., Walter G., Marks J.D., Llewelyn M.B., Winter G.. Structural repertoire of the human VH segments. J. Mol. Biol. 1992; 227:799–817. [DOI] [PubMed] [Google Scholar]
- 6. Foote J., Winter G.. Antibody framework residues affecting the conformation of the hypervariable loops. J. Mol. Biol. 1992; 224:487–499. [DOI] [PubMed] [Google Scholar]
- 7. Tomlinson I.M., Cox J.P., Gherardi E., Lesk A.M., Chothia C.. The structural repertoire of the human V kappa domain. EMBO J. 1995; 14:4628–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Lazikani B., Lesk A.M., Chothia C.. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 1997; 273:927–948. [DOI] [PubMed] [Google Scholar]
- 9. Chothia C., Gelfand I., Kister A.. Structural determinants in the sequences of immunoglobulin variable domain. J. Mol. Biol. 1998; 278:457–479. [DOI] [PubMed] [Google Scholar]
- 10. Decanniere K., Muyldermans S., Wyns L.. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes. J. Mol. Biol. 2000; 300:83–91. [DOI] [PubMed] [Google Scholar]
- 11. Kuroda D., Shirai H., Kobori M., Nakamura H.. Systematic classification of CDR-L3 in antibodies: implications of the light chain subtypes and the VL-VH interface. Proteins. 2009; 75:139–146. [DOI] [PubMed] [Google Scholar]
- 12. Chailyan A., Marcatili P., Cirillo D., Tramontano A.. Structural repertoire of immunoglobulin lambda light chains. Proteins. 2011; 79:1513–1524. [DOI] [PubMed] [Google Scholar]
- 13. North B., Lehmann A., Dunbrack R.L. Jr. A new clustering of antibody CDR loop conformations. J. Mol. Biol. 2011; 406:228–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vargas-Madrazo E., Paz-Garcia E.. Modifications to canonical structure sequence patterns: analysis for L1 and L3. Proteins. 2002; 47:250–254. [DOI] [PubMed] [Google Scholar]
- 15. Shirai H., Kidera A., Nakamura H.. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996; 399:1–8. [DOI] [PubMed] [Google Scholar]
- 16. Morea V., Tramontano A., Rustici M., Chothia C., Lesk A.M.. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 1998; 275:269–294. [DOI] [PubMed] [Google Scholar]
- 17. Shirai H., Nakajima N., Higo J., Kidera A., Nakamura H.. Conformational sampling of CDR-H3 in antibodies by multicanonical molecular dynamics simulation. J. Mol. Biol. 1998; 278:481–496. [DOI] [PubMed] [Google Scholar]
- 18. Kim S.T., Shirai H., Nakajima N., Higo J., Nakamura H.. Enhanced conformational diversity search of CDR-H3 in antibodies: role of the first CDR-H3 residue. Proteins. 1999; 37:683–696. [PubMed] [Google Scholar]
- 19. Shirai H., Kidera A., Nakamura H.. H3-rules: identification of CDR-H3 structures in antibodies. FEBS Lett. 1999; 455:188–197. [DOI] [PubMed] [Google Scholar]
- 20. Whitelegg N.R., Rees A.R.. WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng. 2000; 13:819–824. [DOI] [PubMed] [Google Scholar]
- 21. Marcatili P., Rosi A., Tramontano A.. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008; 24:1953–1954. [DOI] [PubMed] [Google Scholar]
- 22. Messih M.A., Lepore R., Marcatili P., Tramontano A.. Improving the accuracy of the structure prediction of the third hypervariable loop of the heavy chains of antibodies. Bioinformatics. 2014; 30:2733–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Q., Canutescu A.A., Dunbrack R.L. Jr. SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 2008; 3:1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Almagro J.C., Beavers M.P., Hernandez-Guzman F., Maier J., Shaulsky J., Butenhof K., Labute P., Thorsteinson N., Kelly K., Teplyakov A. et al. . Antibody modeling assessment. Proteins. 2011; 79:3050–3066. [DOI] [PubMed] [Google Scholar]
- 25. Almagro J.C., Teplyakov A., Luo J., Sweet R.W., Kodangattil S., Hernandez-Guzman F., Gilliland G.L.. Second antibody modeling assessment (AMA-II). Proteins. 2014; 82:1553–1562. [DOI] [PubMed] [Google Scholar]
- 26. Marcatili P., Olimpieri P.P., Chailyan A., Tramontano A.. Antibody modeling using the prediction of immunoglobulin structure (PIGS) web server [corrected]. Nat. Protoc. 2014; 9:2771–2783. [DOI] [PubMed] [Google Scholar]
- 27. Sircar A., Kim E.T., Gray J.J.. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 2009; 37:W474–W479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maier J.K., Labute P.. Assessment of fully automated antibody homology modeling protocols in molecular operating environment. Proteins. 2014; 82:1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fasnacht M., Butenhof K., Goupil-Lamy A., Hernandez-Guzman F., Huang H., Yan L.. Automated antibody structureprediction using Accelrys tools: results and best practices. Proteins. 2014; 82:1583–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamashita K., Ikeda K., Amada K., Liang S., Tsuchiya Y., Nakamura H., Shirai H., Standley D.M.. Kotai Antibody Builder: automated high-resolution structural modeling of antibodies. Bioinformatics. 2014; 30:3279–3280. [DOI] [PubMed] [Google Scholar]
- 31. Leem J., Dunbar J., Georges G., Shi J., Deane C.M.. ABodyBuilder: automated antibody structure prediction with data-driven accuracy estimation. MAbs. 2016; 8:1259–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chailyan A., Marcatili P., Tramontano A.. The association of heavy and light chain variable domains in antibodies: implications for antigen specificity. FEBS J. 2011; 278:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woloschak G.E., Krco C.J.. Regulation of kappa/lambda immunoglobulin light chain expression in normal murine lymphocytes. Mol. Immunol. 1987; 24:751–757. [DOI] [PubMed] [Google Scholar]
- 34. Chui S.H., Lam C.W., Lai K.N.. Light-chain ratios of immunoglobulins G, A, and M determined by enzyme immunoassay. Clin. Chem. 1990; 36:501–502. [PubMed] [Google Scholar]
- 35. Krogh A., Brown M., Mian I.S., Sjolander K., Haussler D.. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994; 235:1501–1531. [DOI] [PubMed] [Google Scholar]