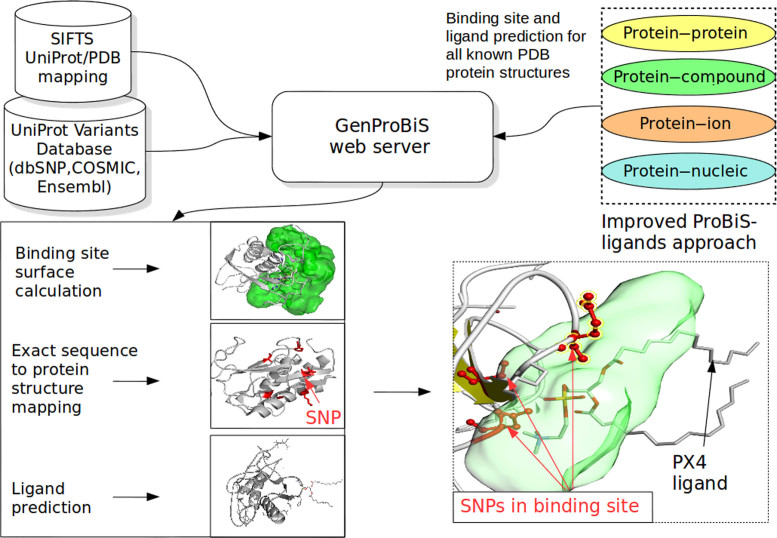
Abstract
Discovery of potentially deleterious sequence variants is important and has wide implications for research and generation of new hypotheses in human and veterinary medicine, and drug discovery. The GenProBiS web server maps sequence variants to protein structures from the Protein Data Bank (PDB), and further to protein–protein, protein–nucleic acid, protein–compound, and protein–metal ion binding sites. The concept of a protein–compound binding site is understood in the broadest sense, which includes glycosylation and other post-translational modification sites. Binding sites were defined by local structural comparisons of whole protein structures using the Protein Binding Sites (ProBiS) algorithm and transposition of ligands from the similar binding sites found to the query protein using the ProBiS-ligands approach with new improvements introduced in GenProBiS. Binding site surfaces were generated as three-dimensional grids encompassing the space occupied by predicted ligands. The server allows intuitive visual exploration of comprehensively mapped variants, such as human somatic mis-sense mutations related to cancer and non-synonymous single nucleotide polymorphisms from 21 species, within the predicted binding sites regions for about 80 000 PDB protein structures using fast WebGL graphics. The GenProBiS web server is open and free to all users at http://genprobis.insilab.org.
INTRODUCTION
Sequence variants that occur in coding regions of genes and alter protein's amino acid sequence presumably affect protein function. Variants can occur in genes of somatic cells, for example mis-sense mutations in cancers or germline cells, such as non-synonymous single nucleotide polymorphisms (nsSNPs). The latter can either substitute amino acids (mis-sense SNPs) or introduce premature stop codons, or nonsense codons resulting in incomplete proteins (nonsense SNPs) (1). Non-synonymous SNPs affect phenotypic diversity, disease development and response to drugs. Both somatic and germline sequence variants have been linked to various cancers (2) and other diseases (3). Sickle-cell anemia is a classic example of a disease caused by a single nsSNP, where a glutamic acid residue is replaced by valine in hemoglobin (4).
Binding sites on proteins interact with various ligands and hence govern the biochemical functions of proteins. It was found that disease-causing nsSNPs are preferentially located at protein–protein interfaces rather than in non-interface regions of protein surfaces (5). Significant enrichments of somatic mis-sense mutations were found within protein–protein, protein–nucleic acid and protein–metal ion binding sites in several proteins involved in tumorigenesis (6). As such, binding site sequence variants are of great interest to drug development chemists and clinicians who seek to predict an individual's response to a drug. A variety of algorithms, web servers and databases have been developed to identify nsSNPs which influence protein function (7–9) and response to drugs (10). Mapping of nsSNPs to Protein Data Bank (PDB) (11) protein structures has been accomplished for human proteins (11–15) as well as for both human and non-human proteins (16) but to our knowledge, mapping of somatic mutations and nsSNPs from many different species to diverse types of binding sites and further, to each site's ligand specifically for all PDB protein structures, does not exist.
Detection of protein binding sites is a challenging task. Proteins typically bind several different ligands, but any single protein structure in the PDB only contains one or a few co-crystallized ligands and thus shows an incomplete state of the actual binding sites. To finesse this problem, we define binding sites on proteins using the ProBiS-ligands approach (17), which has been improved in GenProBiS. This accounts for the co-crystallized ligands from the same binding site, as well as for the ligands binding to similar binding sites in other PDB structures. The approach detects and aligns similar binding sites irrespective of their proteins' similar folding patterns using the ProBiS algorithm (18). In this algorithm, protein structures are represented as graphs, in which vertices represent functional groups of surface amino acids and edges are drawn between pairs of vertices that are <15 Å apart. Two protein graphs are divided into several subgraphs that together completely sample the two protein surfaces. From each pair of protein subgraphs, a product graph is constructed, i.e. an approximate representation of all possible local superimpositions of the two protein structures. Using our maximum clique algorithm (19), the largest complete subgraph is detected within each product graph, which corresponds to the best local superimposition of the two compared protein structures. Ligands, co-crystallized in the superimposed similar binding sites, are then transposed to the query protein based on this superimposition. The transposed ligands are clustered by their spatial proximity and each such cluster represents one binding site. Finally, degrees of structural evolutionary conservation are calculated for each query protein's amino acid residue from the multiple protein structure alignment (18). Recently, a variation of this approach was successfully used for discovery of small-molecule inhibitors of InhA enzyme in Mycobacterium tuberculosis and this resulted in identification of three previously unrecognized inhibitors with novel scaffolds (20).
In this paper we describe a new web server, GenProBiS, which allows mapping of human somatic mis-sense mutations related to cancer and nsSNPs from genome sequences of 21 species to protein binding sites in the PDB. The concept of a binding site is understood in the broadest sense, which includes glycosylation and other post-translational modification sites. These are in GenProBiS classified under protein–compound binding sites. Binding sites are defined as the space occupied by atoms of all co-crystallized ligands transposed to the query protein from PDBs sharing similar binding sites with the query protein. Binding site grids are generated and visualized as solvent accessible molecular surfaces. GenProBiS enables detection of sequence variants within a protein binding site and visual exploration of interactions, or loss of interactions, of a specific mis-sense mutation with a specific ligand. We show the usability of GenProBiS on selected disease-related nsSNPs and somatic mutations whose importance in disease development and potential drug response effects can be explained by their presence in binding sites and their interactions with ligands.
GENPROBIS WEB SERVER
The GenProBiS web server implements a novel approach to the discovery of sequence variants that have potentially deleterious effect on protein function and ligand binding through gain or loss of the binding site (Figure 1). Currently, the web server maps around 550 000 sequence variants to about 5 million amino acid residues in 80 000 PDB protein structures enriched with protein–protein, protein–nucleic acid, protein–compound and protein–metal ion binding sites. The sequence variants were collected from the UniProt variants dataset (21), which contains data from various databases including around 95 000 somatic mis-sense mutations from human cancers from the COSMIC database (2), 460 000 nsSNPs from 14 different species listed according to the decreasing numbers of nsSNPs, including Homo sapiens, Bos taurus, Mus musculus, Sus scrofa, Gallus gallus, Anopheles gambiae, Danio rerio, Canis familiaris, Equus caballus, Macaca mulatta, Oryza indica, Oryza sativa, Ovis aries and Plasmodium falciparum from the dbSNP database (22), around 500 polymorphisms from six plant species, Zea mays, Yitis vinifera, Sorghum bicolor, Solanum lycopersicum, Phytophtora infestans and O. sativa from the EnsemblPlants (23) and around 60 polymorphisms from Aedes aegypti, Ixodes scapularis and A. gambiae species obtained from the EnsemblMetazoa database (23). UniProt amino acid sequence locations of sequence variants were converted to PDB structure locations using the Structure integration with function, taxonomy and sequence (SIFTS) project conversion table (24).
Figure 1.
The GenProBiS web server approach depicted on example of nsSNPs mapping to a compound (low molecular weight ligand) binding site.
Binding sites were predicted by local structural comparisons of whole protein structures using the ProBiS algorithm (18) and transposition of ligands from the similar binding sites found to the query protein using an updated ProBiS-ligands approach (17) with the following major improvements introduced in GenProBiS:
Protein, nucleic acid, compound and metal ion binding sites and ligands are predicted for ∼300 000 protein chains in the PDB. The original ProBiS-ligands approach only enabled prediction of ligands for the 42 000 protein chains in the 95% non-redundant PDB.
Predicted protein or nucleic acid ligands that severely clash with the query protein, i.e. have >10 atoms <1.0 Å from any query protein atom, are now discarded.
The cutoffs for binding site similarity scores (z-scores), originally 1.0 for all ligand types, are now 2.5 for compounds, 3.0 for proteins, 3.0 for nucleic acids and 2.0 for metal ions. While binding site z-scores and whole-sequence identities are not directly comparable, a z-score of 2.0 in GenProBiS, as a rule of thumb corresponds to ∼30% sequence identity.
Ligands have been clustered by their spatial proximity using OPTICS algorithm (25), each cluster containing from a single to hundreds of ligands and representing one binding site, where the measure of distance is now their minimum distance between any two atoms; in an earlier approach we used distance between geometric centers of ligands, which did not cluster protein and nucleic acid ligands well.
Biologically relevant ion and compound ligands are identified using the list of non-specific binders and known crystallization artifacts at http://insilab.org/files/GenProBiS/non-specific.txt. Additionally, ions that belong to clusters with <10 members are considered artifacts.
Binding site grids are now generated as hexagonal close-packed grids with a resolution of 1.5 Å, encompassing the space occupied by atoms of predicted clustered ligands, where grid points had to be <4 Å from any predicted ligand's atom and <8 Å from any query protein atom.
Protein residues <3 Å from any grid point are considered as binding site residues.
A residue and a ligand are considered to interact if the distance between any of their atoms is <5 Å.
Solvent accessible surfaces of binding site grids, which are visualized in the GenProBiS web server, have been precomputed using an in-house algorithm. Structurally mapped somatic mutations and nsSNPs were then assigned to one or more binding sites, and were labeled according to the binding site's ligand type (protein, nucleic acid, compound or ion) and the number of the binding site. To facilitate high-speed access to the binding sites, we precomputed protein binding sites for all protein structures in the PDB, i.e. around 300 000 combinations of PDB and Chain IDs. This binding site prediction across the entire PDB was computationally intensive. It was completed in about 2 months using 1400 CPUs. Future updates of the database will require considerably less time (about a week on a single CPU) since only the difference between the initial and the updated PDBs will need to be recomputed.
INPUT
GenProBiS requires as input the PDB and Chain ID (11). It also can use dbSNP's reference SNP cluster (rs) ID (22), COSMIC's Mutation ID (2), Uniprot ID or Uniprot's Gene Symbol (21). The basic input is a protein structure (PDB and Chain ID) and when these are entered, clicking the »Search« button takes the user directly to the results page. Alternatively, one may enter dbSNP's rs ID, COSMIC's Mutation ID, UniProt ID or Uniprot's Gene Symbol and then the »Conversion tool« opens and displays the list of PDB protein structures corresponding to the input. A user can then choose a specific structure for further exploration. Using the »Custom input« link the user can also upload a list of custom variants with UniProt sequence positions and chooses the PDB structure to which they are to be mapped.
OUTPUT
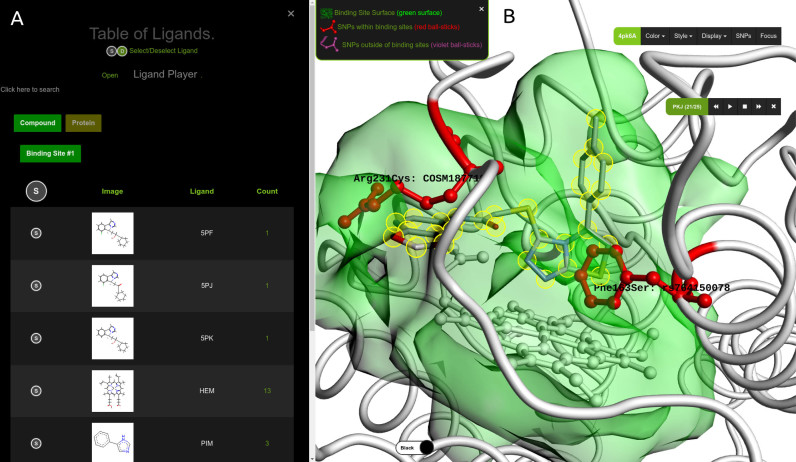
GenProBiS maps sequence variants to protein binding sites for the given query protein (Figure 2). The server allows intuitive visual exploration of mapped sequence variants within the predicted binding site regions using WebGL graphics implemented in the Molmil molecular viewer (26). Molmil allows visualization of large proteins and their multiple ligands in an internet browser. Users can explore three-dimensional (3D) poses of all the transposed ligands within the same query binding site and their potential interactions with mis-sense mutations, a feature not available elsewhere.
Figure 2.
GenProBiS output page for indoleamine 2, 3-dioxygenase as the query protein (PDB and Chain ID: 4pk6A). (A) »Table of Ligands« listing the predicted compound ligands, including co-crystallized inhibitors, transposed from other crystal structures to the query protein. (B) »3D Viewer« zoomed-in on the predicted binding site for compounds (green surface). Polymorphic amino acid residues Arg231Cys and Phe163Ser are red ball-and-sticks and the inhibitor (Ligand ID: PKJ) is CPK-colored sticks model highlighted with yellow halos. Change of arginine to cysteine likely results in the loss of electrostatic interaction between the positively charged arginine and the partially negatively charged oxygen (red stick closest to arginine) of the 1, 3-benzodioxole fragment belonging to the PKJ inhibitor; change of phenylalanine to serine results in loss of the pi–pi stacking interaction between phenylalanine and the imidazothiazole (bicyclic fragment behind the phenylalanine) of PKJ. The native co-crystallized heme is shown as white ball-and-sticks model. The draggable menu and the »Ligand Player« console are in the upper-right corner of the viewer.
GenProBiS results page has a »Vertical Menu« on the left side and the remainder of the browser window is the »3D viewer«. Upon clicking on any of its main links, the vertical menu expands to display tables with sequence variants, binding site and ligands mapping data. Above, there is a camera icon which allows the user to save the current state of the »3D Viewer« as a PNG picture; a play icon to open the »Ligand Player« (discussed in the Table of Ligands section below); a download icon to save mapping of sequence variants to binding sites as a text file; and a link icon to the Evolutionarily Conserved Regions (ECR) genome browser (27) that allows exploration of alignments of the query protein's gene with certain different species. Below these icons, the main links are as follows.
Table of sequence variants
Sequence variants that are within and outside the predicted binding sites are listed in this table in which each row contains: (i) three circular buttons to show (S), label (L) or zoom in (Z) on the sequence variant (e.g. nsSNP) as a stick model on the query protein structure in the »3D Viewer«; (ii) description of the amino acid change, for example Asn78Ser indicates that asparagine changes to serine at the 78th position in the protein sequence according to the UniProt sequence numbering; (iii) if a sequence variant is in one or more binding sites, this is shown as one or more small circles, whose colors indicate the ligand type—brown for metal ions, green for compounds, yellow for proteins and blue for nucleic acids. A number inside each circle is the binding site number; (iv) the variant's accession number, which also serves as an http link that allows exploring the sequence variant in its original database; (v) where available, links to various annotation databases, such as ClinVar (3) and PharmGKB (10).
Table of binding sites
Protein binding sites and sequence variants that are associated with each binding site are listed in this table. Binding sites can be selected according to their ligands' types with buttons labeled »Compound«, »Ion«, »Nucleic«, and »Protein«, and binding site numbers within each ligand type. Selecting a binding site results in a table with its mapped sequence variants. The »Sticks« and »Surface« buttons above this table allow each binding site to be displayed either as sticks or surface models, the latter being the default view.
Table of ligands
Selecting a binding site according to its type and number, prompts a display of a table of its corresponding ligands. Each ligand (or several ligands at once) can be selected using (S) button, resulting in ligands’ 3D structures being displayed in the query protein in the »3D Viewer«. Interactions of ligands with sequence variants can be seen by clicking the (I) button, which opens a table listing the minimum distances between all the ligands with the same name and the sequence variant residues. Clicking on a row in this table zooms in and shows the corresponding interaction as a line in the »3D Viewer«. In the »Ligand« column is the name of the ligand (its PDB code or Ligand ID), which is also an http link to the ligand's PDB web page. The »Count« column provides the number of ligands with the same PDB code or Ligand ID. Clicking the »Ligand Player« near the top, opens a small console on the right side of the screen with »play«, »forward«, »backward« and »stop« buttons which allows the user to browse through the ligand's predicted 3D poses one by one. This allows the user to visually examine interactions as lines between ligands and variant amino acids and determine potential gain or loss of interactions, allowing for estimation of the impact of a sequence variant on protein's function and ligand binding.
Sequence viewer
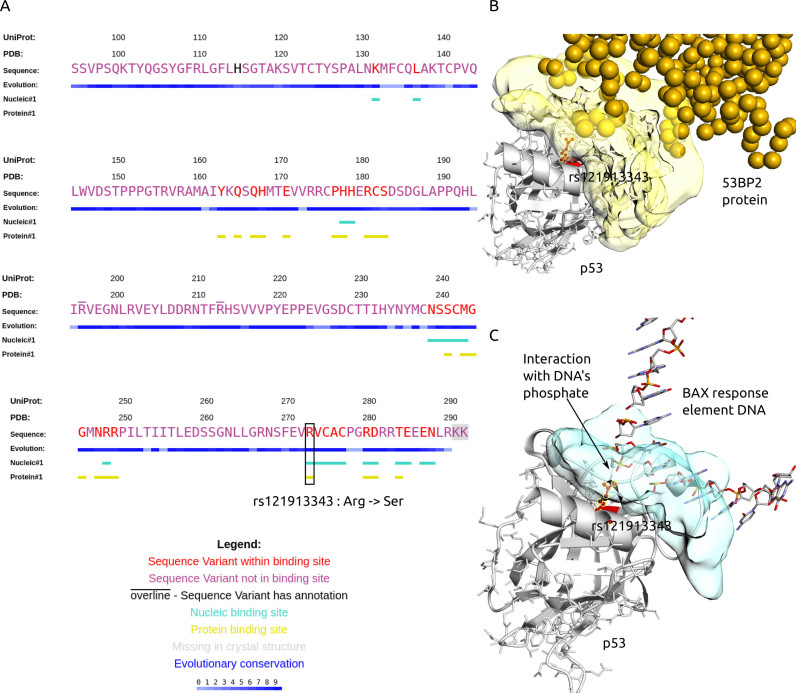
Sequence Viewer allows the user to see, as an alternative to the structural view, PDB protein sequences annotated with binding sites, sequence variants, and degrees of structural evolutionary conservation (Figure 3). The degrees of structural conservation, calculated from multiple structure alignments with ProBiS algorithm (18), often indicate the position of binding sites or other functionally important sites.
Figure 3.
Summary of GenProBiS results for p53 tumor suppressor protein (gene symbol: TP53; PDB and Chain ID: 1gzhC). (A) Sequence view of p53 with mapped nsSNPs, somatic mis-sense mutations and binding sites. Binding site mis-sense mutation rs121913343 Arg273Ser (red) is located in nucleic and protein–protein binding sites. (B–C) Structural view of p53's (gray cartoon) rs121913343 (red ball-and-sticks) interaction with (B) tumor suppressor p53-binding protein 2 (53BP2) ligand (yellow spheres), where each sphere represents one protein residue and protein binding site on p53 is yellow surface; (C) promoter of proapoptotic gene (Bax) ligand (CPK colored sticks), where the nucleic acid binding site on p53 is a blue surface.
Three-dimensional viewer
Most of the browser window is the 3D structural viewer, which initially displays the query protein as a cartoon model with one of the protein binding sites shown as the solvent accessible molecular surface. Mapped sequence variants are ball-and-stick models, variants that are outside the currently selected binding site are purple and binding site variants are red (Figure 2). On the right side is a draggable menu with the PDB and Chain ID of the query protein that allows different coloring schemes and styles to be applied to the query protein, display crystal waters, co-crystallized ligands and hydrogens, and allows the structure to be refocused in the center of the screen.
CASE STUDY 1: nsSNP AND SOMATIC MUTATION EFFECTS ON INHIBITOR BINDING
Indoleamine 2, 3-dioxygenase (IDO1) is an enzyme that catabolizes tryptophan and has been demonstrated to have an immunosuppressive role (28). It is a validated oncotarget and is thought to be involved in one of the possible mechanisms by which cancer cells evade immune response. Developed inhibitors of this enzyme, aside from binding to heme, form several key interactions with binding site amino acids, for example, Phe163, Phe226 and Arg231 (29). Using GenProBiS with the IDO1 query protein structure (4pk6A), we identified two of these amino acids, Phe163 and Arg231, to be polymorphic (red sticks, Figure 2B). To analyze the effects of these polymorphisms on inhibitor binding, we used the »Ligand Player« console (Figure 2B) to browse through all the available co-crystallized inhibitors (listed in the table in Figure 2A). We discuss the recently developed imidazothiazole derivative inhibitors with PDB's Ligand IDs PKJ and PKL (29):
rs764150078 (Phe163Ser) results in loss of favorable pi–pi interaction of the imidazothiazole ring with phenylanine and reduces binding of both PKJ and PKL inhibitors (Figure 2B).
rs774225205 (Arg231Cys) and rs745677091 (Arg231Leu and Arg231His) delete favorable electrostatic interactions of arginine with inhibitor PKJ at the entrance to the binding site cavity.
COSM187719 (Arg231Cys) is a somatic mutation that results in loss of electrostatic interactions with PKJ inhibitor and could lead to drug resistance during cancer therapy with this inhibitor (Figure 2B).
These polymorphisms are likely to result in reduced effectiveness of the inhibitors and should be considered in the design of future inhibitors and in their potential clinical usage.
CASE STUDY 2: SOMATIC MUTATION IN P53 LINKED TO GLIOBLASTOMA MULTIFORME
Glioblastoma multiforme is a most aggressive and malignant subtype of human brain tumor. Variant rs121913343 in the TP53 gene was found in tumor tissue of patients with glioblastoma and was linked to tumor growth (30). The TP53 gene encodes for the tumor suppressor protein p53, which plays an essential role in preventing cancer. Using the gene symbol TP53 as the query (the structure chosen in the »Conversion Tool« was 1gzhC), we observed that the mutation Arg273Ser corresponding to rs121913343 occurs in a nucleic acid binding site for BAX response element (Figure 3) (31). We postulate that the replacement of arginine by serine vitiates the salt bridge interaction of the arginine with DNA's phosphate group. This weakens the p53-DNA interaction and decreases the tumor suppression activity of p53. The importance of this finding could be experimentally tested by comparing the stability of the wild-type to that of the mutated p53–DNA complex.
CASE STUDY 3: INTERPRETATION OF GENOME-WIDE ASSOCIATION STUDIES
Serum concentration levels of intercellular adhesion molecule 1 (ICAM-1) have been associated with diverse conditions. In a genome-wide association study, several nsSNPs, including rs1799969, were associated with lower solubility of this protein in plasma (32). Using the rs1799969 as the query (the structure chosen in the »Conversion Tool« was 1p53B), we suggest that the decreased solubility may be due to this mutation disrupting glycosylation of this protein. Glycosylation has been shown to increase solubility of proteins (33) and indeed, rs1799969 which describes the change Gly241Arg occurs in the N-glycosylation site (binding site #3) on ICAM-1. The substituted arginine (UniProt location: 241; PDB residue ID: 214) could form a salt bridge with the nearby aspartate (UniProt location: 268; PDB residue ID: 241) belonging to the N-glycosylation sequon Asn-Asp-Ser, thereby changing its structure and preventing glycosylation of ICAM-1. This result offers an alternative explanation for the effect of this polymorphism on the solubility of ICAM-1, which was previously thought to be due to the weakened binding to integrin MAC-1 (34).
CONCLUSION
GenProBiS is a web server designed for detection and 3D visualization of sequence variants such as somatic mis-sense mutations and nsSNPs in protein binding sites. Binding sites and their ligands are predicted with no prior knowledge of binding sites, but based on detected local structural similarities in proteins and transposition of ligands between protein structures irrespective of protein folding. GenProBiS allows suggestion of functional effects of mutations on ligand binding and as such represents a key tool in both drug discovery and personalized medicine. The results of the GenProBiS web server could enable focused laboratory experiments based on targeted hypotheses in several research fields including human, veterinary medicine, animal and plant breeding.
FUNDING
The authors acknowledge the financial support from the Slovenian Research Agency (P1-0002 and P4-0220). The authors acknowledge the project (Computational tools development for modeling of pharmaceutically interesting molecules, J1-6743) was financially supported by the Slovenian Research Agency. Funding for open access charge: P1-0002, National Institute of Chemistry, Hajdrihova 19, SI-1000 Ljubljana, SLOVENIA.
Conflict of interest statement. None declared.
REFERENCES
- 1. den Dunnen J.T. Describing sequence variants using HGVS nomenclature. Methods Mol. Biol. 2017; 1492:243–251. [DOI] [PubMed] [Google Scholar]
- 2. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R.. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014; 42:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wishner B.C., Ward K.B., Lattman E.E., Love W.E.. Crystal structure of sickle-cell deoxyhemoglobin at 5 Å resolution. J. Mol. Biol. 1975; 98:179–194. [DOI] [PubMed] [Google Scholar]
- 5. David A., Razali R., Wass M.N., Sternberg M.J.E.. Protein–protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum. Mutat. 2012; 33:359–363. [DOI] [PubMed] [Google Scholar]
- 6. Kamburov A., Lawrence M.S., Polak P., Leshchiner I., Lage K., Golub T.R., Lander E.S., Getz G.. Comprehensive assessment of cancer mis-sense mutation clustering in protein structures. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5486–E5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao N., Han J.G., Shyu C.-R., Korkin D.. Determining effects of non-synonymous SNPs on protein-protein interactions using supervised and semi-supervised Learning. PLOS Comput. Biol. 2014; 10:e1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C.. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40:W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R.. A method and server for predicting damaging missense mutations. Nat. Methods. 2010; 7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hewett M., Oliver D.E., Rubin D.L., Easton K.L., Stuart J.M., Altman R.B., Klein T.E.. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002; 30:163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niknafs N., Kim D., Kim R., Diekhans M., Ryan M., Stenson P.D., Cooper D.N., Karchin R.. MuPIT interactive: webserver for mapping variant positions to annotated, interactive 3D structures. Hum. Genet. 2013; 132:1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang D., Song L., Singh V., Rao S., An L., Madhavan S.. SNP2Structure: a public and versatile resource for mapping and three-dimensional modeling of missense SNPs on human protein structures. Comput. Struct. Biotechnol. J. 2015; 13:514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu H.-C., Herrera Braga J., Fraternali F.. PinSnps: structural and functional analysis of SNPs in the context of protein interaction networks. Bioinformatics. 2016; 32:2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Solomon O., Kunik V., Simon A., Kol N., Barel O., Lev A., Amariglio N., Somech R., Rechavi G., Eyal E.. G23D: online tool for mapping and visualization of genomic variants on 3D protein structures. BMC Genomics. 2016; 17:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gress A., Ramensky V., Büch J., Keller A., Kalinina O.V.. StructMAn: annotation of single-nucleotide polymorphisms in the structural context. Nucleic Acids Res. 2016; 44:W463–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Konc J., Janežič D.. ProBiS-ligands: a web server for prediction of ligands by examination of protein binding sites. Nucleic Acids Res. 2014; 42:W215–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konc J., Janežič D.. ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment. Bioinformatics. 2010; 26:1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konc J., Janezic D.. An improved branch and bound algorithm for the maximum clique problem. MATCH Commun. Math. Comput. Chem. 2007; 58:569–590. [Google Scholar]
- 20. Štular T., Lešnik S., Rožman K., Schink J., Zdouc M., Ghysels A., Liu F., Aldrich C.C., Haupt V.J., Salentin S. et al. Discovery of mycobacterium tuberculosis InhA inhibitors by binding sites comparison and ligands prediction. J. Med. Chem. 2016; 59:11069–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. The UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kersey P.J., Allen J.E., Armean I., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Falin L.J., Grabmueller C. et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016; 44:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Velankar S., Dana J.M., Jacobsen J., van Ginkel G., Gane P.J., Luo J., Oldfield T.J., O’Donovan C., Martin M.-J., Kleywegt G.J.. SIFTS: structure integration with function, taxonomy and sequences resource. Nucleic Acids Res. 2013; 41:D483–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ankerst M., Breunig M.M., Kriegel H.-P., Sander J.. Davidson SB, Faloutsos C. OPTICS: ordering points to identify the clustering structure. Proceedings of the 1999 ACM SIGMOD International Conference on Management of Data. 1999; NY: ACM; 49–60. [Google Scholar]
- 26. Bekker G.-J., Nakamura H., Kinjo A.R.. Molmil: a molecular viewer for the PDB and beyond. J. Cheminform. 2016; 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L.. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004; 32:W280–W286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takamatsu M., Hirata A., Ohtaki H., Hoshi M., Hatano Y., Tomita H., Kuno T., Saito K., Hara A.. IDO1 plays an immunosuppressive role in 2, 4, 6-trinitrobenzene sulfate-induced colitis in mice. J. Immunol. 2013; 191:3057–3064. [DOI] [PubMed] [Google Scholar]
- 29. Tojo S., Kohno T., Tanaka T., Kamioka S., Ota Y., Ishii T., Kamimoto K., Asano S., Isobe Y.. Crystal structures and structure–activity relationships of imidazothiazole derivatives as IDO1 inhibitors. ACS Med. Chem. Lett. 2014; 5:1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Backes C., Harz C., Fischer U., Schmitt J., Ludwig N., Petersen B.-S., Mueller S.C., Kim Y.-J., Wolf N.M., Katus H.A. et al. New insights into the genetics of glioblastoma multiforme by familial exome sequencing. Oncotarget. 2014; 6:5918–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Y., Zhang X., Dantas Machado A.C., Ding Y., Chen Z., Qin P.Z., Rohs R., Chen L.. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res. 2013; 41:8368–8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paré G., Chasman D.I., Kellogg M., Zee R.Y.L., Rifai N., Badola S., Miletich J.P., Ridker P.M.. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6, 578 women. PLoS Genet. 2008; 4:e1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sinclair A.M., Elliott S.. Glycoengineering: The effect of glycosylation on the properties of therapeutic proteins. J. Pharm. Sci. 2005; 94:1626–1635. [DOI] [PubMed] [Google Scholar]
- 34. Ryan M., Diekhans M., Lien S., Liu Y., Karchin R.. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009; 25:1431–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]