Abstract
The secondary metabolism of bacteria, fungi and plants yields a vast number of bioactive substances. The constantly increasing amount of published genomic data provides the opportunity for an efficient identification of gene clusters by genome mining. Conversely, for many natural products with resolved structures, the encoding gene clusters have not been identified yet. Even though genome mining tools have become significantly more efficient in the identification of biosynthetic gene clusters, structural elucidation of the actual secondary metabolite is still challenging, especially due to as yet unpredictable post-modifications. Here, we introduce SeMPI, a web server providing a prediction and identification pipeline for natural products synthesized by polyketide synthases of type I modular. In order to limit the possible structures of PKS products and to include putative tailoring reactions, a structural comparison with annotated natural products was introduced. Furthermore, a benchmark was designed based on 40 gene clusters with annotated PKS products. The web server of the pipeline (SeMPI) is freely available at: http://www.pharmaceutical-bioinformatics.de/sempi.
INTRODUCTION
Polyketide synthases (PKS) are well known for the great variety of their bioactive products. They comprise numerous important antibacterial, anticancer, antifungal, antiviral, antiparasitic and several other significant substances in clinical use (1). Even though polyketides (PK) are a very diverse group of compounds, they are produced by similar synthesis pathways. PKS are multifunctional enzymes that can be divided into three types (2). While the products of type-II and type-III PKS are synthesized in a single reaction cavity, type-I PKS can be divided into structured modules that are passed during product formation.
Each of them consists of a substrate specific ketosynthase, possibly followed by a ketoreductase, dehydratase, enoyl reductase and/or methyltransferase that modify the newly attached PK-segment (3). This highly organized structure allows for prediction of resulting PK-chains based on genome clusters (4). However, the PK chain formation is often ensued by cyclization and additional modifications whose products are hardly predictable with currently available methods (5). The sustained scientific interest in these compounds has led to a remarkable number of identified PK, but also to an increasing rediscovery rate (6). With rapidly decreasing costs, genomic screening has become an important method in the search for new natural drugs. For example, the identification and reactivation of silent gene clusters has resulted in several new discoveries (7). However, activating these clusters is an elaborate task, and predicting the resulting products as precisely as possible supports the determination of the most interesting candidates in advance (8).
Based on sophisticated methods for identification of known gene families and related clusters, the identification of relevant genes for biosynthesis and even multigene modules has significantly improved in recent years (9–11). Even gene clusters of so far unknown classes can be predicted accurately by probabilistic approaches such as ClusterFinder (6). However, the prediction of the resulting secondary metabolites based on novel genomic sequences is still a major challenge.
One of the best studied PKS synthesis routes is the modular type I subclass, due to its well-structured building-block composition (12). The few available tools for secondary structure prediction such as NP.searcher (13), PRISM (14) and antiSMASH 3.0 (9) provide accurate results if the subjected gene cluster has a significant sequence similarity to annotated clusters with known products. However, prediction quality decreases significantly if sequence similarity is low.
SeMPI follows an ab-initio approach and predicts the correct order of the substrates which constitute the secondary metabolite. The predicted PK-structure represents the initial PKS biosynthesis without any cyclization or further post-modifications. In order to limit the possible structures of PKS products, we developed an automated workflow which compares the predicted polyketide chain to annotated natural products in a large database of natural products (StreptomeDB 2.0 (1)). The pipeline reversely transforms annotated metabolites to their initial biosynthesis products without post-modifications. This allows for adequate comparison of predicted PK-chain and reported compounds. As a consequence, the determination of the product structure based on genome information will be strongly supported.
MATERIALS AND METHODS
Database pipeline
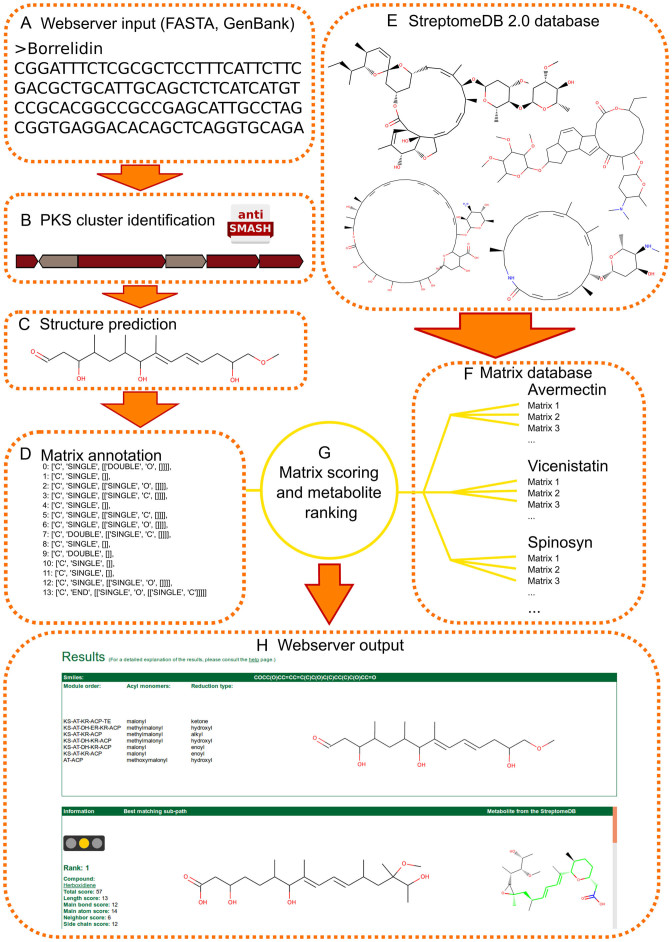
The prediction pipeline consists of two parts, the prediction of the PK-chain (which is performed on the fly for a submitted gene cluster), and the identification of putative molecule scaffolds that fit to the predicted PK-chain. For the comparison, possible paths through natural products annotated in the StreptomeDB 2.0 have been pre-processed. The comparisons are scored according to the maximum common sub-paths within each molecule. A flowchart of the pipeline is shown in Figure 1.
Figure 1.
Flowchart of the SeMPI pipeline. (A) Data can be submitted as FASTA, GenBank files, or raw DNA code. (B) Gene cluster is identified with antiSMASH 3.0. (C) Structure of the carbon chain is predicted. (D) Carbon chain is translated into the matrix annotation. (E) StreptomeDB 2.0 provides the natural compounds for the path similarity search. (F) A set of matrices was computed for each compound. (G) The matrix from the prediction is compared with each matrix set of the database. (H) The web server output displays the prediction and domain information as well as the 10 best matching compounds.
Polyketide chain prediction
Initially, the submitted DNA sequence is screened for gene clusters of PKS type I using antiSMASH 3.0 (9). Identified gene clusters are screened for domain signatures of modular KS and lack of other types of PKS or NRPS signatures. Exact starting and ending positions of the domains in the gene cluster are then identified by a sequence similarity search using Hidden Markov Model profiles specific for modular PKS type I. HMM profiles were built from multiple sequence alignments generated with Clustal Omega (15) and processed with HMMER 3 (16). Sequences of experimentally characterized domains were retrieved from DoBISCUIT (17) (Supplementary Table S1).
Based on this method, the following domains are classified: ketoacyl synthase (KS), acetyltransferase (AT), acyl carrier protein (ACP), keto reductase (KR), dehydratase (DH), enoylreductase (ER) and thioesterase (TE). KSQ domains are distinguished from those KS domains in which the reactive cysteine has been mutated to glutamine (3).
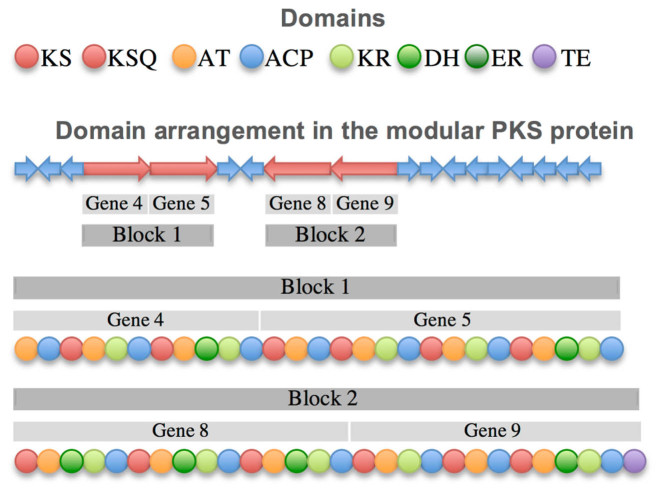
The different genes that a PKS may contain are predicted by using GeneMark (18). Domains are assigned to the related genes based on their position in the sequence. The start codon is selected from its distance to the initial codon of the first KS domain, which is considered to be 30–40 residues, the length of the N-terminal docking domain (19). The arrangement of the genes is then predicted by the following method: Contiguous genes are clustered in blocks. The first block is characterized by the presence of a KSQ domain or an AT-ACP loading module, and the final block by containing a TE domain. The order of the non-flanking blocks is determined by a pair of specific interactions between the docking domains. N-terminal docking domains are located between a start codon and the first KS domain of a block, whereas C-terminal docking domains are located between the last ACP domain of a block and the stop codon. The residues involved in the interaction between two docking domains are derived from the molecular structure of the docking domains of the modular PKS 6-deoxyerythronolide B synthase (DEBS) (PDB: 1PZR (19)). Interacting residues of homologous sequences are defined by related positions in a sequence alignment performed with Clustal Omega. All permutations of block arrangements are calculated, and possible interactions between the docking domains are classified as favorable, neutral, or unfavorable according to the interacting residue pairs (5). The permutation with the highest number of favorable interactions indicates the predicted arrangement of the blocks. If multiple combinations share the best score, the order of the genes in the genome is favored. The domain arrangement is illustrated in Figure 2.
Figure 2.
The prediction algorithm preserves contiguous genes in the PKS and collects them in blocks. These blocks are combined according to the most favored interaction of their docking domains. The final domain arrangement is translated into the PK-chain.
Substrate specificity of the AT domains is predicted based on position probability matrices (PPMs) of substrate specific AT domains (Supplementary Formula S2). A PPM has been calculated for each substrate (malonyl, methylmalonyl, ethylmalonyl and methoxymalonyl) from a multiple sequence alignment of experimentally characterized substrate specific AT domains retrieved from DoBISCUIT. The active site residues are defined by a sequence alignment with a structurally resolved AT domain of DEBS (PDB: 2HG4 (20)). Five residues were considered as interacting with the substrate: Q643, L671, Y742, S744 and L795. The best scored substrate specific PPM for a subjected AT domain determines the selection of the next building-block in the elongation of the PK chain.
The ß-keto group of the acyl-monomer is subsequently modified to a hydroxyl, methine, or methylene according to the presence of a KR, DH or ER domains in the following module.
Generation of polyketide chains from described natural products
Whereas the initial biosynthesis steps of modular PKS I metabolites are well predictable due to their modular chain elongation pattern, post-modifications like cyclization, phosphorylation and glycosylation are difficult to determine. However, the initially synthesized carbon chain is normally conserved in the final metabolite (3,12). To identify this primary PK-chain in already described natural products we developed a new algorithm. After the termination of the modular biosynthesis, the last link of the carbon chain is released from the thioesterase, leaving a carboxylic group. In tailoring reactions, this group can be involved in cyclization, methylation, amination or reduction steps (21,22).
Acetyl-, carboxyl-, lactone- and amide-groups were identified with substructure searches, and used as starting units for the path calculation. The longest possible paths to an ending carbon were then calculated. These paths also include side chains up to the second atom to allow a more detailed path comparison. Sulfur and oxygen atoms, as well as peptide bonds lead to a termination of a path, as the initially synthesized chain is not expected to include non-carbon atoms in the main path. This approach automatically cleaves glycosides, phosphates, amino acids and other post-modifications.
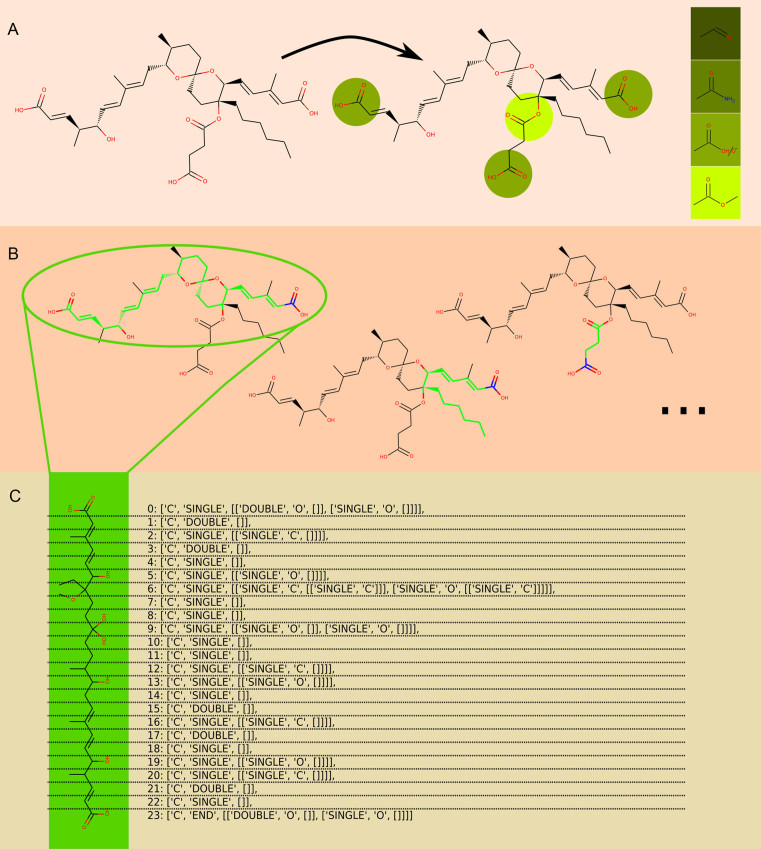
All calculated paths of a molecule are stored in a format referred as matrix, which allows uniform and fast comparison. Within a matrix, each element of a path is numbered beginning with the starter unit, and is described by atom, bond to the next element, and neighbor atoms/bonds. A scheme of the path generation is shown in Figure 3. Matrix sets were calculated for all molecules in StreptomeDB 2.0 and stored in a database.
Figure 3.
PK-chain generation shown for reveromycin. (A) Possible starter units of the path are identified in the molecule. (B) Unique atom-chains are calculated within the molecule (green), beginning with the starter units (blue). (C) The path are extracted and transformed into the matrix annotation. The dotted lines show how each segment is translated.
Path comparison
A matrix annotation of the predicted molecule is created via the same algorithm as mentioned above. The matrices of compounds from the database are step-wise compared to the matrix of the prediction, assigning points for each matching property. The scoring system is explained in detail in Supplementary Table S3. Database compounds are then ranked based on the achieved points using the standard competition ranking.
Website
The website allows the uploading of genomes or gene clusters as FASTA (*.fna, *.fasta) or GenBank (*.gbk) files, but also the pasting of raw sequence data. Results are generated for modular PKS type I clusters. An error report provides feedback if a cluster cannot be processed. The result page for each cluster comprises the predicted PK-chain, including information about module order within the PKS, building blocks and reduction types. The 10 best matches from the path comparison are listed below the predicted PK-chain. The rank and total score are displayed. Sub-scores give more details about the similarity with the predicted chain. A traffic light provides a quick visual evaluation of the matches. A red light shows a score <50, yellow 50–100 and green >100. The corresponding natural compounds from the database are shown next to their matched path, including a link to their entry in StreptomeDB 2.0, allowing for a more thorough investigation.
Implementation
The entire pipeline is written in python, including the web framework, which is based on the Django package (https://djangoproject.com). Molecular operations are performed using the rdkit module (http://www.rdkit.org), which allows for very fast calculations due to its C++ core data structure. The path algorithm is based on rdkit and is itself written as a python module. By introducing new classes which focus on PK specific molecular operations, this project provides further functionalities, which can be easily extended due to the object-oriented code. The algorithm for the path extraction can be downloaded from GitHub: (https://github.com/paulzierep/flp). A PostgreSQL database is connected via the psycopg2 wrapper for database access. The molecules in the matrix database can be complemented with more collections of natural compounds from other sources, which will give the SeMPI pipeline more comprehensiveness with future updates.
Benchmark design
A test dataset of gene clusters with already annotated products was collected from MiBIG (23). In order to establish a conclusive comparison with antiSMASH 3.0, the benchmark was adjusted accordingly. Certain predictions of antiSMASH 3.0 did not contain a starting unit that could be identified by the path algorithm. These molecules were excluded from the test dataset. Additionally, antiSMASH 3.0 can predict nitrogens as part of the main chain. This is not the case for the SeMPI software, due to its focus on pure PKS type I modules. In order to increase the comparability of the results, we included nitrogen heteroatoms as a possible element of the path algorithm. The dataset comprised 40 PKSs type I modular. Their products were predicted using SeMPI as well as antiSMASH 3.0. For each prediction, a ranking was calculated using the above described path algorithm. A modified competition ranking was applied, where in the case of a set of equally ranked compounds, the worst rank was assigned to all the compounds of the set. Based on the ranking of the actual product of the gene cluster, the true positive rates (TPRs) and false positive rates (FPRs) were calculated and plotted in a receiver operating characteristic curve (ROC-curve).
Additionally, the rankings were performed with all compounds from StreptomeDB 2.0 to illustrate the individual performances in a big dataset.
RESULTS AND DISCUSSION
The evaluation of structure prediction tools depends on the algorithm used to measure structural similarity and the minimal threshold which is required to consider a molecule from the test dataset as putative match to a subjected gene cluster. The evaluation by using a ROC curve based on ranking has the advantage of being independent of specific thresholds (24).
The structure prediction algorithm implemented in antiSMASH 3.0 was chosen as a tool for comparison to SeMPI, as both applications predict exactly one carbon chain for each submitted gene cluster as distinguished from PRISM, that gives the choice between multiple possible predictions or NP.searcher that supplies simulated tailoring reaction in some cases.
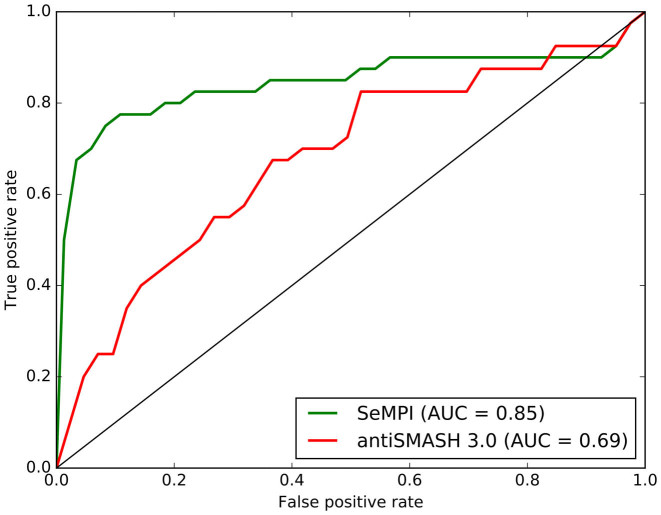
Based on the 40 gene clusters from the test dataset, SeMPI could reach an AUC-value of 0.85, opposed to antiSMASH 3.0 with an AUC-value of 0.69 (Figure 4). Obviously, we could improve the predictive power for the products of modular PKS type I gene clusters significantly.
Figure 4.
ROC curve for a test dataset of 40 genome clusters with annotated products. The AUC values for SeMPI and antiSMASH 3.0 are given in the legend. The diagonal line would indicate a random ranking.
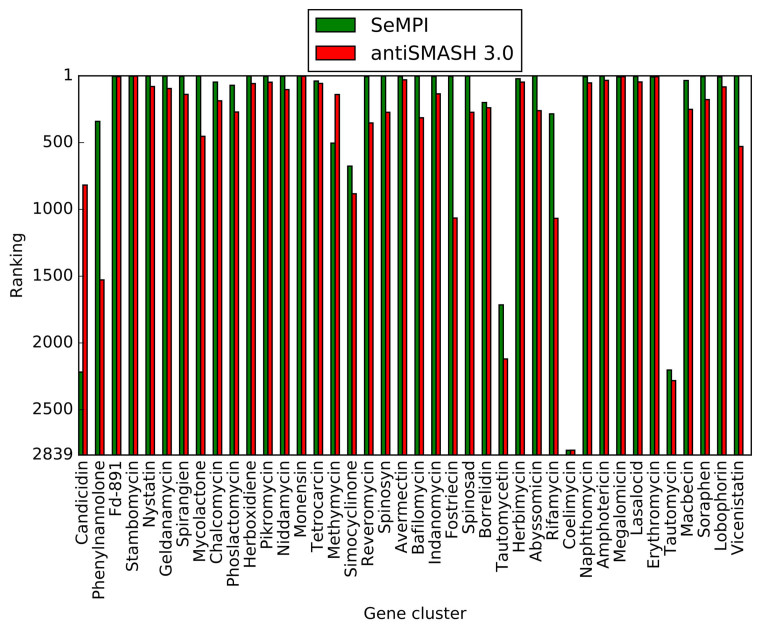
In the individual ranking of the gene cluster products among natural products from the StreptomeDB 2.0, SeMPI could rank 27 actual gene cluster products (antiSMASH 3.0: 5) within the first ten of 2839 possible ranks (Figure 5). This demonstrates the efficiency of the algorithm to detect the correct paths in a large number of molecules.
Figure 5.
Bar chart representation of the individual ranking for the test dataset, matched with all the suitable compounds from the StreptomeDB 2.0 (2839).
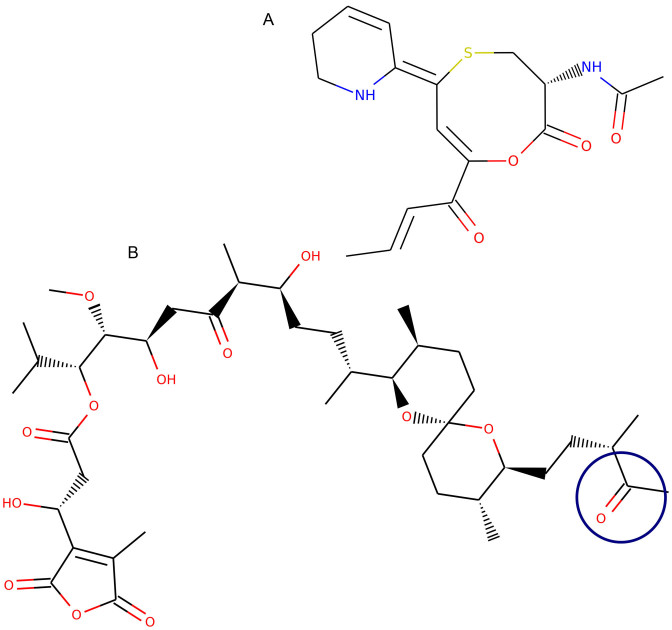
In the case of a poor ranking we could work out two main reasons. If the prediction was made for a gene cluster that produces a compound with multiple tailoring reactions, the path algorithm reaches its limitations. This explains for example the very weak ranking of coelimycin, where at least 10 tailoring reactions are proposed after the modular biosynthesis (25) (Figure 6A). Similar difficulties occur if the starting unit for the path algorithm is wrongly identified, due to an unexpected tailoring reaction of the unit. This could be observed, for example, for tautomycin, where the starting unit reacted to a ketone (26), which is currently not identified by the path algorithm (Figure 6B).
Figure 6.
Examples for gene cluster products which were difficult to predict. (A) The initial polyketide chain of coelimycin is modified by multiple tailoring reactions, resulting in a strongly modified scaffold. (B) The path algorithm identified the wrong starting unit for tautomycin, the correct starting unit is circled in blue.
A ranking within the first 10 possible ranks would lead to an entry in the shown SeMPI result page for the given gene cluster. The rationale of presenting several molecules is, that compounds having the same rank can in many cases be explained with the occurrence of multiple derivatives of the same natural product in the database. These derivatives differ only slightly among themselves for example in their stereochemistry or the type of glycosidic residue. It is desired to rank those compounds on the same position, as this allows a very detailed investigation of all possible products of a submitted gene cluster.
Even though SeMPI does rank natural products for any given prediction, the resulting ten predictions do not always match, as was already shown by the benchmark. Especially for novel gene clusters, the metabolite might not be annotated in the database yet. In order to give the user a basic estimation of the quality of the shown molecules, the traffic light was introduced. A green light indicates a very good match. In this case, the chances are high that the predicted metabolite of the submitted gene cluster has at least a scaffold that is very similar to the proposed molecules. Molecules with a yellow traffic light can still lead to a basic understanding of the overall structure of the prediction, especially when the 10 best matches show similar features. But it is unlikely that the exact secondary metabolite was identified in the resulting list. The interpretation of a red traffic light, allows two possibilities. If the predicted carbon chain is very short, there are only few distinctive features, which restricts the comparison algorithm significantly. If a long carbon chain is predicted, a red light would indicate that this compound is comprised of a scaffold which is not described in the database, but could also be the first hint for a newly discovered PK.
In future, updates we will extend SeMPI by implementing additional database search features, for example by increasing the library of possible starting units for the path algorithm. Another improvement will be the integration of other databases for natural compounds in order to apply a maximum number of putative annotated molecules that come into question for subjected gene clusters.
CONCLUSIONS
The SeMPI web server successfully combines and complements available polyketide prediction methods with a unique database matching algorithm. Whereas other tools focus on the simulation of possible tailoring reaction in order increase the information value about the predicted metabolites (PRISM, NP.searcher), the SeMPI path algorithm tries to identify putative patterns derived from already annotated secondary metabolites. Both approaches do not completely overcome the difficulty in predicting post-modifications based on genome cluster mining. However, the alternative approach of SeMPI provides a new insights into putative molecule structures, and will help researchers to understand the syntheses steps of the gene clusters much better. With the increasing number of sequenced genomes, SeMPI will enable researchers to identify promising gene clusters more efficiently, but also prevent them from investing great efforts in structure determination of a cluster product although it has already been described in literature.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Simon Nicklaus, Michael Kaiser and Lea Purschke for assistance in testing and refinement of the prediction pipeline. Furthermore, we would like to thank Robert A. Burrows for carefully reading the manuscript. Lastly, we want to thank all users who contributed to the development of SeMPI by giving us helpful suggestions and critical feedback.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German National Research Foundation [DFG, Research Training Group 1976].
Conflict of interest statement. None declared.
REFERENCES
- 1. Klementz D., Doring K., Lucas X., Telukunta K.K., Erxleben A., Deubel D., Erber A., Santillana I., Thomas O.S., Bechthold A. et al. . StreptomeDB 2.0–an extended resource of natural products produced by streptomycetes. Nucleic Acids Res. 2016; 44:D509–D514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Lanen S.G., Shen B.. Advances in polyketide synthase structure and function. Curr. Opin. Drug Discovery Dev. 2008; 11:186–195. [PubMed] [Google Scholar]
- 3. Keatinge-Clay A.T. The structures of type I polyketide synthases. Nat. Prod. Rep. 2012; 29:1050–1073. [DOI] [PubMed] [Google Scholar]
- 4. Hopwood D.A. Complex Enzymes in Microbial Natural Product Biosynthesis, Part B: Polyketides, Aminocoumarins and Carbohydrates. 2009; 1st edn, San Diego: Academic Press Inc. [DOI] [PubMed] [Google Scholar]
- 5. Yadav G., Gokhale R.S., Mohanty D.. Towards prediction of metabolic products of polyketide synthases: an In Silico Analysis. PLoS Comput. Biol. 2009; 5:e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomes E.S., Schuch V., de Macedo Lemos E.G.. Biotechnology of polyketides: new breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Braz. J. Microbiol. 2013; 44:1007–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ochi K., Hosaka T.. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013; 97:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu X.M., Hackl S., Thaker M.N., Kalan L., Weber C., Urgast D.S., Krupp E.M., Brewer A., Vanner S., Szawiola A. et al. . Biosynthesis of the fluorinated natural product nucleocidin in Streptomyces calvus is dependent on the bldA-Specified Leu-tRNA(UUA) molecule. Chembiochem. 2015; 16:2498–2506. [DOI] [PubMed] [Google Scholar]
- 9. Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Muller R., Wohlleben W. et al. . antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015; 43:W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Medema M.H., Takano E., Breitling R.. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 2013; 30:1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dutta S., Whicher J.R., Hansen D.A., Hale W.A., Chemler J.A., Congdon G.R., Narayan A.R., Håkansson K., Sherman D.H., Smith J.L. et al. . Structure of a modular polyketide synthase. Nature. 2014; 510:512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li M.H., Ung P.M., Zajkowski J., Garneau-Tsodikova S., Sherman D.H.. Automated genome mining for natural products. BMC Bioinf. 2009; 10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L., Wyatt M.A., Magarvey N.A.. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 2015; 43:9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sievers F., Higgins D.G.. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014; 1079:105–116. [DOI] [PubMed] [Google Scholar]
- 16. Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R.. HMMER web server: 2015 update. Nucleic Acids Res. 2015; 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ichikawa N., Sasagawa M., Yamamoto M., Komaki H., Yoshida Y., Yamazaki S., Fujita N.. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013; 41:D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Besemer J., Borodovsky M.. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005; 33:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Broadhurst R.W., Nietlispach D., Wheatcroft M.P., Leadlay P.F., Weissman K.J.. The structure of docking domains in modular polyketide synthases. Chem. Biol. 2003; 10:723–731. [DOI] [PubMed] [Google Scholar]
- 20. Tang Y., Kim C.Y., Mathews II, Cane D.E., Khosla C.. The 2.7-Å crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11124–11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rix U., Fischer C., Remsing L.L., Rohr J.. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 2002; 19:542–580. [DOI] [PubMed] [Google Scholar]
- 22. Olano C., Mendez C., Salas J.A.. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat. Prod. Rep. 2010; 27:571–616. [DOI] [PubMed] [Google Scholar]
- 23. Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. et al. . Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015; 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greiner M., Pfeiffer D., Smith R.D.. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 2000; 45:23–41. [DOI] [PubMed] [Google Scholar]
- 25. Gomez-Escribano J.P., Song L., Fox D.J., Yeo V., Bibb M.J., Challis G.L.. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 2012; 3:2716–2720. [Google Scholar]
- 26. Li W., Ju J., Rajski S.R., Osada H., Shen B.. Characterization of the tautomycin biosynthetic gene cluster from Streptomyces spiroverticillatus unveiling new insights into dialkylmaleic anhydride and polyketide biosynthesis. J. Biol. Chem. 2008; 283:28607–28617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.