Abstract
Background.
Biomarkers can improve clinical trial efficiency, but designing and interpreting biomarker-driven trials require knowledge of relationships among biomarkers, clinical covariates, and endpoints. We investigated these relationships across genomic subgroups of glioblastoma (GBM) within our institution (DF/BWCC), validated results in The Cancer Genome Atlas (TCGA), and demonstrated potential impacts on clinical trial design and interpretation.
Methods.
We identified genotyped patients at DF/BWCC, and clinical associations across 4 common GBM genomic biomarker groups were compared along with overall survival (OS), progression-free survival (PFS), and survival post-progression (SPP). Significant associations were validated in TCGA. Biomarker-based clinical trials were simulated using various assumptions.
Results.
Epidermal growth factor receptor (EGFR)(+) and p53(−) subgroups were more likely isocitrate dehydrogenase (IDH) wild-type. Phosphatidylinositol-3 kinase (PI3K)(+) patients were older, and patients with O6-DNA methylguanine-methyltransferase (MGMT)–promoter methylation were more often female. OS, PFS, and SPP were all longer for IDH mutant and MGMT methylated patients, but there was no independent prognostic value for other genomic subgroups. PI3K(+) patients had shorter PFS among IDH wild-type tumors, however, and no DF/BWCC long-term survivors were either EGFR(+) (0% vs 7%, P = .014) or p53(−) (0% vs 10%, P = .005). The degree of biomarker overlap impacted the efficiency of Bayesian-adaptive clinical trials, while PFS and OS distribution variation had less impact. Biomarker frequency was proportionally associated with sample size in all designs.
Conclusions.
We identified several associations between GBM genomic subgroups and clinical or molecular prognostic covariates and validated known prognostic factors in all survival periods. These results are important for biomarker-based trial design and interpretation of biomarker-only and nonrandomized trials.
Keywords: biomarkers, clinical trial design, glioblastoma
Importance of the study
GBM and other cancers frequently have genetic aberrations in canonical signaling pathways that are currently being targeted in clinical trials. Genomic biomarkers offer the potential for personalized medicine by identifying patient populations that may be more likely to respond to a therapeutic agent targeting an associated pathway. Effective design and interpretation of biomarker-driven clinical trials require an understanding of the frequency of biomarker subgroups, their overlap, and the intrinsic association with various clinical trial endpoints, however. Here we demonstrate the value of large-scale genomic/clinical data correlation and identify several associations between genomic subgroups in GBM and clinical and molecular prognostic factors that will be useful for clinical trial design and interpretation. We then show how the data pertaining to biomarker frequency, overlap, and endpoint relationships impact the design and simulation of Bayesian clinical trials and discuss the impact on non-adaptive clinical trial design.
Biomarker-driven studies, in which experimental agents are tested within specific genomic or molecular subpopulations, offer a promising approach to trial design to improve efficiency and deliver precision medicine.1 There are many different ways to design such trials—from tumor-specific trials like the biomarker-selected, randomized, controlled Lung Master Protocol (LUNG-MAP) in squamous cell lung cancer2 and the adaptively randomized I-SPY 2 trial in breast cancer,3to the National Cancer Institute’s cross-tumor “basket” trial Molecular Analysis for Therapy Choice (NCI-MATCH).4 The design and interpretation of such trials benefit from biomarker-specific data regarding the relative frequency and degree of overlap between biomarker subgroups, a priori prognostic capacity of subgroups, and the relationship between endpoints for each subgroup. These data can impact decisions regarding eligibility criteria, endpoints, accrual estimates, and selection of appropriate controls. Additionally, this information is essential to generate assumptions for simulations of Bayesian clinical trials to elucidate operating characteristics. The potential to abstract foundational data for biomarker-driven clinical trial design is an underappreciated value of large clinically annotated datasets such as The Cancer Genome Atlas (TCGA),5 as the relative frequency and natural history of genetically defined subgroups with respect to various trial endpoints may not be well characterized.
Despite over 1400 published trials and an increasing number of potential therapies, glioblastoma (GBM) continues to confer poor outcomes with limited therapeutic progress. Work by TCGA and others has identified 3 canonical pathways with recurrent aberrations, including receptor tyrosine kinase signaling and the p53 and retinoblastoma tumor suppressor pathways.5–7 There is much interest in targeting these pathways and the ability to screen for multiple biomarkers with genomic sequencing makes platform trials that test multiple therapies under one master protocol attractive. In fact, the NCI’s Brain Malignancy Steering Committee Targeted Therapies Working Group recommended the development of a multi-arm adaptively randomized clinical trial to efficiently test targeted agents with associated genomic biomarkers,8 and such a trial has recently opened in response: the INdividualized Screening trial for Innovative Glioblastoma Therapy (INSIGhT; NCT02977780). Additionally, there are other biomarker-selected trials that match therapies to tumors with specific aberrations in nonrandomized, uncontrolled studies. Designing biomarker-driven trials in general and interpretation of uncontrolled, single-arm trials, however, may be complicated by the association of specific biomarker subgroup tumor biology and natural history.8,9 This may impact overall survival (OS) and progression-free survival (PFS) times or influence the association between endpoints. The relationship between PFS and OS may differ between therapeutic classes,10 and this potential may exist among biomarker-defined classes.11 To better inform our design choices and simulations for INSIGhT and create a resource to interpret single-arm biomarker-based trial results, we collected and analyzed clinical and genomic data from patients with newly diagnosed GBM from Dana-Farber/Brigham and Women’s Cancer Center (DF/BWCC) for associations with relevant clinical covariates, known molecular prognostic factors, and potential clinical trial endpoints and validated significant associations in GBM data from TCGA. Relevant biomarker categories were prospectively hypothesized based on potential interactions with intended agents targeted to epidermal growth factor receptor (EGFR), phosphatidylinositol-3 kinase (PI3K), p53, and cyclin-dependent kinase (CDK) pathways. Furthermore, we characterized the frequency and degree of overlap between biomarker categories to be included on INSIGhT and demonstrated how variations in those factors and survival times would impact clinical trial design through simulation.
Materials and Methods
Datasets and Genomic Assays
The DF/BWCC cohort consisted of patients ≥18 years old with a newly diagnosed GBM and clinical molecular profiling.12 Each patient underwent at least 1 of 3 genotyping assays for genomic profiling: OncoCopy,13 a multiplexed copy number assay based on whole genome array comparative genomic hybridization; OncoMap,14 a targeted and multiplexed mass spectrometry–based mutation genotyping (Sequenom) covering 471 mutations from 41 cancer genes (version 4); and OncoPanel,15 a targeted exome sequencing platform covering 275 cancer genes and 91 select introns across 30 genes to detect somatic mutations, copy number alterations, and structural rearrangements. We disregarded mutations with low (<5%) allelic fraction, with <20 reads of mutant allele on OncoPanel, previously unreported in the COSMIC database, and previously known to be single nucleotide polymorphisms to reduce classification error. OncoCopy data from clinical testing reports were obtained from the medical record under consented and waiver of consent research protocols approved by the Dana-Farber Harvard Cancer Center (DF/HCC) institutional review board (IRB). Somatic mutational profiling was performed with consent for DF/BWCC Profile clinical research studies approved by the DF/HCC IRB. All tests were performed within the Cytogenetics Division (OncoCopy) and Molecular Diagnostics (OncoMap) Divisions of the Brigham and Women’s Hospital Center for Advanced Molecular Diagnostics, a Clinical Laboratory Improvement Amendment (CLIA)–certified laboratory environment. Central histopathologic review was performed on all genotyped tumor specimens at DF/BWCC using standard World Health Organization criteria.16 O6-DNA methylguanine-methyltransferase (MGMT)–promoter methylation status in this cohort was generally assessed using methylation-specific polymerase chain reaction (MS-PCR). Clinical, demographic, pathologic, and follow-up data were collected retrospectively from the medical record following approval from the DF/HCC IRB.
Significant findings from our institutional cohort were validated in data generated by TCGA Research Network available on the TCGA website5,17 and cBioPortal.18 The provisional TCGA GBM dataset was queried for levels 1 and 2 clinical and genomic data related to DNA copy number (HG-CGH-244A), whole-genome next-generation sequencing (Illumina®), conventional sequencing (Sanger), and DNA methylation arrays (Illumina Infinium Human Methylation-27 [HM-27] and HM-450), as previously described.17 Clinically relevant MGMT-promoter methylation status was estimated from cytosine-phosphate-guanine islands using a logistic regression of 2 clinically relevant methylation probes, as previously described.19
Biomarker Subgroups
We prespecified biomarker categories on the basis of genetic aberrations for subsequent analyses based on the Targeted Therapies Working Group recommendations:
EGFR: (+) defined as patients with EGFR amplification or mutation;
PI3K: (+) defined as patients with PIK3CA mutation/amplification, PIK3R1 mutation, AKT3 amplification, PIK3C2B >1 copy gain, or PTEN dual loss through either homozygous deletion or deletion plus mutation;
p53: (+) defined as patients with TP53 mutation;
CDK: (+) defined as patients with RB1 wild-type (WT) and CDK4 amplification, CDK6 amplification, or CDKN2A >1 copy loss.
Assignment to biomarker categories was contingent on sufficient data from available molecular analyses. For example, p53(+) could be assessed with either OncoPanel or OncoMap if a relevant mutation was present. P53(−) could only be reliably determined from OncoPanel, since the entire coding region was sequenced, however. Similarly, PI3K(+) could be determined based on OncoMap or OncoPanel if an activating mutation was present, but positivity determined based on copy number and mutation required overlap with OncoCopy.
Statistical Analysis
Patients with isocitrate dehydrogenase (IDH)1 and 2 mutations and/or 1p/19q codeletions were excluded from biomarker subgroup analyses based on the intended eligibility criteria of INSIGhT.8 We evaluated associations between clinical factors and biomarker subgroups using Fisher’s exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Survival outcomes, including PFS, survival post-progression (SPP), and OS, were estimated for the genomic biomarker, MGMT, and IDH subgroups using the Kaplan–Meier method and compared using the log-rank test. PFS/OS ratio was calculated for patients who had a progression and death. SPP was calculated from time of progression for those patients who had a progression prior to death and censored identically to OS. Progression was defined independently by TCGA and retrospectively by the DF/BWCC cohort through clinical note assessments integrating imaging and clinical status. Pearson correlation coefficients were used to characterize the relationship between PFS, an auxiliary endpoint, and OS to assess for differences in the natural history of disease across biomarker groups among patients who achieved death and progression.
Univariable and multivariable Cox regressions were used to identify clinical factors independently predicting for OS. We then created new models for each biomarker category with significant clinical variables to assess whether biomarker groups independently predicted for OS. All P-values are 2-sided, and analyses were performed using RStudio (version 0.98.1028) running R (version 3.1.0)20 with the survival package.21
To assess the impact of pre-trial genomic biomarker data on clinical trial planning, we assumed various scenarios related to biomarker frequency, overlap, and endpoint distributions into early clinical trial simulations for INSIGhT, a multi-arm, Bayesian adaptively randomized clinical trial currently in development. For the purposes of these comparisons, we assumed the 3 experimental arms above compared against a control arm, with 1 arm showing survival benefit compared with control.
Results
Baseline Patient Characteristics and Frequency of Biomarkers
The DF/BWCC cohort consisted of 265 patients with a median follow-up of 15.4 months overall and 16.8 months among survivors (range: 0.2–197.3 mo). OncoMap data were available for 78 patients, OncoPanel for an additional 157 (no overlap), and OncoCopy for 157 patients (90 patient overlaps with OncoPanel, 37 patient overlaps with OncoMap). Median age was 60 years, 54% were male, and median KPS was 80. MGMT-promoter status was methylated in 95 patients (36%), unmethylated in 82 (31%), and untested in 88 (33%). IDH1/2 were WT in 234 (88%), mutant in 28 (11%), and untested in 3 (1%).
The provisional TCGA dataset consisted of 549 patients with primary GBM diagnosed between 1988 and 2013 with a median follow up of 11.1 months overall and 6.7 months among survivors (range: 0.1–127.5 mo). Overall, median age was 60 years, 61% were male, and median KPS was 80. MGMT-promoter status was methylated in 170 patients (31%), unmethylated in 200 (36%), and unknown in 179 (33%). IDH1/2 were WT in 526 (96%), mutant in 23 (4%).
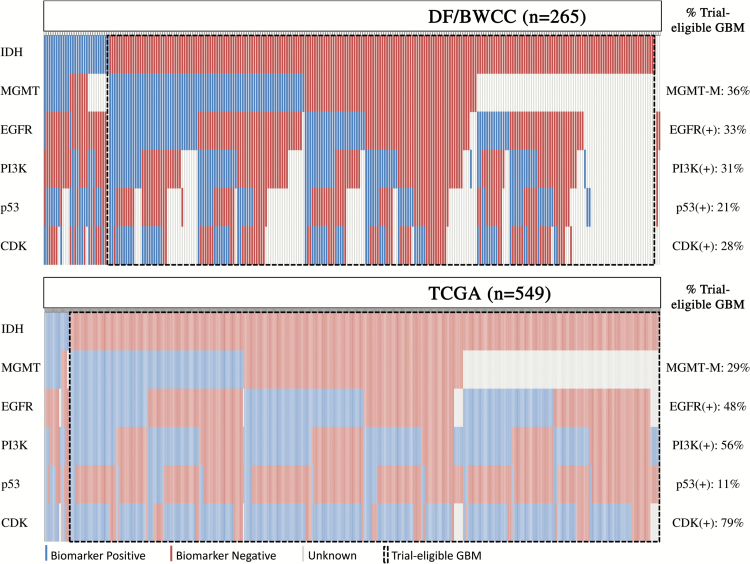
Fig. 1 illustrates the distribution of biomarker signatures based on IDH1/2 mutation, MGMT-promoter methylation, EGFR, PI3K, p53, and CDK group statuses. Among trial-eligible patients, considered as those without IDH1/2 mutation, p53(+) subclass occurred least commonly, although the 4 primary biomarker groups were otherwise fairly balanced in size. EGFR(+) and CDK(+) seldom occurred in isolation and were more often accompanied by inclusion in other biomarker(+) groups. Specifically, among trial-eligible patients in the DF/BWCC cohort, 33% of patients were EGFR(+) (n = 76), 31% were PI3K(+) (n = 73), 21% were p53(+) (n = 50), and 28% were CDK(+) (n = 66); in TCGA, 48% were EGFR(+) (n = 255), 56% were PI3K(+) (n = 293), 11% were p53(+) (n = 60), and 79% were CDK(+) (n = 413).
Fig. 1.
Biomarker status by individual in the DF/BWCC cohorts and TCGA cohorts. Status of IDH, MGMT, EGFR, PI3K, p53, and CDK biomarker groups for each individual patient are arranged in columns in both the DF/BWCC and TCGA cohorts. Trial-eligible GBM patients include those without IDH mutation or 1p/19q codeletion.
Patients with tumors harboring MGMT-promoter methylation were more often female than their unmethylated counterparts in our cohort (54% vs 35%, P = .02) and in that of TCGA (49% vs 34%, P = .005). MGMT-promoter methylated tumors were also more likely to be “multifocal” by imaging report in our dataset (20% vs 8%; P = .04), but this parameter was not available in the dataset of TCGA in order to validate the finding.
Association with Biomarker Subgroups with Known Prognostic Molecular Markers
EGFR(+) (odds ratio [OR] = 7.8, P < .001) and p53(−) (OR = 5.5, P = .002) subgroups were more likely to be IDH WT, and these associations were validated in the TCGA cohort (EGFR OR = 20.1, P < .001; p53 OR = 8.4, P < .001). There were no associations between the genetic biomarker groups and MGMT-promoter methylation status.
Association of Biomarker Subgroups with Clinical Covariates
Overall clinical and prognostic factors and significant associations with biomarker groups are shown in Table 1, excluding patients with IDH1/2 mutations or 1p/19q codeletions. With respect to clinical covariates, patients in the PI3K(+) subgroup were older (median 64.3 y vs 59.7 y, P = .015) and this was also validated in TCGA (median 62.3 y vs 58.8 y, P = .002). P53(+) had smaller contrast-enhancing tumors than the p53(−) group (median 3.8 cm in largest dimension vs 4.2 cm, P = .029) but this could not be validated in TCGA due to data limitations in that dataset. Patients with PI3K(+) tumors also had lower KPS in our cohort (P = .014), but this was not found in the dataset of TCGA. It should be noted, however, that in our dataset the KPS was consistently measured just prior to adjuvant chemoradiotherapy, while the timing of KPS measurements in TCGA was highly variable, limiting the utility.
Table 1.
Baseline clinical characteristics and associations with biomarker groups in DF/BWCC
| Patient Cohort | DF/BWCC | |||
|---|---|---|---|---|
| Biomarker Association* | ||||
| Total | 233 | |||
| Age , y | Median (IQR) | 60.1 (53.2–67.7) |
PI3K: (+)64.3 vs (−)59.7; P = .015 | |
| Gender | Male | 126 (54%) | MGMT: Male (M)46% vs (U)65%; P = .02 | |
| Female | 107 (46%) | |||
| KPS | PI3K(+) | PI3K(-); P = .014 | ||
| <60 | 11 (5%) | 2 (3%) | 4 (4%) | |
| 60–80 | 105 (45%) | 46 (65%) | 41 (42%) | |
| 90–100 | 97 (41%) | 23 (32%) | 52 (54%) | |
| Multifocal (%) | No | 199 (85%) | ||
| Yes | 34 (15%) | MGMT: (M)20% vs (U)8%; P = .04 | ||
| Size (cm) | Median (IQR) | 4.2 (3.1–5.3) | p53: (+)3.8 vs (−)4.2; P = .029 | |
| Resection (%) | Biopsy | 17 (7%) | ||
| STR/GTR | 207 (88%) | |||
| Temozolomide (%) | Received | 214 (92%) | ||
| Not received | 12 (5%) | |||
| Alive at 5 y (%) | No | 223 (96%) | ||
| Yes | 10 (4%) | EGFR: (+)0% vs (−)7%; P = .014; p53: (+)10% vs (−)0%; P = .005 | ||
| OS (mo) | Median (IQR) | 20.8 (11.4–50.8) |
MGMT: (M)20.0 vs (U)12.8; P = .036 | |
| PFS (mo) | Median (IQR) | 9.7 (5.7–18.1) |
MGMT: (M)11.6 vs (U)6.9; P = .022; PI3K: (+)9.3 vs (−)10.4; P = .049 |
|
Abbreviations: IQR, interquartile range; STR, subtotal resection; GTR, gross total resection, M, methylated; U, unmethylated.
Empty values under biomarker associations indicate no significant clinical associations for any biomarker group.
Note: Table includes only patients with wild-type IDH-1/2 and without 1p/19q codeletion.
Association Between Biomarkers and Endpoints
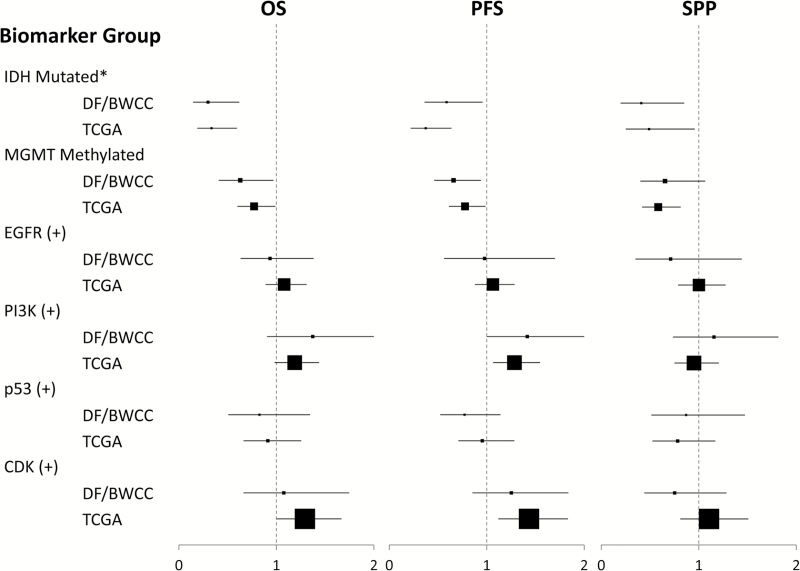
IDH mutant and MGMT-promoter methylated patients demonstrated increased OS, PFS, and SPP in both our cohort and the dataset of TCGA (Table 2, Fig. 2). PI3K(+) patients had shorter PFS in both our cohort (HR 1.42 [95% CI: 1.001–2.00] P = .049) and TCGA’s (HR 1.28 [95% CI: 1.07–1.55], P = .009). EGFR(+) patients were less likely to live 5 years (0% vs 7%, P = .014) as were p53(−) patients (0% vs 10%) in our cohort, but these results could not be recapitulated in TCGA, potentially due to the extremely low rate of 5-year survivors in that dataset (1%). Age, KPS, MGMT-promoter methylation, and the use of temozolomide were all independently associated with OS on multivariate analysis in the DF/BWCC (Table 3). After controlling for these factors, there were no independent associations of genetic biomarker subgroups with survival (Table 4) or with PFS (Supplementary Tables 1 and 3). Notably, the association between PI3K and PFS was no longer significant after correcting for any clinical prognostic factors.
Table 2.
Survival and auxiliary endpoints by biomarker group in the DF/BWCC
| Biomarker | OS, mo [IQR] | PFS, mo [IQR] | PFS/OS [IQR] | SPP, mo [IQR] |
|---|---|---|---|---|
| IDH^ N | 262 | 262 | 118 | 188 |
| Mut | 74.9 [34.8–NA] | 15.5 [7.8–38.3] | 0.65 [0.42–0.82] | 67.34 [12.8-NA] |
| WT | 20.1 [11.1–50.8] | 9.5 [5.6–18.1] | 0.59 [0.42–0.70] | 10.75 [4.6–82.6] |
| P-value | 0.001 | 0.032 | 0.59 | 0.017 |
| MGMT* N | 156 | 156 | 73 | 109 |
| M | 20.9 [11.4–NA] | 11.6 [6.8–21.8] | 0.67 [0.48–0.75] | 12.8 [5.3–NA] |
| U | 16.0 [9.1–28.7] | 6.9 [5.0–12.5] | 0.61 [0.52–0.75] | 7.7 [2.7–25.8] |
| P-value | 0.036 | 0.022 | 0.61 | 0.089 |
| EGFR* N | 198 | 198 | 97 | 142 |
| (+) | 21.3 [12.6–30.5] | 10.0 [6.0–15.3] | 0.59 [0.53–0.69] | 12.8 [6.2–NA] |
| (−) | 19.0 [9.8–103.4] | 10.0 [5.8–18.0] | 0.61 [0.50–0.72] | 8.9 [3.4–82.6] |
| P-value | 0.74 | 0.42 | 0.816 | 0.3 |
| PI3K* N | 177 | 177 | 89 | 132 |
| (+) | 17.0 [10.2–31.3] | 9.3 [6.4–13.4] | 0.66 [0.60–0.70] | 8.0 [3.4–NA] |
| (−) | 21.0 [12.8–75.0] | 10.4 [5.8–19.2] | 0.57 [0.40–0.73] | 11.5 [5.3–82.6] |
| P-value | 0.14 | 0.049 | 0.082 | 0.53 |
| p53* N | 142 | 142 | 70 | 105 |
| (+) | 20.1 [11.4-NA] | 10.1 [6.0–15.4] | 0.64 [0.50–0.69] | 11.5 [4.2–NA] |
| (−) | 19.9 [10.9–29.9] | 10.1 [5.6–14.9] | 0.60 [0.52–0.71] | 10.2 [4.8–NA] |
| P-value | 0.44 | 0.19 | 0.77 | 0.61 |
| CDK* N | 134 | 134 | 61 | 98 |
| (+) | 21.3 [11.4–29.9] | 10.8 [5.6–14.5] | 0.61 [0.55–0.66] | 12.8 [5.6–NA] |
| (−) | 19.0 [10.8–75.0] | 10.1 [6.4–18.5] | 0.64 [0.52–0.74] | 7.9 [3.6–NA] |
| P-value | 0.77 | 0.25 | 0.39 | 0.30 |
Abbreviations: IQR, interquartile range, M, methylated; U, unmethylated
IDH survival analyses include all patients with primary GBM.
MGMT, EGFR, PI3K, p53, and CDK survival analyses include only patients with wild-type IDH-1/2 and without 1p/19q codeletion.
Fig. 2.
Hazard ratios for OS, PFS, and SPP by biomarker subgroups in TCGA and DF/BWCC patient cohorts. *Outcomes across IDH subgroups were compared across the entire cohort. Outcomes across remaining biomarker subgroups were compared across only trial-eligible GBM-patients (IDH WT). Hazard ratios are displayed for positive biomarker status relative to negative status as the baseline, with HR <1 representing a favorable endpoint. Point estimates for the HR are displayed by a square box, scaled to the representative sample size of biomarker (+) patients, with 95% CIs displayed in horizontal bars.
Table 3.
Univariate and multivariate analyses for clinical predictors of overall survival in DF/BWCC
| DF/BWCC | ||||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR [95% CI] |
P-value | AHR [95% CI] | P-value | |
| Age | 1.05 [1.03–1.07] | <.001 | 1.04 [1.01–1.07] | .003 |
| Gender | ||||
| Male | 1 | |||
| Female | 0.96 [0.95–1.17] | .28 | ||
| KPS | 0.96 [0.95–0.98] | <.001 | 0.97 [0.95–0.99] | .009 |
| Multifocal | ||||
| No | 1 | 1 | ||
| Yes | 1.85 [1.17–2.91] | .008 | 1.75 [0.95–3.21] | .071 |
| Size | 0.98 [0.87–1.09] | .69 | ||
| Resection | ||||
| STR/GTR | 1 | 1 | ||
| Bx | 1.98 [1.06–3.68] | .032 | 1.47 [0.44–4.90] | .53 |
| Temozolomide | ||||
| Yes | 1 | 1 | ||
| No | 5.17 [2.73–9.79] | <.001 | 2.55 [1.05–6.19] | .038 |
| MGMT | ||||
| Unmethylated | 1 | 1 | ||
| Methylated | 0.63 [0.41–0.97] | .037 | 0.53 [0.33–0.85] | .009 |
Abbreviations: AHR, adjusted hazard ratio; STR, subtotal resection; GTR, gross total resection; Bx, biopsy.
Note: Table includes only patients with wild-type IDH-1/2 and without 1p/19q codeletion.
Table 4.
Multivariate analyses for overall survival with biomarker groups in DF/BWCC
| Multivariate | EGFR (+) | PI3K (+) | p53 (+) | CDK (+) | |
|---|---|---|---|---|---|
| Biomarker | AHR [95% CI] | 1.02 [0.64–1.61] | 0.98 [0.59–1.64] | 1.04 [0.54–2.01] | 0.87 [0.47–1.61] |
| P-value | .94 | .95 | .91 | .65 | |
| Age | AHR [95% CI] | 1.04 [1.02–1.07] | 1.04 [1.01–1.06] | 1.04 [1.01–1.08] | 1.05 [1.01–1.08] |
| P-value | .001 | .008 | .007 | .007 | |
| KPS | AHR [95% CI] | 0.97 [0.95–0.99] | 0.97 [0.95–0.99] | 0.96 [0.93–0.99] | 0.97 [0.94–0.99] |
| P-value | .003 | .005 | .003 | .01 | |
| TMZ | AHR [95% CI] | 2.19 [0.92–5.20] | 1.25 [0.41–3.82] | 1.18 [0.37–3.73] | 1.18 [0.37–3.77] |
| P-value | 0.076 | 0.69 | 0.78 | 0.78 | |
| MGMT | AHR [95% CI] | 0.57 [0.35–0.91] | 0.52 [0.31–0.86] | 0.61 [0.33–1.13] | 0.52 [0.28–0.99] |
| P-value | .02 | .01 | .12 | .048 |
Abbreviations: AHR, adjusted hazard ratio; TMZ, temozolomide chemotherapy received.
Note: Table includes only patients with wild-type IDH-1/2 and without 1p/19q codeletion.
Clinical Trial Simulations Using Biomarker Data
There were 3 major areas for which our genomic biomarker analysis on retrospective cohorts was applicable to the design and simulations for INSIGhT—the overall frequencies for different biomarker groups, the overlap of various biomarker categories, and the relationship of endpoints within given biomarker subgroups. To illustrate the potential impact of these data on design elements of the trial, we first assumed disparate scenarios for each area (frequency, overlap, endpoint) to have a robust comparison of scope and then compared operating characteristics. Our simulations showed the sensitivity of the power (log-rank test) in detecting positive treatment effects for biomarker subgroups and the sensitivity of the resulting biomarker-specific treatment effect confidence intervals.
Independently of the biomarker correlations with each other and the randomization assignment algorithm (Bayesian adaptive, non-adaptive), we observed direct proportionality of the minimum sample size requirements to achieve (60%, 80%, or 90%) power with treatment hazard ratios (HRs) (0.7 and 0.5) in all of our simulations. In these power analyses, the sample size requirement varied between 191 and 920 patients, and the maximal deviation from direct proportionality (sample size = constant/prevalence) with fixed biomarker correlations, treatment effects, and power thresholds that we observed was equal to 13 patients.
We then conducted simulations to compare operating characteristics under varying assumptions of biomarker correlation. The goal of these simulations was complementary to those described for biomarker frequency in the previous paragraph. In this instance, however, we fixed the biomarker prevalence (25% or 50%) and specified scenarios with different correlations between −0.7 and +0.7. The goal was to evaluate the robustness of Bayesian adaptive designs to the biomarker subgroup overlap. In this case, we observed that Bayesian adaptive randomization was sensitive to differences in the degree of biomarker overlap, with power variations up to 6%, demonstrating the importance of appropriate biomarker overlap estimates for design purposes of such trials.
In the last set of simulations, we used the Dana-Farber Cancer Institute biomarker prevalence estimates and defined a set of sensitivity scenarios with different PFS and OS baseline distributions for the control arm, with positive and negative scale variations up to 40% for both PFS times and SPP times. We also included variations limited to single biomarker subgroups. The operating characteristics of the Bayesian adaptive design with fixed treatment effects, obtained using identical time-scale multiplicative constants for control and treatment arms, was not very sensitive to PFS/OS variations. By multiplying PFS and OS by scale factors equal to 40%, we obtained power reductions at most equal to 3%.
Discussion
A growing interest in precision medicine and the availability of targeted agents has heightened interest in genomic biomarker-based clinical trials. Our findings of biomarker category associations with relevant prognostic covariates have some implications for design and interpretation of clinical trials. We found an association of PI3K(+) patients with older age and lower performance status, older age being validated in TCGA. Since age is an independent prognostic factor for adult patients with GBM, single-arm bucket trials using PI3K definitions as eligibility may be erroneously interpreted as poorly performing compared with historical controls if this is not taken into account. PI3K(+) in fact had worse PFS on univariate analysis in our dataset. P53(−) and EGFR(+) genomic subgroups were also associated with being IDH WT. If a single-arm phase II trial designed with a PFS or OS endpoint for agents targeted at these genomic subgroups were conducted and compared with unselected historical controls, we might erroneously conclude a negative result, as the historical control may have included IDH mutant tumors while our genetic selection effectively excluded them without our knowledge. Also of note, both cohorts showed MGMT-promoter methylation to be associated with female gender, which is a novel association to our knowledge that should be validated in future studies.
But while genomics is presumed to be a key determinant of the biology and behavior of tumor growth, we found no independent associations of genomic biomarkers and survival-based endpoints. This suggests that there may be few confounding molecular variables in clinical trials outside of the known factors of IDH mutation, MGMT-promoter methylation, and 1p/19q status. Therefore, while control groups should always be used when evaluating survival endpoints such as PFS and OS, comparison to unselected historical controls in genomic biomarker-selected studies may not have any additional confounding factors based on the biomarker selection as long as clinical covariates and known molecular prognostic factors are considered. Furthermore, should an uncontrolled single-arm study of a targeted agent show a strong prognostic signal related to a genomic biomarker previously shown to have no prognostic value, this may suggest that the biomarker is behaving in a predictive capacity given the new therapeutic context and suggest hypotheses for further testing.
Some studies have identified genomic alterations in EGFR, TP53, PTEN, and CDK4 as negative prognostic biomarkers,22–24 but this has not been consistently replicated or systematically evaluated in other studies.22 Controlling for known clinical covariates is also important when reporting such data. For example, a recently published analysis of TCGA’s glioblastoma and lower-grade glioma datasets identified PI3K mutations to be negatively prognostic for survival.25 Similarly, our study demonstrated an association between PI3K(+) tumors and poorer PFS on univariate analysis of both cohorts. However, significant associations were also seen between PI3K(+) tumors and lower KPS in our cohort and older age in both cohorts, and after controlling for clinical prognostic covariates PI3K was no longer associated with OS or PFS. These findings underscore the need for combined assessment of clinical and biomarker prognostic information.
The prognostic value of any given biomarker may additionally depend on the precise characteristics of its diagnostic assay. For illustration, in the DF/BWCC cohort, MGMT-promoter methylation was assessed primarily by MS-PCR and demonstrated significant associations with female gender, tumor multifocality, and improved PFS and OS. After multivariate adjustment for prognostic clinical factors, MGMT status remained highly prognostic for OS and PFS. In the validation set of TCGA, MGMT status showed similar univariate associations with gender, PFS, and OS; however, survival associations were not significant after multivariate adjustment. This may be due to the use by TCGA of methylation arrays (Illumina HM27K and HM450K) and the potentially imperfect concordance between these assays and the MGMT-STP27 logistic regression model used to categorize MGMT-promoter methylation status,19 a point of importance given that many pathology laboratories are considering adoption of methylation arrays and replacement of MS-PCR MGMT-based assays. More validation and comparison data of these 2 methods are likely needed based on our results. In addition, it is possible that MGMT status was less prognostic overall in TCGA, as fewer patients were treated with temozolomide in TCGA versus the DF/BWCC cohort (52% vs 92%). Finally, the prognostic signal from MGMT may have been overshadowed by more heterogeneous annotation of clinical prognostic factors in TCGA.
Designing clinical trials using an auxiliary endpoint requires knowledge of the relationship between that auxiliary endpoint and more clinically relevant ones, as these relationships may differ according to molecular subtype. Perhaps the best example of this is the relationship between pathologic complete response rate (pCR) and recurrence-free survival in breast cancer where the predictive capability of pCR varies by biologic subtype,26 the knowledge of which was helpful when designing I-SPY 2.3 Having knowledge of the relationships between biomarker-defined subgroups and various clinical trial endpoints would significantly aid clinical trial design by informing specific design choices (such as endpoints or the need for control groups) and by providing biomarker-specific data for operating characteristic analysis such as power and sample size. Past studies and meta-analyses have illustrated correlation between PFS effects and OS effects in GBM,27–29 largely driven by the results from the European Organisation for Research and Treatment of Cancer/National Cancer Institute of Canada CE.3 study.10,30 But this relationship did not hold in trials of bevacizumab in which effect on progression was not associated with effect on survival.31,32 Our study identified no significant differences in the relationship between PFS/OS ratios or SPP across genomic biomarker subgroups. Additionally, our trial simulations determined that variations in PFS and SPP times would have only a small impact on INSIGhT, and our data could be helpful to support using PFS or a longitudinal model incorporating PFS as an endpoint to inform randomization.33 In contrast, we observed significantly better OS, PFS, and SPP among IDH mutant patients in both cohorts and among MGMT-promoter methylated patients in TCGA. The longer SPP suggests that the prognostic capacity of IDH mutation and MGMT-promoter methylation is retained following recurrence, a result that is particularly relevant for interpreting results of basket trials in patients with recurrent GBM such as NCI-MATCH.4 Furthermore, the lack of evidence supporting differential relationships of endpoints among the subclasses is important for the interpretation of nonrandomized trials with newly diagnosed patients using PFS as a primary endpoint like the Neuro Master Match (N2M2) trial.34
Finally, the relative frequency of biomarker subgroups and their degree of overlap is important for clinical trial design and planning. For non-adaptive studies, knowledge of biomarker frequency is important to estimate accrual rate of specific genomic subgroups, and the frequency and degree of overlap of biomarker categories inform choices with regard to treatment-arm assignment rules. For example, if biomarker groups are relatively frequent and mutually exclusive, assignment rules may simply be to match biomarker groups with agents targeting those aberrations. Conversely, if there is substantial overlapping of subgroups and some that are relatively rare, algorithms to prioritize or randomize specific treatment arms may be needed. For Bayesian adaptively randomized trials like INSIGhT, the frequency and overlap between biomarker categories directly impact the results of clinical trial simulations that illustrate operating characteristics and how the trial might proceed in the real world. For example, GBM genomic subgroup categories as defined in this study and for INSIGhT are relatively frequent, enabling our preferred design of equal randomization across treatment arms, independent of biomarker subgrouping. This would not be logistically possible if biomarker subgroup frequencies were too low, as randomization to control or a nontargeted treatment arm of a patient with a rare biomarker would make completing the trial for that subgroup challenging. Aside from that design choice, knowledge of biomarker frequency would be used similarly to non-adaptive studies—in both cases the sample size and power are directly related to the biomarker frequency. Furthermore, Bayesian adaptive trial designs such as INSIGhT are impacted by the degree of biomarker overlap, as we found significant variations in power depending on the hypothesized correlations between subgroups. In this manner, the data abstracted from our current study are directly applicable in determining operating characteristics of INSIGhT.
Several limitations exist for this study. First, there are limited preclinical data with targeted agents in GBM to suggest the ideal biomarker categorizations, a priori, and our pathway model may oversimplify the elaborate interconnections and cross-regulation of these pathways in GBM. Additionally, potential differences in the 2 study populations may limit comparison and combined analysis across the sets of TCGA and DF/BWCC. Clinical and molecular factors were also not uniformly defined across our cohort and the validation set of TCGA; for example, in TCGA, KPS could have been defined at multiple time points, while the DF/BWCC cohort uniformly defined KPS postoperatively and prior to radiation therapy. Similarly, inconsistencies in the assessment of MGMT-promoter methylation were discussed above. Progression endpoint identification was also not standardized in either cohort. Nonetheless, these datasets are highly complementary to one another, and the large TCGA dataset was useful for validating our initial findings. Finally, it should be noted that the biomarker subgroupings that were of interest in the current study were not intended to be globally applicable to agents that might target alternative pathways. If there were other biomarkers that were of interest, however, we feel that the general approach and implications of retrospective biomarker analysis for prospective trial planning still hold and should be applied.
In summary, we identified relevant associations between 4 a priori defined genetic subgroups of GBM and known clinical and molecular prognostic factors. After controlling for these factors, there was no association between the genomic biomarker groups and OS, although the PI3K(+) group may have shorter PFS on univariate analysis. Both IDH and MGMT status were not only found to be prognostic initially, but also associated with longer SPP, illustrating a potential differential relationship between endpoints. Clinical trial simulations of both balanced and Bayesian adaptively randomized trials showed the impact of biomarker frequency, overlap, and endpoint relationships on design and operating characteristics. These data represent a foundation to plan and develop innovative genomic biomarker-driven clinical trial designs to accelerate discovery in GBM (currently being used in the development of INSIGhT), and to help interpret findings from genomic biomarker-based basket studies.
Supplementary Material
Supplementary data are available at Neuro-Oncology online.
Funding
This work was supported by the Burroughs Wellcome Innovations in Regulatory Science Award and The Ben and Catherine Ivy Foundation.
Conflict of interest statement. Dr Alexander reports personal fees from Bristol-Myers Squibb, Abbvie, Schlesinger Associates and Precision Health Economics outside of the submitted work. Dr Beroukhim reports stock/ownership in AstraZeneca, grants from Novartis, Merck, and Gilead, and personal fees from Novartis outside of the submitted work. Dr Ligon reports personal fees from Midatech and nonfinancial support from Agilent Technologies outside of the submitted work and has a cancer diagnostics patent issued. Dr Parmigiani reports personal fees from Dainippon Sumitomo Pharma Co Ltd, HughesRiskApps, Metamark, Counsyl, and Springer, is on the Scientific Advisory Board with associated equity from HughesRiskApps and Counsyl, and provides editorial services for Springer and Science, all outside of the submitted work. Dr Wen reports personal fees from Roche, Novartis, Regeneron, Vascular Biogenics, Cavion, Foundation Medicine, Monteris, Novocure; grants from Novartis, Vascular Biogenics, Merck, Angiochem, Agios, AstraZeneca, Exelixis, Roche, GlaxoSmithKline, Karyopharm, and Sanofi Aventis, outside the submitted work.
Supplementary Material
Acknowledgments
The results published here are in part based upon data generated by the TCGA Research Network and the Dana-Farber Cancer Institute/Brigham and Women’s Hospital Profile Project.
References
- 1. Rodón J, Saura C, Dienstmann R, et al. Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol. 2012;9(6):359–366. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Gandara DR, Hirsch FR, et al. Lung master protocol (Lung-MAP)-A biomarker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin Cancer Res. 2015;21(7):1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker AD, Sigman CC, Kelloff GJ, Hylton NM, Berry DA, Esserman LJ. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86(1):97–100. [DOI] [PubMed] [Google Scholar]
- 4. Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41(3):297–299. [DOI] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. [DOI] [PubMed] [Google Scholar]
- 7. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexander BM, Galanis E, Yung WK, et al. Brain Malignancy Steering Committee clinical trials planning workshop: report from the Targeted Therapies Working Group. Neuro Oncol. 2015;17(2):180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oldenhuis CN, Oosting SF, Gietema JA, de Vries EG. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. 2008;44(7):946–953. [DOI] [PubMed] [Google Scholar]
- 10. Alexander BM, Trippa L. Progression-free survival: too much risk, not enough reward? Neuro Oncol. 2014;16(5):615–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep. 2015;15(1):506. [DOI] [PubMed] [Google Scholar]
- 12. Ramkissoon SH, Bi WL, Schumacher SE, et al. Clinical implementation of integrated whole-genome copy number and mutation profiling for glioblastoma. Neuro Oncol. 2015;17(10):1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craig JM, Vena N, Ramkissoon S, et al. DNA fragmentation simulation method (FSM) and fragment size matching improve aCGH performance of FFPE tissues. PLoS One. 2012;7(6):e38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4(11):e7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. R Core Team. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 21. Therneau TM. A Package for Survival Analysis in S. 2015. [Google Scholar]
- 22. England B, Huang T, Karsy M. Current understanding of the role and targeting of tumor suppressor p53 in glioblastoma multiforme. Tumour Biol. 2013;34(4):2063–2074. [DOI] [PubMed] [Google Scholar]
- 23. Idoate MA, Echeveste J, Diez-Valle R, Lozano MD, Aristu J. Biological and clinical significance of the intratumour heterogeneity of PTEN protein expression and the corresponding molecular abnormalities of the PTEN gene in glioblastomas. Neuropathol Appl Neurobiol. 2014;40(6):736–746. [DOI] [PubMed] [Google Scholar]
- 24. Labussière M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83(13):1200–1206. [DOI] [PubMed] [Google Scholar]
- 25. Draaisma K, Wijnenga MM, Weenink B, et al. PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients. Acta Neuropathol Commun. 2015;3:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Esserman LJ, Berry DA, DeMichele A, et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL–CALGB 150007/150012, ACRIN 6657. J Clin Oncol. 2012;30(26):3242–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ballman KV, Buckner JC, Brown PD, et al. The relationship between six-month progression-free survival and 12-month overall survival end points for phase II trials in patients with glioblastoma multiforme. Neuro Oncol. 2007;9(1):29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han K, Ren M, Wick W, et al. Progression-free survival as a surrogate endpoint for overall survival in glioblastoma: a literature-based meta-analysis from 91 trials. Neuro Oncol. 2014;16(5):696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polley MY, Lamborn KR, Chang SM, Butowski N, Clarke JL, Prados M. Six-month progression-free survival as an alternative primary efficacy endpoint to overall survival in newly diagnosed glioblastoma patients receiving temozolomide. Neuro Oncol. 2010;12(3):274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 31. Chinot OL, Wick W, Mason W, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–722. [DOI] [PubMed] [Google Scholar]
- 32. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trippa L, Wen PY, Parmigiani G, Berry DA, Alexander BM. Combining progression-free survival and overall survival as a novel composite endpoint for glioblastoma trials. Neuro Oncol. 2015;17(8):1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hertenstein A, Jones D, Sahm F, et al. Umbrella protocol for phase I/IIa trials of molecularly matched targeted therapies plus radiotherapy in patients with newly diagnosed glioblastoma without MGMT promoter methylation Neuro Master Match (N2M2). Paper presented at: ASCO Annual Meeting Proceedings2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.