Abstract
The structural and conformational organization of chromosomes is crucial for gene expression regulation in eukaryotes and prokaryotes as well. Up to date, gene expression data generated using either microarray or RNA-sequencing are available for many bacterial genomes. However, differential gene expression is usually investigated with methods considering each gene independently, thus not taking into account the physical localization of genes along a bacterial chromosome. Here, we present WoPPER, a web tool integrating gene expression and genomic annotations to identify differentially expressed chromosomal regions in bacteria. RNA-sequencing or microarray-based gene expression data are provided as input, along with gene annotations. The user can select genomic annotations from an internal database including 2780 bacterial strains, or provide custom genomic annotations. The analysis produces as output the lists of positionally related genes showing a coordinated trend of differential expression. Graphical representations, including a circular plot of the analyzed chromosome, allow intuitive browsing of the results. The analysis procedure is based on our previously published R-package PREDA. The release of this tool is timely and relevant for the scientific community, as WoPPER will fill an existing gap in prokaryotic gene expression data analysis and visualization tools. WoPPER is open to all users and can be reached at the following URL: https://WoPPER.ba.itb.cnr.it
INTRODUCTION
There is a complex interaction between chromosome organization and gene expression in bacteria, influencing the transcription of large sets of genes. Global changes in the bacteria transcriptional program are elicited in response to different conditions and stresses. These changes depend on a complex regulatory network, as well as on the physical organization of the chromosome (1–4). Transcription regulation and the genome structural organization have been defined by evolution allowing adaptation to multiple environmental pressures (5,6). Recently, the use of chromosome conformation capture (3C)-based techniques allowed exploring the architecture of some bacterial genomes, thus relating 3D genome structure and transcription profiles in bacteria (7–10).
Despite numerous evidences of correlation between expression patterns and physical localization of genes along the chromosomes ever since the early days of genome-wide studies (11–14), still transcriptional data are mostly analyzed using methods considering each gene independently. Only a few methods integrate quantitative transcriptional data and structural information for the identification of differentially expressed chromosomal regions (15–17).
Here, we present WoPPER, a web tool for the analysis of differential gene expression in bacteria that integrates transcriptional data and gene localization. WoPPER detects groups of physically contiguous genes characterized by similar, differential transcriptional profiles. The analysis procedure is based on our previously published R-package PREDA (18). The core of the method is the Locally Adaptive Procedure (LAP) algorithm (17), which was developed to identify clusters of genes with coordinated expression pattern. Differently from other methods used to detect chromosome co-expression maps (19), LAP integrates gene location and local gene density with differential expression data. This is achieved by applying a locally adaptive smoothing to the statistical score for differential expression over the positional coordinates of genes along the genome. This procedure, on the one hand takes into account local variations in gene density, while at the same time it is independent of any pre-defined assumptions on genome size, global gene density and distribution. LAP has already been used to analyze bacterial time-course gene expression data (20) and to compare transcriptomic data from high- and low-antibiotic producers (21). In both cases the analysis gave a clear view of how extended genomic regions were almost completely up- or downmodulated during the shift between exponential and stationary growth phases, highlighting the induction or repression of primary and secondary metabolism along the time course, and revealing major differences in chromosomal transcription between high- or low-producers strains (20,22).
To facilitate the application of PREDA to transcriptional data from prokaryotes, we developed WoPPER a user-friendly web tool for Position Related data analysis of gene Expression in Prokaryotes. The tool analyzes both RNA-seq and microarray gene expression data, to identify clusters of genes with coordinated expression pattern along the chromosomes, considering the two genome strands either separately or together. WoPPER contains a pre-compiled internal database of genome annotations for 2780 bacterial strains (including all their annotated plasmids). It can work also on custom genomic annotations provided by the user. The analysis returns as output a list of genomic regions comprising genes with a coordinated trend of differential expression. Furthermore, multiple graphical representations allow intuitive and user-friendly browsing of the results.
To the best of our knowledge, no other web tool is currently available for the integrative analysis of transcriptional expression data and genomic positions in bacteria. Two other web tools, i.e. NuST (1) and GREAT (23), can in principle analyze positional patterns of bacterial genes. Nevertheless, they perform completely different types of analysis as compared to WoPPER.
In particular, NuST (Nucleoid Survey Tools) is a web tool for the analysis of Escherichia coli genomics features (genes or other loci). NuST analyzes a pre-selected user-defined list of genomic features at multiple scales of observation to assess their clustering along the genome (1).
The Genome Regulatory Architecture Tools (GREAT), is a set of online tools for the analysis of user-defined lists of genomics features (23). A tool within GREAT can also analyze DNA sequences to identify transcription factors binding sites. However, the focus of GREAT is on the identification of periodicity patterns in the positions of genomics features (24). GREAT in principle can work on any genome, as it requires just the genome size as input annotation. However, the online tool has pre-loaded annotations only for E. coli and Bacillus subtilis.
Differently from both NuST and GREAT, WoPPER: (i) does not analyze just the position of genes, but also uses quantitative expression data for each gene (Log2-fold-change across conditions); (ii) does not use as input a pre-filtered list of genes, considering instead the entire genome-wide expression profile; (iii) can perform the analysis also on separated strands, to identify strand-specific patterns; (iv) can analyze data from virtually every bacteria with a sequenced genome, as it contains the genomic annotations for all of the NCBI complete bacterial genomes pre-loaded in its internal database and it allows users to load custom annotations.
In principle WoPPER can be used to analyze any prokaryotic microarray or RNA-seq dataset, thus helping the scientific community to get a comprehensive view of how prokaryotic gene expression is related to chromosomes structure, and filling an existing gap in prokaryotic gene expression data analysis and visualization tools.
WEB SERVER DESCRIPTION
WoPPER architecture is composed by: (i) an input data web manager; (ii) a database containing all the complete bacteria genomes annotations from NCBI; (iii) a backend based on PREDA R package for the analysis; and (iv) a web module for the visualization of the results and interactive graphics.
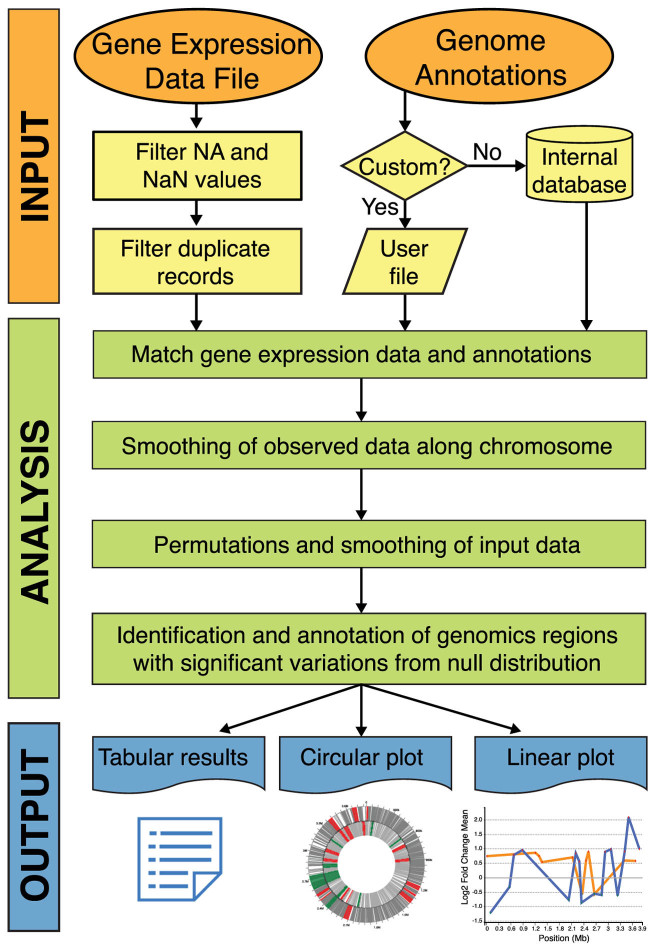
Figure 1 shows WoPPER workflow steps.
Figure 1.
Overview of WoPPER data analysis workflow. In the orange ellipses the user-defined input data are reported: the Genome Annotation File can be selected from the internal database or, alternatively, a custom annotation can be provided by the user. The yellow boxes report the main steps for GED file pre-processing. The analysis steps and computations performed by PREDA algorithm are represented by green boxes. For each execution, WoPPER provides one Tabular output reporting the list of identified gene clusters with significant variations, and two visual representations where the clusters and genes are plotted on bacterial chromosomes in circular (Circular plot) and in linear view (Linear plot), represented at the bottom.
Input description
Gene Expression Data file
WoPPER requires as input at least one Gene Expression Data file (GED file) containing the genome-wide differential expression levels across two conditions of interest. Expression data are in the form of Log2 ratios, also called Log2-fold-change values (Log2FC). Log2FC values can be easily obtained from most widely used gene expression analysis tools for RNA-seq (24,25) or microarrays (26). Most importantly, the GED file must contain the Log2FC for all available genes, i.e. no gene has to be filtered out even if its transcriptional modulation is marginal. The GED is a plain-text file containing at least two columns: one for the gene names and one for the Log2FC values. Gene names must match those provided in the genome annotation, as WoPPER will use this information to connect expression data and gene coordinates.
Genome annotation database
A large collection of annotations for bacterial genomes is available in the WoPPER internal database. It includes all completely sequenced bacteria genomes and annotations derived from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_refseq/Bacteria/), for a total of 2780 bacterial strains, including 2970 chromosomes and 2188 plasmids (Supplementary Table S1). Organisms with multiple chromosomes are reported with multiple records, i.e. one per chromosome, which implies that WoPPER analysis on these bacteria must be conducted one chromosome at a time.
Custom genome annotation
In case the genome of interest is not present in the internal database, a custom genome annotation file can be provided. Standard BED6 or GFF formats can be used (see https://genome.ucsc.edu/FAQ/FAQformat for format specifications), as well as delimiter-separated text files containing at least five columns: chromosome name, gene coordinates (start–end), gene names and strand of transcription. WoPPER online help provides full description of the accepted formats.
Analysis description
WoPPER is based on PREDA (Position RElated genomics Data Analysis), a Bioconductor R package to identify clusters of genomic features with coordinated patterns of expression in genomic data. The core of the PREDA analysis method is the LAP. LAP uses a locally adaptive kernel smoothing function (lokern) to integrate gene expression data and the physical position of genes along the genome. This smoothing function accounts for the heterogeneous distribution of genes along the chromosome. The significance of local maxima and minima in the smoothed expression data is estimated against the expected null distribution. Namely, the observed smoothed data are compared to random expectations (null distribution) empirically estimated by repeated permutations of expression values associated to each gene, followed by application of the smoothing function. Within WoPPER, PREDA functions are used to identify regions (clusters) of genes with increased (upregulated) or decreased (downregulated) expression between the compared conditions of interest. As compared to the original PREDA package, WoPPER introduces several major improvements, as: (i) a user-friendly web interface that does not require any programming skills; (ii) the integration with a large database containing all the prokaryotic complete genome sequences available at NCBI; (iii) the ability to manage gene expression data generated by both RNA-seq or microarray and to analyze them considering the two genome strands of transcription either separately or together; and (iv) a user-friendly and browsable graphical visualization of the results.
Separated versus non-separated strands analysis
WoPPER can analyze the transcriptional data considering separately or not the position of genes on the positive and negative strand. The ‘separated strands’ analysis is functional to identify strand-specific patterns of expression. This is particularly useful in bacteria where multiple genes on the same strand might be transcribed together as operons. However, this type of analysis is recommended only for genomes having more than 2000 genes, as on smaller genomes the smoothing and permutation steps of WoPPER would be based on few data points, thus yielding noisy statistics.
Output description
WoPPER provides three types of output: Tabular Results, Circular Plots and Linear Plots (Figure 2). Results are stored for 15 days and are retrievable using a web link which is reported at the top of the WoPPER::Results page and, upon completion of the analysis, is sent by email to the user-provided email address (optional).
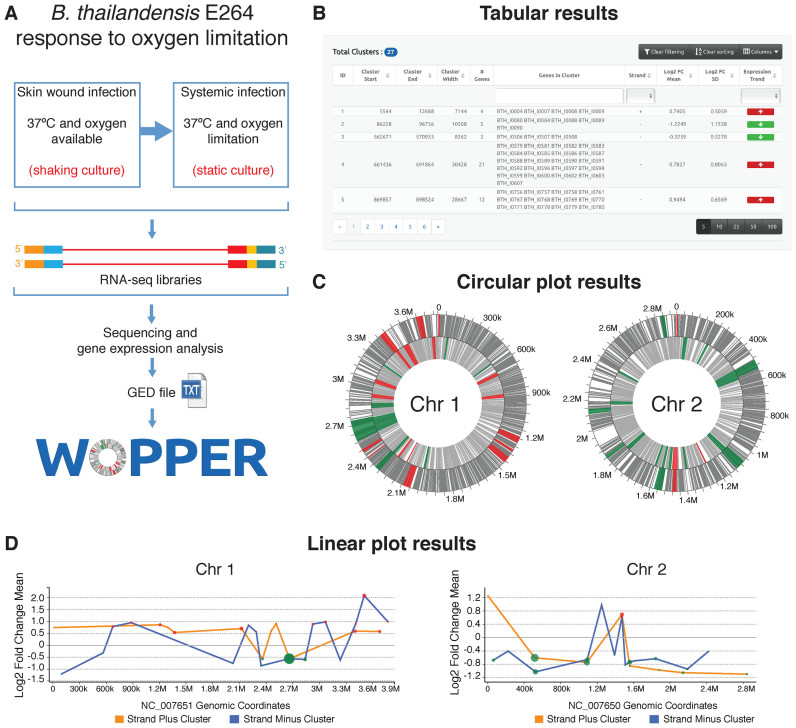
Figure 2.
Schematic representation of WoPPER analysis of RNA-seq Gene Expression Data (GED) by Peano et al. (29). (A) Gene Expression Data are obtained from an RNA-seq experiment in which Burkholderia thailandensis (E264 strain) response to growth in oxygen limiting condition was analyzed: i.e. growth at 37°C in static compared to shaking culture conditions. The experimental design and workflow are schematically represented. (B) Example of tabular output layout obtained by WoPPER. (C) Circular plots representing the two B. thailandensis chromosomes with genes (gray lines) and clusters plotted over chromosomal coordinates. Clusters identified by WoPPER analysis on separated strands are colored in RED (upregulated clusters) and GREEN (downregulated clusters). (D) Linear plot representations of the two B. thailandensis chromosomes in linear coordinates. Each colored red or green dot represents a cluster up- or downregulated, respectively: X-axis coordinate represents its middle position along the chromosome, Y-axis coordinate reports the average Log2FC for genes within each cluster, the dot size is proportional to the cluster width. An orange or blue line connects clusters identified on the plus or minus strands, respectively. Alternative color blind friendly color schemes for both the circular and linear plots can be interactively selected in the online interface.
Tabular results
WoPPER reports a table with a list of identified gene clusters (i.e.: groups of spatially associated and similarly regulated genes). For each cluster the table contains: start and end coordinates; its size and strand (if strand-specific analysis was performed); the number and names of genes belonging to the cluster; the expression trend (red or green arrows indicating up- or downregulated clusters, respectively); the average Log2FC value of the genes in the cluster and its standard deviation. By default, clusters are sorted by start coordinate, but users can browse the results by ordering and filtering them according to any field. The tabular output can be downloaded as a plain-text file.
Circular plots
The circular plot represents the bacterial chromosome as a circle in which genomic features and gene clusters are visualized. This is particularly useful to highlight cluster densities, location and strand specificity all in one intuitive plot. Pop-up windows allow the users to retrieve information for clusters of interest. The circular plot can be downloaded either as Portable Network Graphics (PNG) or Scalable Vector Graphics (SVG) files for high-quality images.
Linear plots
The linear graph represents the gene clusters (colored dots) along bacterial chromosome coordinates (X-axis) with their respective average Log2FC values (on the Y-axis). This plot can help users in the identification of stretches of similarly regulated clusters. When performing a ‘separated strands’ analysis, two different lines connect the cluster dots, allowing the distinction between the clusters belonging to different strands. Linear graphs can be saved as PNG or SVG files.
More detailed descriptions of the input and output data, analyses and interactive visualizations are provided in the online Tutorials and Help pages.
Implementation
The web server is based on a lightweight and flexible PHP Content Management System (Typesetter—https://www.typesettercms.com/). A specialized RESTful module is responsible for asynchronous communication with the web interface that manages: (i) file upload and validation; (ii) execution monitoring; and (iii) analysis results and interactive graphs visualization.
The web front-end, on the client side, is CSS3 and HTML5 compliant. It adopts a Bootstrap Framework (http://getbootstrap.com/) for responsive and mobile-first web design features. The JavaScript libraries are implemented in ECMAScript6 and the web pages built upon Google AngularJS (http://angularjs.org/). For the interactive visualization of the results we used C3.js (http://c3js.org/) and GenomeD3Plot (https://github.com/brinkmanlab/GenomeD3Plot), both based on D3 JavaScript library (https://d3js.org/).
Session management and results availability
For each analysis a unique 16-characters Experiment ID and associated folders on the server, to store input data and analysis results, are created. The random key, used in the results web link, will guarantee a safe and stable user access to the analysis results.
Server and browser compatibility
The whole system is deployed on a local server with 64 GB RAM and 16-core CPUs which ensures an efficient performance also with a heavy user access.
Thanks to the standards adopted (CSS3, HTML5, ECMAScript6 JavaScript), WoPPER can work on any recently updated browser.
CASE STUDY RESULTS
The tutorials available online on WoPPER website provide representative case studies, including ready to execute working examples and sample input (GED files) derived from previously published microarray or RNA sequencing datasets (27–30).
Among the examples provided we will herein describe the results obtained by re-analyzing with WoPPER the GED file of an RNAseq experiment on Burkholderia thailandensis in response to various environmental cues (29). Burkholderia thailandensis is considered a model organism for the human pathogen Burkholderia pseudomallei, and is itself a pathogen for invertebrates (31,32). Thus, the RNAseq analysis aimed at investigating the gene expression response to environmental stimuli associated to the host infection, such as low oxygen availability (29). The complete step-by-step tutorial for this case study is provided as Supplementary File 1.
Figure 2A summarizes the experimental workflow and the first steps of the analysis of gene expression response to oxygen limitation. Figure 2B–D show the different types of output returned by WoPPER.
The circular plots (Figure 2C) provide an overview of the location of differentially expressed gene (DEG) clusters. A prominent trend of chromosome 1 upregulation (318 genes in 18 upmodulated clusters and only 187 genes in 9 downmodulated clusters) and the general downregulation of chromosome 2 (only 22 genes in 4 upmodulated clusters and 168 genes in 17 downmodulated clusters) can be noted. This result is in agreement with previous observations (29) that oxygen limitation increases expression of primary metabolism genes, mostly located on chromosome 1.
In this case study, WoPPER analysis was performed on separated strands for both chromosomes. However, it can be noted that DEGs on chromosome 1 (mainly upregulated, RED) show a more evident strand-specific pattern of expression in comparison to those on chromosome 2. The genes included in the clusters without a strand-specific pattern of expression (mainly DOWN regulated, GREEN) are genes prevalently encoding for secondary metabolites or belonging to functional categories linked to secondary metabolism (data not shown). The observation that clusters of genes involved in secondary metabolism appears to be regulated in a strand-independent manner is in agreement with previous works (20,22). This is also consistent with the hypothesis that these genomic regions could represent a dynamic portion of the chromosome, which can readily acquire novel genes. Moreover, unidirectional transcriptional regions are typical instead of chromosome portions in which genes involved in primary metabolism are clustered (33). Thus, the simple visual inspection of WoPPER circular output plots allows extracting relevant information on the main effect of growth in oxygen limiting conditions on B. thailandensis transcriptome regulation.
In order to assess the relevance of WoPPER in characterizing B. thailandensis response to anoxia, we compared the list of DEGs published by Peano et al. (29) to the genes clustered by WoPPER, as listed in the tabular output file. This highlighted how WoPPER analysis improves the detection of complete operons within the genomic regions resulting significantly up- or downregulated, both on chromosome 1 and 2 (Supplementary Table S2). In this comparison the list of previously published DEGs was filtered imposing either stringent parameters (adjusted P-value ≤ 0.01 and Log2FC ≥ 1 or Log2FC ≤ -1) or more relaxed ones (adjusted P-value ≤ 0.05 and Log2FC ≥ 0.866 or Log2FC ≤ -0.722). The last two Log2FC filtering parameters are the average values of log2FC calculated by WoPPER respectively for up- or downregulated clusters. We noted that WoPPER enhances the numbers of complete operons both in up- or downregulated regions, despite a slight difference between the total number of clustered genes and the number of previously published DEGs (29). The total number of modulated complete operons is increased on average about 2-fold. Whereas if considering separately up- or downregulated operons the increase is 1.4- and 3.5-fold, respectively.
The results obtained with WoPPER provide a more complete view of how anoxia induces a genome-wide effect on gene expression deregulation of complete operons. This might induce to some extent an ‘opening’ of wide portions of chromosome 1, for a fast activation of operons involved in primary metabolism, to facilitate quick adaptation to the adverse conditions that the bacterium could encounter during systemic host infection.
To determine if WoPPER analysis could add useful information from a functional point of view, the same lists of previously reported DEGs were also analyzed by performing a functional enrichment analysis. Three of the functional categories previously found to be significantly enriched with adjusted P-value < 0.05 (29) (i.e. ‘translation ribosomal structure and biogenesis,’ ‘energy production and conversion’ and ‘nucleotide transport and metabolism’) were confirmed in our new analysis (Supplementary Table S2 reports results of Fisher exact test after Bonferroni's multiple testing correction). Moreover, WoPPER analysis was able to generate one more functional category significantly enriched (i.e. ‘defense mechanisms’). Thus the analysis performed with WoPPER reinforces the observations derived from the standard gene expression analysis. It also adds new insights into the pathways involved in the response to oxygen limiting conditions and governing the mechanisms tangled in systemic host infection.
CONCLUSION
WoPPER is, to our knowledge, the first web tool that integrates transcriptional expression data and genomic annotations to identify groups of physically contiguous genes characterized by regional differential expression in bacterial genomes.
It can analyze any RNA-seq or microarray-based gene expression dataset from any microorganism with a sequenced and annotated genome. Through a user-friendly web interface that does not require any programming skills, WoPPER returns meaningful graphical visualizations of the results.
We believe that WoPPER will provide researchers with novel and informative insights regarding the correlation between gene expression and chromosomal organization in bacterial genomes.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Prof. Paolo Landini from the University of Milan for the help in interpreting the results. We would like to thank Giada Caredda and Maria Vurchio (Institute of Biomedical Technologies, National Research Council, Milan) for technical support, Nicola Losito (Institute of Biomedical Technologies, National Research Council, Bari) for server management support, Arianna Consiglio (Institute of Biomedical Technologies, National Research Council, Milan) for the help in the graphic layout design.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Education and University, PRIN Project [2010P3S8BR_002 to C.P.]; National Research Council (CNR) Flagship Project InterOmics; AIRC Start-up grant 2015 [N.16841 to F.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Scolari V.F., Zarei M., Osella M., Cosentino M.. NuST: analysis of the interplay between nucleoid organization and gene expression. Bioinformatics. 2012; 28:1643–1644. [DOI] [PubMed] [Google Scholar]
- 2. Rimsky S., Travers A.. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr. Opin. Microbiol. 2011; 14:136–141. [DOI] [PubMed] [Google Scholar]
- 3. Srinivasan R., Scolari V.F., Lagomarsino M.C., Seshasayee A.S.N.. The genome-scale interplay amongst xenogene silencing, stress response and chromosome architecture in Escherichia coli. Nucleic Acids Res. 2015; 43:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie T., Fu L.-Y., Yang Q.-Y., Xiong H., Xu H., Ma B.G., Zhang H.Y.. Spatial features for Escherichia coli genome organization. BMC Genomics. 2015; 16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wright M.A., Kharchenko P., Church G.M., Segre D.. Chromosomal periodicity of evolutionarily conserved gene pairs. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:10559–10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scolari V.F., Bassetti B., Sclavi B., Lagomarsino M.C.. Gene clusters reflecting macrodomain structure respond to nucleoid perturbations. Mol. Biosyst. 2011; 7:878–888. [DOI] [PubMed] [Google Scholar]
- 7. Le T.B.K., Imakaev M. V., Mirny L.A., Laub M.T.. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013; 342:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cagliero C., Grand R.S., Jones M.B., Jin D.J., O'Sullivan J.M.. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res. 2013; 41:6058–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W., Li G.W., Chen C., Xie X.S., Zhuang X.. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011; 333:1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Umbarger M.A., Toro E., Wright M.A., Porreca G.J., Baù D., Hong S.H., Fero M.J., Zhu L.J., Marti-Renom M.A., McAdams H.H. et al. . The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell. 2011; 44:252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen B.A, Mitra R.D., Hughes J.D., Church G.M.. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 2000; 26:183–186. [DOI] [PubMed] [Google Scholar]
- 12. Caron H., Schaik B., van der Mee M., van der Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A. et al. . The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001; 291:1289–1292. [DOI] [PubMed] [Google Scholar]
- 13. Versteeg R., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H.. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003; 13:1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen T.E., Price N.D., Joyce A.R., Palsson B.. Long-range periodic patterns in microbial genomes indicate significant multi-scale chromosomal organization. PLoS Comput. Biol. 2006; 2:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levin A.M., Ghosh D., Cho K.R., Kardia S.L.R.. A model-based scan statistic for identifying extreme chromosomal regions of gene expression in human tumors. Bioinformatics. 2005; 21:2867–2874. [DOI] [PubMed] [Google Scholar]
- 16. Toedling J., Schmeier S., Heinig M., Georgi B., Roepcke S.. MACAT–microarray chromosome analysis tool. Bioinformatics. 2005; 21:2112–2113. [DOI] [PubMed] [Google Scholar]
- 17. Callegaro A., Basso D., Bicciato S.. A locally adaptive statistical procedure (LAP) to identify differentially expressed chromosomal regions. Bioinformatics. 2006; 22:2658–2666. [DOI] [PubMed] [Google Scholar]
- 18. Ferrari F., Solari A., Battaglia C., Bicciato S.. PREDA: an R-package to identify regional variations in genomic data. Bioinformatics. 2011; 27:2446–2447. [DOI] [PubMed] [Google Scholar]
- 19. Reyal F., Stransky N., Bernard-Pierrot I., Vincent-Salomon A., De Rycke Y., Elvin P., Cassidy A., Graham A., Spraggon C., Désille Y. et al. . Visualizing chromosomes as transcriptome correlation maps: evidence of chromosomal domains containing co-expressed genes—a study of 130 invasive ductal breast carcinomas. Cancer Res. 2005; 65:1376–1383. [DOI] [PubMed] [Google Scholar]
- 20. Peano C., Bicciato S., Corti G., Ferrari F., Rizzi E., Bonnal R.J., Bordoni R., Albertini A., Bernardi L.R., Donadio S. et al. . Complete gene expression profiling of Saccharopolyspora erythraea using GeneChip DNA microarrays. Microb. Cell Fact. 2007; 6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peano C., Damiano F., Forcato M., Pietrelli A., Palumbo C., Corti G., Siculella L., Fuligni F., Tagliazucchi G.M., De Benedetto G.E. et al. . Comparative genomics revealed key molecular targets to rapidly convert a reference rifamycin-producing bacterial strain into an overproducer by genetic engineering. Metab. Eng. 2014; 26:1–16. [DOI] [PubMed] [Google Scholar]
- 22. Peano C., Tala A., Corti G., Pasanisi D., Durante M., Mita G., Bicciato S., De Bellis G., Alifano P.. Comparative genomics and transcriptional profiles of Saccharopolyspora erythraea NRRL 2338 and a classically improved erythromycin over-producing strain. Microb. Cell Fact. 2012; 11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bouyioukos C., Bucchini F., Elati M., Képès F.. GREAT: a web portal for Genome Regulatory Architecture Tools. Nucleic Acids Res. 2016; 44:W77–W82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Junier I., Hérisson J., Képès F.. Periodic pattern detection in sparse boolean sequences. Algorithms Mol. Biol. 2010; 5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maciag A., Peano C., Pietrelli A., Egli T., De Bellis G., Landini P.. In vitro transcription profiling of the sigmaS subunit of bacterial RNA polymerase: re-definition of the sigmaS regulon and identification of sigmaS-specific promoter sequence elements. Nucleic Acids Res. 2011; 39:5338–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rossi E., Longo F., Barbagallo M., Peano C., Consolandi C., Pietrelli A., Jaillon S., Garlanda C., Landini P.. Glucose availability enhances lipopolysaccharide production and immunogenicity in the opportunistic pathogen Acinetobacter baumannii. Future Microbiol. 2016; 11:335–349. [DOI] [PubMed] [Google Scholar]
- 29. Peano C., Chiaramonte F., Motta S., Pietrelli A., Jaillon S., Rossi E., Consolandi C., Champion O.L., Michell S.L., Freddi L. et al. . Gene and protein expression in response to different growth temperatures and oxygen availability in Burkholderia thailandensis. PLoS One. 2014; 9:e93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McClure R., Balasubramanian D., Sun Y., Bobrovskyy M., Sumby P., Genco C.A., Vanderpool C.K., Tjaden B.. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013; 41:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu Y., Kim H.S., Chua H.H., Lin C.H., Sim S.H., Lin D., Derr A., Engels R., DeShazer D., Birren B. et al. . Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 2006; 6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wand M.E., Muller C.M., Titball R.W., Michell S.L.. Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 2011; 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Challis G.L., Hopwood D.A.. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14555–14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.