Abstract
Microbial natural products represent a rich resource of pharmaceutically and industrially important compounds. Genome sequencing has revealed that the majority of natural products remain undiscovered, and computational methods to connect biosynthetic gene clusters to their corresponding natural products therefore have the potential to revitalize natural product discovery. Previously, we described PRediction Informatics for Secondary Metabolomes (PRISM), a combinatorial approach to chemical structure prediction for genetically encoded nonribosomal peptides and type I and II polyketides. Here, we present a ground-up rewrite of the PRISM structure prediction algorithm to derive prediction of natural products arising from non-modular biosynthetic paradigms. Within this new version, PRISM 3, natural product scaffolds are modeled as chemical graphs, permitting structure prediction for aminocoumarins, antimetabolites, bisindoles and phosphonate natural products, and building upon the addition of ribosomally synthesized and post-translationally modified peptides. Further, with the addition of cluster detection for 11 new cluster types, PRISM 3 expands to detect 22 distinct natural product cluster types. Other major modifications to PRISM include improved sequence input and ORF detection, user-friendliness and output. Distribution of PRISM 3 over a 300-core server grid improves the speed and capacity of the web application. PRISM 3 is available at http://magarveylab.ca/prism/.
INTRODUCTION
Microbial secondary metabolism has historically represented a rich resource of evolved, bioactive small molecules, which form the foundations of many therapeutic regimens (1). Despite a decline in natural product discovery from a ‘golden age’ in the middle of the 20th century, genome sequencing indicates that the majority of genetically encoded natural products remain unknown (2,3). Methods that connect biosynthetic gene clusters to their corresponding natural products therefore have the potential to facilitate the targeted discovery of genetically encoded compounds. In this context, a central challenge is to define the structures of genetically encoded natural products and not solely the clusters themselves. Meeting this challenge requires the development of cheminformatic algorithms capable of accounting for the complete catalog of chemical reactions promoted by enzymatic catalysts. Nonetheless, early methods were designed primarily to facilitate detection of biosynthetic gene clusters (4–11), whereas few methods exist to facilitate the prediction of natural product structures from microbial genomes (6,11).
Central to the challenge of finding new chemical entities is to create accurate algorithms that can predict genetically encoded molecular structures from detected biosynthetic genes. In 2015, we presented PRISM (PRediction Informatics for Secondary Metabolomes), a Java application and web server for the chemical structure prediction of genetically encoded nonribosomal peptides (NRPs) and type I and II polyketides (PKs) (12). PRISM takes as input a microbial nucleotide sequence in FASTA or GenBank format, searches the sequence with a library of hidden Markov models (HMMs) associated with secondary metabolism, clusters the identified biosynthetic genes and leverages identified biosynthetic information for structure prediction (Figure 1A). Because the exact site of tailoring reactions is not always unambiguously predictable, PRISM generates combinatorial libraries of predicted structures to account for variability in the action of tailoring enzymes or in the permutation of monomers that forms the natural product backbone. We have additionally detailed the extension of structure prediction to ribosomally synthesized and post-translationally modified peptides (RiPPs) (13) and described the addition of a library of 257 HMMs to identify genes associated with antimicrobial resistance (14).
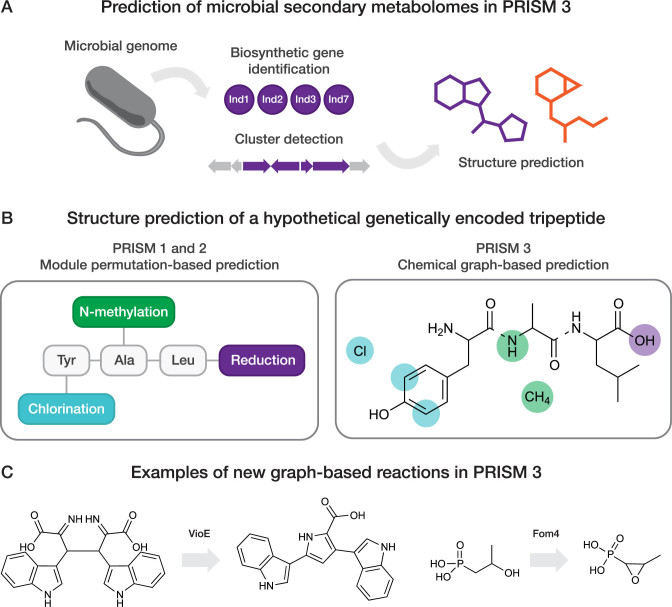
Figure 1.
(A) Schematic overview of microbial secondary metabolome prediction in PRISM 3. Following ORF detection in a microbial genome sequence, protein sequences are analyzed and clustered using a library of hidden Markov models for secondary metabolite biosynthesis genes. Identified biosynthetic information is subsequently leveraged for combinatorial prediction of secondary metabolite chemical structures. (B) Overview of chemical graph-based secondary metabolite structure prediction in PRISM 3. Modeling a natural product as a chemical graph, rather than a linear permutation of monomers, facilitates manipulation of the predicted structure at the level of individual atoms or bonds rather than at the level of the monomers. In PRISM 3, individual sets of atoms, rather than individual sets of modules, are tagged as potential sites of tailoring reactions before combinatorialization. Linkages between residues within the same subgraph are indicated as dashed lines. (C) Examples of new virtual reactions facilitated by graph-based structure prediction in PRISM 3.
Both accurate chemical structure predictions and comparison of genetically encoded chemistry to known compounds are required to reveal which evolved natural products should be targeted for isolation and testing. Recently, we described GARLIC, an algorithm to compare known and genetically encoded non-ribosomal peptides and polyketides (15). As an immediate next step, we sought to expand cluster detection and structure prediction within PRISM to a wider collection of natural product classes, with an eye toward further definition of clusters encoding novel products. A ground-up rewrite of the PRISM structure prediction algorithm was envisioned to permit prediction of more diverse reaction sequences and to effect the transformations that occur within natural product biosynthetic pathways. Here, we describe major improvements to the biosynthetic scope and functionality of PRISM in version 3 of the PRISM web server, permitting the prediction of non-modular natural product assemblies and a broader set of natural product tailoring enzymes.
CHEMICAL GRAPH-BASED STRUCTURE PREDICTION IN PRISM 3
The original iteration of the PRISM web server was designed to predict the structures of modular genetically encoded secondary metabolites: in particular, thiotemplated natural products (NRPs and type I PKs) (12,16). In these natural products, operational modules composed of multiple domains are responsible for extending the growing natural product by addition of a single residue. For example, in NRPSs, the prototypical module is composed of a condensation domain, an adenylation domain and a thiolation domain. These domains work in concert to elongate the growing peptide chain. This approach permitted increased predictive accuracy relative to existing software (12). However, this framework represented a barrier to extending PRISM to non-modular classes of natural products.
We therefore undertook a ground-up rewrite of the PRISM structure prediction algorithm, modeling the structure of the natural product scaffold as a chemical graph, rather than a linear permutation of modules (Figure 1B). In this paradigm, individual residues, or combinations of residues with a fixed pattern of connectivity (such as amino acids activated by adjacent modules on a NRPS), are represented as subgraphs within a complete chemical graph. Functional moieties introduced by tailoring enzymes, such as methyl groups added by methyltransferases, are also modeled as subgraphs. Each tailoring enzyme is associated with a reaction that adds or removes one or more bonds between or within subgraphs: for instance, a methyltransferase adds a bond between the carbon atom in the methyl subgraph and a methylation site on another subgraph within the natural product. For modular natural products, linkages between backbone residues are additionally created based on the biosynthetically rational permutations of the identified modules, as previously described (12). This approach facilitates manipulation of predicted structures at the level of individual bonds or atoms, rather than at the level of the monomer.
PRISM 3 demonstrates the utility of this approach by extending structure predictions to four classes of natural products which cannot be effectively modeled as a linear sequence of residues: aminocoumarins, bisindoles, phosphonate-containing natural products and antimetabolites (including 3-methylarginine, bacilysin/anticapsin, cycloserine, dapdiamide and indolmycin) (Table 1). We compiled 145 new HMMs and developed 131 new virtual reactions to comprehensively enable structure prediction for natural products of these biosynthetic classes. Supplementary Table S1 describes the rules for cluster detection of the new families in PRISM 3, while Supplementary Data S1 provides all HMMs and virtual tailoring reactions added in PRISM 3. We validated structure predictions by calculating the Tanimoto coefficient (Tc) of predicted structures to the known products of 31 biosynthetic gene clusters, using the ECFP6 fingerprint (17). Because more than one structure may be generated for each input biosynthetic gene cluster, we calculated both the median and maximum Tc within each predicted structure library. PRISM 3 structure libraries had an average median Tc of 0.67 (Figure 2A) and an average maximum Tc of 0.81 (Supplementary Figure S1A) to the corresponding known cluster products. This is comparable to the average median Tc of 0.69 reported for RiPPs (13) and considerably higher than the average median Tc of ∼0.25 for thiotemplated natural products (12).
Table 1. Summary of biosynthetic analyzes included in PRISM 3.
| Structure prediction |
| Non-ribosomal peptides |
| Type I polyketides |
| except iterative type I polyketides, enediynes |
| Type II polyketides |
| Ribosomally synthesized and post-translationally modified peptides (RiPPs) |
| Deoxy and hexose sugar moieties |
| Aminocoumarins |
| Antimetabolites |
| Bisindoles |
| Phosphonate-containing natural products |
| Biosynthetic gene cluster detection |
| Type I polyketides |
| iterative type I polyketides, enediynes |
| Acyl homoserine lactone |
| Aryl polyene |
| Butyrolactone |
| Ectoine |
| Furan |
| Isopropylstilbene |
| Ladderane |
| Melanin |
| Phenazine |
| Phosphoglycolipid |
| Resorcinol |
| Gene detection |
| Resistance genes |
| Rare monomer biosynthesis genes |
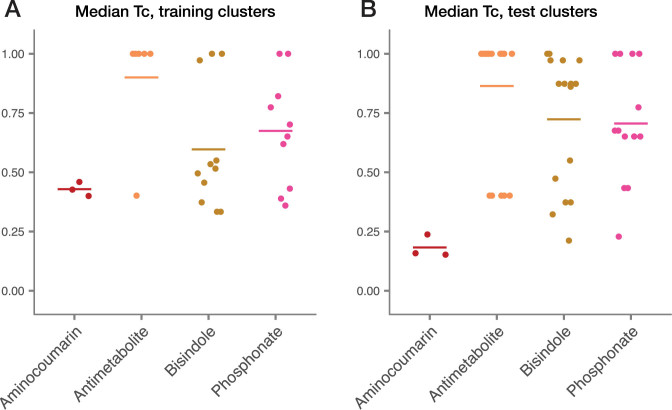
Figure 2.
Validating the accuracy of genomic structure predictions for four new classes of natural products in PRISM 3. (A) Median Tanimoto coefficients (Tc) within predicted structure libraries for clusters associated with the biosynthesis of known natural products (training set). (B) Median Tc within predicted structure libraries for clusters excluded from the training set (test set).
We additionally validated structure prediction for a second set of 54 biosynthetic gene clusters which were excluded during the development of structure prediction algorithms. For antimetabolites, bisindoles and phosphonate-containing natural products, we conducted a homology-based search using the JGI-IMG browser (18) and manually generated structure predictions based on homology to known clusters. For aminocoumarins, we included three clusters whose biosynthesis is incompletely understood (cacibiocin, rubradirin and simocyclinone) and which were therefore excluded during the development of structure prediction. PRISM 3 predicted structure libraries for this test dataset had an average median Tc of 0.75 (Figure 2B) and an average maximum Tc of 0.87 (Supplementary Figure S1B); the average Tc is likely higher for this dataset due to the inclusion of a greater number of antimetabolite biosynthetic gene clusters, which display relatively less chemical diversity than the other three classes. Thus, the design of a chemical graph-based algorithm for structure prediction in PRISM 3 enables accurate prediction of four new classes of natural products, which cannot accurately be modeled as linear permutations of monomers.
OTHER NEW FEATURES IN PRISM 3
The rapid uptake of the PRISM web application by the natural products and microbial genomics communities exceeded the capacity of the original PRISM server, leading to server downtime and restricting access for users without the technical proficiency to run PRISM on a local Tomcat server or from the command line. Consequently, we have undertaken modifications to the algorithm to permit distribution of PRISM over a 300-core server grid. We anticipate that this major investment in the infrastructure underlying the PRISM 3 web server will improve the speed and capacity of the web application.
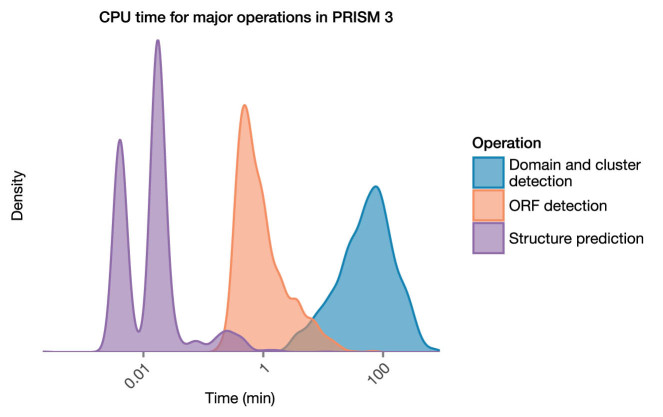
We benchmarked the speed of PRISM by using the program to analyze 2314 prokaryotic genomes from the Human Microbiome Project, with all searches enabled and default parameters. Figure 3 displays the amount of time dedicated to each of the major steps of the PRISM algorithm for each of the 2314 genomes (open reading frame (ORF) detection, domain and cluster detection, and structure prediction). The median PRISM 3 runs on a microbial genome finished in 58.8 min (interquartile range, 29.1–107.8 min). A small number of outliers (161 genomes or 7.0%) required more than 4 h to process. The longest step was domain and cluster detection (median CPU time 57.5 min), with ORF detection accounting for a median CPU time of 0.72 min. Structure prediction in PRISM 3 was highly efficient, with a median CPU time of only 0.70 s. PRISM 3 thus produces genome-wide analyzes of secondary metabolism and structure predictions for select classes of secondary metabolites within a reasonable computational time; users who wish to expedite analysis may wish to disable select libraries of HMMs in the web application interface.
Figure 3.
Benchmarking the speed of PRISM 3. Total CPU time required for the three major components of the PRISM algorithm (ORF finding, domain and cluster detection and structure prediction) for analysis of 2314 prokaryotic genomes is shown.
In addition to generating genomic structure predictions for four new classes of natural products, PRISM 3 extends cluster detection (but not structure prediction) to 11 new classes, including acyl homoserine lactones, aryl polyenes, butyrolactones, ectoines, furans, isopropylstilbenes, ladderanes, melanins, phenazines, phosphoglycolipids and resorcinols (Supplementary Table S1). Thus, the biosynthetic scope of the application has been expanded in PRISM 3 from the original three classes to predict eight and detect 22, in addition to detecting resistance genes (Table 1). We have also developed an additional 61 HMMs for the identification of antibiotic resistance determinants (Supplementary Data S1).
PRISM 3 includes several improvements to sequence input and ORF detection. An improved sequence file parser, implemented in BioJava (19), automates sequence file type detection and facilitates GBFF file input. In addition to finding all possible coding sequences between start and stop codons, the option to read ORFs directly from a GenBank file is provided. PRISM 3 also implements Prodigal (20) for prokaryotic gene recognition. All options for ORF identification are enabled by default. When ORFs identified by more than one method overlap, coordinates read from GenBank are prioritized over Prodigal, which are in turn prioritized over potential coding sequences. The option to upload open reading frame coordinates in GTF format is also provided. PRISM output in JSON format has been adjusted to reflect the changes to the web application since its inception and to provide compatibility with downstream analysis platforms, including GRAPE and GARLIC (15).
The user-friendliness of the PRISM interface has also been improved in an effort to make the web application fully accessible to users without any experience in bioinformatics or sequence analysis. A redesigned home page features ‘one-click’ submission, making PRISM analysis accessible to any user wishing to analyze a microbial genome sequence. Alternatively, a sample input can be automatically loaded. Advanced settings are hidden by default, but remain accessible to users with specific needs: in particular, users have the option to adjust the base pair window for cluster detection (set by default to 10 000), adjust the maximum size of combinatorial structure libraries generated by PRISM (set by default to 50) and adjust the methods for open reading frame prediction or input (as described above). Users additionally have the option of disabling HMM searches for individual families of biosynthetic or resistance domains. PRISM 3 includes a module to render libraries of predicted SMILES for each cluster within the browser using RDKit, increasing the accessibility of chemical structure predictions to users without use of desktop structure rendering software. Finally, a more clear and concise help page is included within PRISM 3, in addition to a fully annotated sample output from a PRISM search to facilitate result interpretation.
Some limitations of PRISM 3 should be highlighted. PRISM relies on homology to known biosynthetic gene clusters and enzymes whose biosynthetic transformations are experimentally characterized and therefore cannot identify novel biosynthetic paradigms or predict unknown enzymatic reactions. PRISM was designed primarily for prokaryotic genome analysis and consequently cannot identify biosynthetic gene clusters for families of secondary metabolites thought to be specific to eukaryotes. Finally, despite the expansion of structure prediction in PRISM 3, it is not yet possible to predict a chemical structure for every known biosynthetic pathway type.
CONCLUSION
With the development of a chemical graph-based paradigm for structure prediction and extension to eight classes of natural products, PRISM 3 represents a uniquely comprehensive resource for automated prediction of the chemical structures of genetically encoded secondary metabolites. However, despite the increased biosynthetic scope of PRISM 3, the chemical structures of a considerable range of microbial secondary metabolites remain difficult to predict except via manual annotation by specialists. Future improvements to PRISM will leverage chemical graph-based prediction to extend structure prediction to new classes of genetically encoded secondary metabolites, with the ultimate goal of integrating all available biosynthetic knowledge concerning secondary metabolism into a single framework.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science and Engineering Research Council [RGPIN 371576–2009, 101997–2006 and 388681–2011]; Canada Research Chair (CIHR) New Investigator Award (to N.A.M.); Ontario Early Investigator Award (to N.A.M.); Canada Research Chairs Program (to N.A.M.); CIHR Operating Grant (to N.A.M.); CIHR–Joint Programming Initiative on Antimicrobial Resistance Research Grant (to N.A.M.). Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. N.A.M. is a founder of Adapsyn Bioscience. M.A.S. is a consultant to Adapsyn Bioscience.
REFERENCES
- 1. Newman D.J., Cragg G.M.. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016; 79:629–661. [DOI] [PubMed] [Google Scholar]
- 2. Nett M., Ikeda H., Moore B.S.. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009; 26:1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doroghazi J.R., Albright J.C., Goering A.W., Ju K.S., Haines R.R., Tchalukov K.A., Labeda D.P., Kelleher N.L., Metcalf W.W.. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014; 10:963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Starcevic A., Zucko J., Simunkovic J., Long P.F., Cullum J., Hranueli D.. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008; 36:6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachmann B.O., Ravel J.. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009; 458:181–217. [DOI] [PubMed] [Google Scholar]
- 6. Li M.H., Ung P.M., Zajkowski J., Garneau-Tsodikova S., Sherman D.H.. Automated genome mining for natural products. BMC Bioinformatics. 2009; 10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim J., Yi G.S.. PKMiner: a database for exploring type II polyketide synthases. BMC Microbiol. 2012; 12:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziemert N., Podell S., Penn K., Badger J.H., Allen E., Jensen P.R.. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012; 7:e34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Heel A.J., de Jong A., Montalban-Lopez M., Kok J., Kuipers O.P.. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013; 41:W448–W453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cimermancic P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014; 158:412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Muller R., Wohlleben W. et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015; 43:W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L., Wyatt M.A., Magarvey N.A.. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 2015; 43:9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skinnider M.A., Johnston C.W., Edgar R.E., Dejong C.A., Merwin N.J., Rees P.N., Magarvey N.A.. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6343–E6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnston C.W., Skinnider M.A., Dejong C.A., Rees P.N., Chen G.M., Walker C.G., French S., Brown E.D., Berdy J., Liu D.Y. et al. Assembly and clustering of natural antibiotics guides target identification. Nat. Chem. Biol. 2016; 12:233–239. [DOI] [PubMed] [Google Scholar]
- 15. Dejong C.A., Chen G.M., Li H., Edwards M.R., Rees P.N., Skinnider M.A., Webster A.L.H., Magarvey N.A.. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol. 2016; 12:1007–1014. [DOI] [PubMed] [Google Scholar]
- 16. Walsh C.T., Chen H., Keating T.A., Hubbard B.K., Losey H.C., Luo L., Marshall C.G., Miller D.A., Patel H.M.. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 2001; 5:525–534. [DOI] [PubMed] [Google Scholar]
- 17. Rogers D., Hahn M.. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010; 50:742–754. [DOI] [PubMed] [Google Scholar]
- 18. Markowitz V.M., Chen I.M.A., Palaniappan K., Chu K., Szeto E., Pillay M., Ratner A., Huang J., Woyke T., Huntemann M. et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2013; 42:D560–D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prlic A., Yates A., Bliven S.E., Rose P.W., Jacobsen J., Troshin P.V., Chapman M., Gao J., Koh C.H., Foisy S. et al. BioJava: an open-source framework for bioinformatics in 2012. Bioinformatics. 2012; 28:2693–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.