Abstract
Minepath (www.minepath.org) is a web-based platform that elaborates on, and radically extends the identification of differentially expressed sub-paths in molecular pathways. Besides the network topology, the underlying MinePath algorithmic processes exploit exact gene-gene molecular relationships (e.g. activation, inhibition) and are able to identify differentially expressed pathway parts. Each pathway is decomposed into all its constituent sub-paths, which in turn are matched with corresponding gene expression profiles. The highly ranked, and phenotype inclined sub-paths are kept. Apart from the pathway analysis algorithm, the fundamental innovation of the MinePath web-server concerns its advanced visualization and interactive capabilities. To our knowledge, this is the first pathway analysis server that introduces and offers visualization of the underlying and active pathway regulatory mechanisms instead of genes. Other features include live interaction, immediate visualization of functional sub-paths per phenotype and dynamic linked annotations for the engaged genes and molecular relations. The user can download not only the results but also the corresponding web viewer framework of the performed analysis. This feature provides the flexibility to immediately publish results without publishing source/expression data, and get all the functionality of a web based pathway analysis viewer.
INTRODUCTION
Contemporary bioinformatics focuses on enhanced methods that integrate heterogeneous types of established biological knowledge (e.g. -omics data). Gene expression (GE) experiments and molecular pathways with their respective Gene Regulatory Networks (GRNs) present two of the most utilised data and knowledge sources. One of the major research lines in the field, called pathway analysis, is the identification of phenotype differentially expressed GRNs or GRN sub-paths. Most of the current pathway analysis approaches are mainly based on over representation, functional class scoring and topology analysis in order to identify differentially expressed pathways or sub-paths (1). While the methodologies underlying these approaches varies significantly, most of them provide as output just a simple list of statistical significance differential pathways or genes/proteins (2). Other approaches use maps or topology from pathway databases and color gradients, similarly to the heatmap methodology, or assign different colors to genes in order to visually indicate phenotype differential sub-paths (3). The main drawback of such visualization approaches is their gene-oriented view, which in practice is unable to handle differentially expressed sub-paths. These tools visualize only one out of the two phenotypes for downregulated and upregulated genes but, downregulation of genes for a specific phenotype does not necessarily mean that these relations are differentially expressed and upregulated for the other phenotype. One has to view two times the same (very complex) network with up/down regulated genes per phenotype in order to come up to a safe conclusion. Pathway analysis with visualization capabilities like CellWhere (4), KeyPathwayMinerWeb (5), PathAct (6), Graphite Web (7), NetworkTrial (8), ReactomeFIViz (9), EnrichNet (10), Paradigm (11) or the commercial Ingenuity Pathway Analysis IPA use such color-coding schemas for genes to highlight the differential power of the underlying molecular relations. GAM (12) and PATHiWAYS (13) visualize genes and the corresponding relations with the same color coding, which essentially suffers from the same drawback. PathAct (6) provides a relations color coding schema for simulating the effect of a drug in a network, while the differentially expressed sub-paths are again visualized using a gene color-coding schema. Another limitation of current pathway analysis approaches is that they neglect to rank and visualize relations that are functional for both target phenotypes (i.e. not differential). Even though such relations exhibit no differential information, the composition of them with functional sub-paths for target phenotypes may reveal biologically relevant and important pathway routes and regions. This is crucial in order to efficiently serve the researcher's’ exploratory needs and requirements.
Here, we present the MinePath web-server, a free interactive pathway analysis web tool armed with a powerful pathway analysis algorithm, aiming to facilitate and ease the identification and visualization of differentially active paths and able to overcome the aforementioned visualization limitations. The algorithm behind the MinePath web-server takes into account all possible functional interactions in the respective pathway GRNs. Gene-expression profiles and their phenotype assignments are integrated with targeted GRN sub-paths and evaluated for the identification of the most discriminant and informative ones. A short description of the algorithmic approach can be found in Material and Methods section.
Furthermore, MinePath web-server offers the ability to download not only the results but also the interactive web viewer of the analysis which facilitates immediate publication of analysis without publishing source data (i.e. gene expression).
MATERIALS AND METHODS
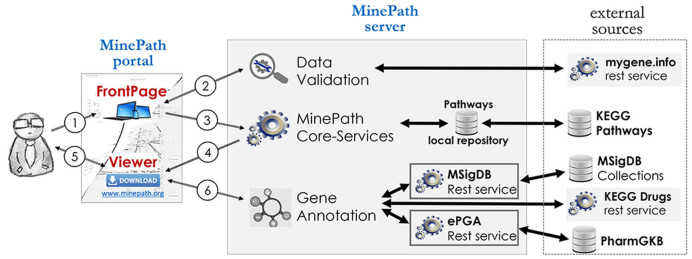
The MinePath web-server has been implemented as a Web 2.0 application, with no installation required and the utilized (private) data are not viewable by anyone other than the user. The architecture behind the server is based on the frontend–backend software design while AJAX calls are used for communication between the different server components. The user interface of the MinePath web-site is a javascript implementation that takes advantage of the open source version of Ext-JS (www.sencha.com/products/extjs/) and the Cytoscape.js (http://js.cytoscape.org) libraries. The backend of MinePath incorporates five interoperable and interacting rest-based services, which are fed with data and information from various established biological databases such as: mygene.info (14), KEGG DBs (15), PharmGKB (16) and MSigDB (17). The high-level architecture and the flow of operations for the MinePath server are shown in Figure 1.
Figure 1.
Minepath Architecture and flow of operations diagram.
The use of MinePath is relatively simple and straightforward. As it can be observed in Figure 1, the user interacts only with the MinePath FrontPage or viewer, while computations, integration and linkage with external biological data sources and the data validation are hidden to the user. The MinePath pathway analysis flow of operations is unfold into six steps. Step 1: from the MinePath web-page (www.minepath.org) the user may select or upload a gene expression dataset as well as the target pathway GNRs he/she wants to focus his/her analysis (in its current version MinePath supports the KEGG pathways). Step 2: if the user uploads a private dataset the system will validate the data and if needed will invoke the mygene.info rest service to annotate the gene names/probesets with the respective EntrezIDs entries. Step 3: the selected pathways along with the selected/uploaded gene expression data are sent to the MinePath core-services that perform the algorithmic pathway analysis processes, with the differentially expressed sub-paths to be computed in real-time. Step 4: the results are sent to the MinePath Viewer. Step 5: the researcher may select and view any pathway he/she wants and interact with the system in an interactive and user-friendly mode, which offers immediate visualization of differentially expressed relations/sub-paths, as well as dynamic annotations for the engaged genes and relations. Step 6: information related to genes or gene-groups are served on demand by calling respective rest services, while the user is able to explore the differentially expressed pathway sub-paths. A detailed description of the FrontPage, the MinePath core service and the MinePath Viewer follows.
MinePath FrontPage
The FrontPage of MinePath provides details about the usage of the web-server, fast run examples (one click away), the MinePath user manual, and the pathway analysis parameterization options. The user can select one of the public gene expression datasets available in the system (31 datasets in the current release) or upload his/her own dataset. The uploaded dataset is private, viewable just by the uploader, with the data to be deleted as soon as the processing of MinePath ends, while the analysis results are discarded after the user exits the platform using session cookies. If the user uploads a dataset that needs to be annotated (i.e. the gene names are not in the expected keggID or EntrezID format) the system will try to automatically annotate the probesets using the mygene.info web-server (http://mygene.info). If the two phenotypes (classes) assigned to the samples could not be automatically identified by the system, a new window will appear that allows to the user to set the proper phenotype to the samples. The user may categorize each sample to two phenotypes or choose to ignore the sample. From the FrontPage the user may optionally set parameters regarding: the thresholds for the differential significance of sub-paths, the threshold for sub-paths to be considered as active for both phenotypes, and the activation or not of the P-value or FDR adjusted P-value filters.
MinePath core service
The MinePath core pathway analysis takes advantage of the topology and the underlying regulatory mechanisms of GRNs, including the direction and the type of the engaged interactions. The algorithmic process includes five modular steps:
Discretization of gene expression data into binary states for up-regulated and down-regulated genes (details regarding the discretization process can be found in (18) and (19)).
Decomposition of the targeted and selected GRNs into their constituent sub-paths including the overlapping sub-paths (e.g. A → B —| C is decomposed into three sub-paths, A → B, B —| C and A → B —| C)
Computation of the functional state for each sub-path as a binary ordered-vector (e.g. A → B —| C is considered functional when A↑ and B↑ are up-regulated and C↓ is down-regulated resulting to the binary vector <1,1,0>)
Matching of sub-paths’ binary vectors against the discretized gene-expression sample profiles
Computation of the differential power (polarity rank) of each sub-path along with their respective p-value and Benjamini & Hochberg FDR scores.
The results contain phenotype discriminant pathways and sub-paths, which are passed to the MinePath Viewer for visualization and exploration of regulatory mechanisms. A detailed description of the methodology and thorough validation of the algorithm using various independent train and test datasets, including microarray and RNA-seq expression data, can be found in (20).
MinePath viewer
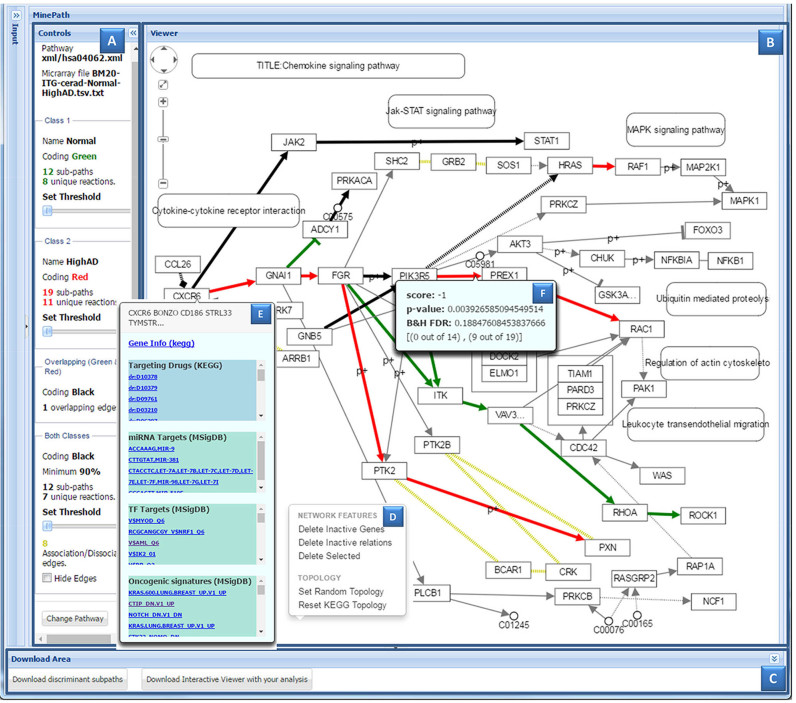
Contrary to other pathway analysis visualization tools, MinePath calculates and visualizes differentially expressed relations and sub-paths instead of just differential genes. To our knowledge this is the first pathway analysis server that introduces and offers visualization of the underlying pathway regulatory machinery. The MinePath Viewer contains three panels, ‘Controls’ (as shown in Figure 2 part A), ‘Viewer’ (Figure 2B), and ‘Download’ (Figure 2C). The ‘Viewer’ panel visualizes the differentially expressed sub-paths on the selected pathway while the KEGG layout topology is preserved. Green edges encode sub-path relations that are functional for phenotype 1 (‘Normal’ for this experiment); red edges encode relations that are functional for phenotype 2 (‘HighAD’, high-risk for Alzheimer disease), black edges encode relations that are functional and active in both phenotypes; and grey edges encode non-functional and inactive relations. The ‘Control’ panel (Figure 2A) supports active interaction and immediate visualization of sub-paths when the user sets new thresholds (using the respective sliders) for the most significant sub-paths or for the sub-paths that are functional for both phenotypes. It further supports the option to hide/show the overlapping relations and the underlying pathway association-dissociation relations (coloured in yellow; these relations are just visualized and are not taken in consideration in the computations for differential sub-paths). In addition, MinePath is equipped with network layout adjustment functionality, enabling the reduction of network's complexity (deletion of genes, relations and/or parts of the network) as well as re-orientation of its topology—a menu with these options appears when right-clicking in the ‘Viewer’ window (Figure 2D). When the user selects (clicks on) a gene or a gene group (a node in the visualized pathway) custom-made and external rest-based interfaces are triggered and respective annotation information is provided (Figure 2E) regarding the corresponding: drug targets (the KEGG DRUG DB rest-service is utilized); miRNA targets, transcription factor targets and gene signatures from MSigDB (http://software.broadinstitute.org/gsea/msigdb); and gene variants and associated drugs from PharmGKB (www.pharmgkb.org). When the user selects a relation the viewer shows information about its respective polarity score, P-value, FDR value and the number of phenotype samples that the specific sub-path is functional and active (Figure 2F).
Figure 2.
The MinePath viewer for BM20 region normal versus high Alzheimer's disease based on the CERAD classification. (A) The ‘Controls’ panel. (B) The ‘Viewer’ that visualizes the Chemokine signalling pathway for the specific analysis. (C) The download area. (D) The network layout adjustment functionality. (E) Gene's annotation information from KEGG, MSigDB and PharmGKB. (F) Relation scoring (polarity, P-value, FDR, coverage).
Apart from the rich visualization features, MinePath gives the option to the user to download not only the results but the whole analysis web viewer framework (Figure 2C). This feature provides to the users the flexibility to immediately publish results without publishing source/expression data and get all the functionality (interactive options) of the MinePath viewer. In an attempt to enable and support ‘microattribution’ services (21), the downloaded viewer contains a watermark of the expression dataset name. A link to the results, a tab-delimited file of the differential and discriminant sub-paths, is also provided—this file could be downloaded for further analysis (e.g. utilizing machine learning methods) inquiries. MinePath Viewer source code is freely available and licenced under GLPv3 (https://bitbucket.org/koumakis/minepathviewer).
RESULTS
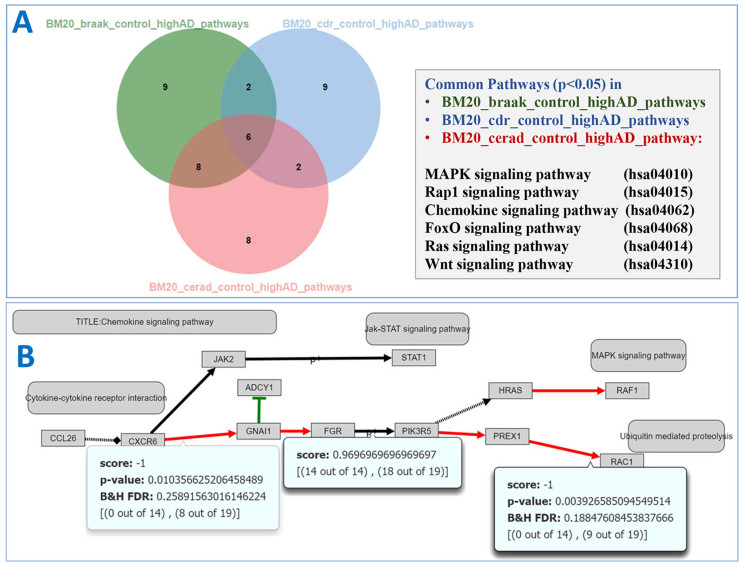
We applied MinePath to analyse a large cohort of gene expression data for Alzheimer disease (AD) coming from the Mount Sinai Brain Bank Expression Array Data (22). The dataset contains 1053 postmortem brain samples of 125 individuals across 19 cortical regions. We selected four brain regions based on the strongest association with gene expression changes in cases with AD compared to controls (22): inferior temporal gyrus (Brodmann area 20 or BM20), middle temporal gyrus (BM21), inferior frontal gyri (BM44) and superior frontal gyri (BM8). For the phenotype inclination, we followed the same procedure with the authors of the original paper and divided samples into three groups, normal, low disease severity and high disease severity, based on one clinical (clinical dementia rating or CDR) and two neuropathological (Braak tangle staging and amyloid plaques) phenotypes. This procedure generated 36 paired datasets (three pairwise group comparisons across four brain regions and three phenotypes). We applied MinePath on the 36 datasets and downloaded the results along with the MinePath viewer. For ease of them we developed a web page that shows all MinePath analyses results, accompanied with respective and dynamically generated Venn diagrams for the phenotype differential and highly discriminant sub-paths (P-value < 0.05) and pathways; for this the open source javascript library jvenn is utilized (23). The web-site with the MinePath AD experiments present an example for immediate publication of MinePath results using the MinePath Viewer (accessed at http://www.minepath.org/Alzheimer). We provide an illustrative example for the intersection of the significant pathways and sub-paths across all three phenotypes discovered in the BM20 of controls and cases with high disease severity (Figure 3A). The chemokine signalling pathway, one of the six significant pathways found common across the three phenotypes, is associated with AD and this is consistent with multiple previous studies that emphasize the importance of cytokines in inflammatory and anti-inflammatory processes in AD (24). In the chemokine signaling pathway, RAC1 is one of the genes that exhibits increased activity in cases with AD as shown in Figure 3B (PREX1 → PAC1 in red, denotes an activation relation for the AD phenotype). Interestingly, RAC1 inhibition negatively regulates transcriptional activity of the amyloid precursor protein gene (25), which have been associated with AD. The Viewer in Figure 2 shows the Chemokine signalling pathway for BM20 region, Normal vs high disease severity based on the CERAD classification, while Figure 3B shows the same pathway, simplified using the MinePath network layout adjustment functionality. MinePath identified as functional for high AD severity the sub-paths CXCR6 → GNAI1 → FGR and PIK3R5 → PREX1 → RAC1, while the FGR → PIK3R5 has been identified as functional for both normal and high AD severity samples. The unique feature of MinePath for identification and visualization of sub-paths that are functional for both target phenotypes revealed a complete functional route for high AD severity from the CXCR6 (C-X-C chemokine receptor type 6) protein to the RAC1 gene (CXCR6 → GNAI1 → FGR → PIK3R5 → PREX1 → RAC1). Other pathway analysis tools would fail to identify such a route since FGR → PIK3R5 has no differential power and would be rejected as non-significant.
Figure 3.
Venn diagram and Chemokine signalling pathway for BM20 region Normal vs high disease severity. (A) Venn diagram for significant pathways of BM20 region Normal versus high disease severity as characterized by Braak (green), CDR (blue) and CERAD (pink). (B) Chemokine signalling pathway for BM20 Normal versus high disease severity (CERAD classification).
DISCUSSION
MinePath introduces a pathway analysis methodology that directly exploits the topology as well as the underlying pathway regulatory mechanisms, including the direction and the type (activation, inhibition) of the engaged regulatory relations. This is in contrast with the traditional pathway analysis methodologies that employ ‘gene set enrichment’ approaches. But pathways are richer and encompass much more knowledge than just a plain list of genes, such as the topology and the involved gene regulatory relations recorded in the respective pathway networks. The web-based server deployment of MinePath overcomes the fundamental limitations of current pathway analysis methodologies and offers a productive environment with efficient, interactive and user-friendly visualization capabilities. It supports live interaction, immediate visualization of phenotype differential regulatory relations (a simple color-coding schema is employed for this), and it is equipped with special topological and network-adjustment functionality. Armed with the aforementioned features and functionality, MinePath may effectively serve the exploratory needs of biomedical researchers to gain insight into regulatory mechanisms that underlie and putatively govern the expression of target phenotypes.
MinePath has been thoroughly tested for its stability. Additional functionality is foreseen in planned future releases of the platform, such as automated uploading of microarray data from public sources (e.g. GEO), merging of gene-expression datasets (to serve meta-analysis needs), as well as mechanisms that enable on-the-fly merging and visualization of two or more pathways in order to enrich exploratory quests.
FUNDING
This work has been partially supported by the FP7-ICT-2011.7 No 288048 EURECA project and the National Institutes of Health [R01AG050986 to P.R., R01MH109677 to P.R.]; Brain Behavior Research Foundation [20540 to P.R.]; Alzheimer's Association [NIRG-340998 to P.R.]; Veterans Affairs [Merit grant BX002395 to P.R.]. Funding for open access charge: European Union under the FP7-ICT programme [FP7-ICT-2011.7 No 288048 EURECA project (Enabling information re-Use by linking clinical REsearch and CAre)], National Institutes of Health [R01AG050986 to P.R., R01MH109677 to P.R.]; Brain Behavior Research Foundation [20540 to P.R.]; Alzheimer's Association [NIRG-340998 to P.R.]; Veterans Affairs [Merit grant BX002395 to P.R.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement. None declared.
REFERENCES
- 1. Khatri P., Sirota M., Butte A.J.. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput. Biol. 2012; 8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Habermann B., Villaveces J., Koti P.. Tools for visualization and analysis of molecular networks, pathways, and -omics data. Adv. Appl. Bioinforma. Chem. 2015; 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garc’ia-Campos M.A., Espinal-Enr’iquez J., Hernández-Lemus E.. Pathway analysis: state of the art. Front. Physiol. 2015; 6:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu L., Malatras A., Thorley M., Aghoghogbe I., Mer A., Duguez S., Butler-Browne G., Voit T., Duddy W.. CellWhere: Graphical display of interaction networks organized on subcellular localizations. Nucleic Acids Res. 2015; 43:W571–W575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. List M., Alcaraz N., Dissing-Hansen M., Ditzel H.J., Mollenhauer J., Baumbach J.. KeyPathwayMinerWeb: online multi-omics network enrichment. Nucleic Acids Res. 2016; 44:gkw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salavert F., Hidago M.R., Amadoz A., Çubuk C., Medina I., Crespo D., Carbonell-Caballero J., Dopazo J.. Actionable pathways: interactive discovery of therapeutic targets using signaling pathway models. Nucleic Acids Res. 2016; 10.1093/nar/gkw369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sales G., Calura E., Martini P., Romualdi C.. Graphite Web: Web tool for gene set analysis exploiting pathway topology. Nucleic Acids Res. 2013; 41:W89–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stöckel D., Müller O., Kehl T., Gerasch A., Backes C., Rurainski A., Keller A., Kaufmann M., Lenhof H.-P.. NetworkTrail—a web service for identifying and visualizing deregulated subnetworks. Bioinformatics. 2013; 29:1702–1703. [DOI] [PubMed] [Google Scholar]
- 9. Wu G., Dawson E., Duong A., Haw R., Stein L.. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Research. 2014; 3:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glaab E., Baudot A., Krasnogor N., Schneider R., Valencia A.. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012; 28:i451–i457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sedgewick A.J., Benz S.C., Rabizadeh S., Soon-Shiong P., Vaske C.J.. Learning subgroup-specific regulatory interactions and regulator independence with PARADIGM. Bioinformatics. 2013; 29:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sergushichev A.A., Loboda A.A., Jha A.K., Vincent E.E., Driggers E.M., Jones R.G., Pearce E.J., Artyomov M.N.. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res. 2016; 44:gkw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sebastian-Leon P., Vidal E., Minguez P., Conesa A., Tarazona S., Amadoz A., Armero C., Salavert F., Vidal-Puig A., Montaner D. et al. . Understanding disease mechanisms with models of signaling pathway activities. BMC Syst. Biol. 2014; 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xin J., Mark A., Afrasiabi C., Tsueng G., Juchler M., Gopal N., Stupp G.S., Putman T.E., Ainscough B.J., Griffith O.L. et al. . High-performance web services for querying gene and variant annotation. Genome Biol. 2016; 17:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016; 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whirl-Carrillo M., McDonagh E.M., Hebert J.M., Gong L., Sangkuhl K., Thorn C.F., Altman R.B., Klein T.E.. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012; 92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M. a, Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Potamias G., Koumakis L., Moustakis V.. Gene Selection via Discretized Gene-Expression Profiles and Greedy Feature-Elimination. Methods and Applications of Artificial Intelligence, Third Helenic Conference on AI, {SETN} 2004, Samos, Greece, May 5–8, 2004, Proceedings. 2004; 256–266. [Google Scholar]
- 19. Koumakis L., Moustakis V., Zervakis M., Kafetzopoulos D., Potamias G.. Coupling regulatory networks and microarays: Revealing molecular regulations of breast cancer treatment responses. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2012; 7297:239–246.LNCS. [Google Scholar]
- 20. Koumakis L., Kanterakis A., Kartsaki E., Chatzimina M., Zervakis M., Tsiknakis M., Vassou D., Kafetzopoulos D., Marias K., Moustakis V. et al. . MinePath: mining for phenotype differential sub-paths in molecular pathways. PLoS Comput. Biol. 2016; 12:e1005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patrinos G.P., Cooper D.N., van Mulligen E., Gkantouna V., Tzimas G., Tatum Z., Schultes E., Roos M., Mons B.. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum. Mutat. 2012; 33:1503–1512. [DOI] [PubMed] [Google Scholar]
- 22. Wang M., Roussos P., McKenzie A., Zhou X., Kajiwara Y., Brennand K.J., De Luca G.C., Crary J.F., Casaccia P., Buxbaum J.D. et al. . Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer's disease. Genome Me. 2016; 8:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C.. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014; 15:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rubio-Perez J.M., Morillas-Ruiz J.M.. A review: inflammatory process in Alzheimer's disease, role of cytokines. Sci. World J. 2012; 2012:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang P.-L., Niidome T., Akaike A., Kihara T., Sugimoto H.. Rac1 inhibition negatively regulates transcriptional activity of the amyloid precursor protein gene. J. Neurosci. Res. 2009; 87:2105–2114. [DOI] [PubMed] [Google Scholar]