Abstract
Many biological processes are mediated by complex interactions between DNA and proteins. Transcription factors, various polymerases, nucleases and histones recognize and bind DNA with different levels of binding specificity. To understand the physical mechanisms that allow proteins to recognize DNA and achieve their biological functions, it is important to analyze structures of DNA–protein complexes in detail. DNAproDB is a web-based interactive tool designed to help researchers study these complexes. DNAproDB provides an automated structure-processing pipeline that extracts structural features from DNA–protein complexes. The extracted features are organized in structured data files, which are easily parsed with any programming language or viewed in a browser. We processed a large number of DNA–protein complexes retrieved from the Protein Data Bank and created the DNAproDB database to store this data. Users can search the database by combining features of the DNA, protein or DNA–protein interactions at the interface. Additionally, users can upload their own structures for processing privately and securely. DNAproDB provides several interactive and customizable tools for creating visualizations of the DNA–protein interface at different levels of abstraction that can be exported as high quality figures. All functionality is documented and freely accessible at http://dnaprodb.usc.edu.
INTRODUCTION
Interactions between proteins and DNA play key roles in many biological processes. Gene regulation and transcription, chromatin formation and organization, as well as DNA replication, repair and recombination are driven by proteins that bind DNA through various mechanisms and at varying levels of binding specificity. Through the structural analysis of proteins bound to DNA binding sites, researchers gain insight into the physical mechanisms that underlie protein biological functions. A number of studies which survey DNA–protein complexes have been performed that classify DNA binding proteins based on the structure of the complexes that they form with DNA (1–6) or to probe mechanisms of DNA recognition based on structure analysis (7–10). Other studies have analyzed structures of DNA–protein complexes to understand the binding mechanisms of individual proteins or protein families (11–15).
The number of DNA–protein complexes available in the Protein Data Bank (PDB) (16) continues to increase; at present there are 3868 such complexes. Consequently, automated tools are needed that can quickly analyze and compare such large structural datasets. These tools should be capable of producing high-quality visualizations automatically to highlight how proteins in the complex interact with and bind to DNA.
Many databases and web servers have been developed that provide information on structural aspects of DNA–protein complexes. For example, PDIdb (17) provides detailed information about each DNA–protein interface in a complex and classifies proteins by function and structure. Users can search the database for entries based on features of the interface, DNA or protein. WebPDA (18) is a web server that analyzes DNA–protein contacts in PDB structures and depicts minor groove and major groove interactions with three-dimensional (3D) visualizations. OnTheFly (19) is a database of transcription factors (TFs) from Drosophila melanogaster and their DNA-binding sites. The DNA–protein interface is annotated by using the MarkUs function annotation server (20). DOMMINO (21) provides data on macromolecular interactions between protein subunits and DNA. This database also includes protein–protein and protein–RNA interactions. The 3D-footprint database (22) provides structure-based binding specificities for all DNA–protein complexes in the PDB and figures that display DNA–protein interactions in the complexes.
Other databases and web servers have been published, but many are out of date or no longer functional. Available web tools and analysis methods are often centered solely on the protein, DNA or interface, with few tools providing information on a wide variety of features. Moreover, few tools allow users to search for structures based on extracted information, produce high-quality customizable visualizations and upload unpublished structures for analysis with the same toolset as is available for published structures.
The DNAproDB web server can be used to perform structure analysis of DNA–protein complexes and extract structural features from the complex via an automated structure-processing pipeline. This pipeline was custom built using a software stack that incorporates many commonly used structure analysis tools and combines generated information in the JavaScript Object Notation (JSON) data format. Users can upload structures to the DNAproDB web server in a secure and private fashion for automatic processing, thereby simplifying the task of structure analysis. In addition, we retrieved DNA–protein complexes from the PDB and processed these complexes using our processing pipeline. Processed data were used to construct the DNAproDB database, which users can search based on features of the DNA, protein or DNA–protein interactions. At present, the database contains 2441 DNA–protein complexes and will be updated regularly with newly released PDB structures. We provide in-browser tools for producing unique, high quality, interactive and customizable visualizations for any structure in our database or that the user has uploaded. These functionalities are available at our website, http://dnaprodb.usc.edu, which has accompanying documentation. We describe the processing pipeline, database and visualization tools below.
STRUCTURE PROCESSING PIPELINE
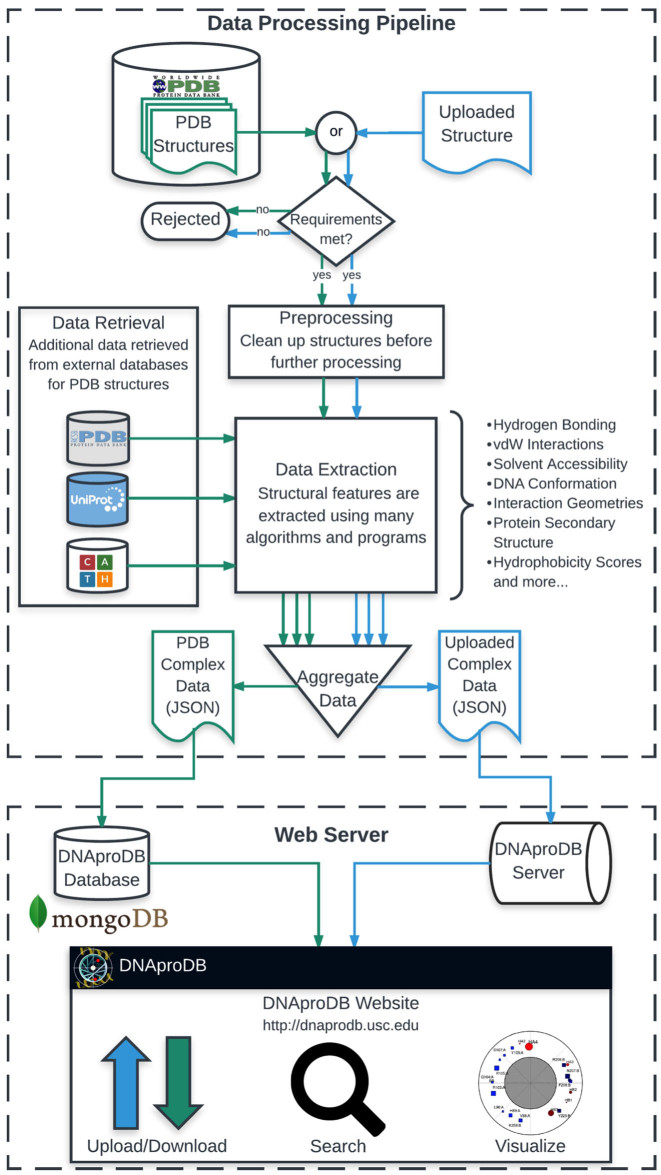
The DNAproDB structure-processing pipeline (Figure 1) takes as input the coordinates of DNA–protein complex structures in PDB or mmCIF format (23) and extracts structural features. The pipeline, which is implemented in Python, relies on well-established published libraries and software. The pipeline has two primary functions: (i) to automate many of the common tasks involved in extracting features for structure analysis of DNA–protein complexes or features that are useful when searching for these complexes, and (ii) to organize extracted features in a consistent and meaningful way. The pipeline consists of five major stages as outlined below.
Figure 1.
Schematic overview of the DNAproDB structure processing pipeline. The main stages are structure pre-processing, feature extraction, data retrieval and data aggregation. The DNAproDB database stores processed structural data for more than 2400 DNA–protein complexes. Users can search the database using features of the DNA, protein or DNA–protein interactions, can generate reports for the returned results and can upload their own structures for private analysis. The report page contains functionality for downloading extracted features as a JSON file and for visualizing data using interactive visualization tools, which can be used to explore interactions between the DNA and protein and can be exported for use as static figures.
Structure requirements
The DNAproDB pipeline processes structures of DNA–protein complexes that contain one or more protein chains bound to a single helical region of double-stranded DNA (dsDNA). Structures containing single-stranded DNA, multiple double helices or DNA forms such as Holliday junctions or G-quadruplexes are currently not supported. The total molecular weight of the structure must be no larger than 201 000 Da, and the dsDNA must contain at least five base pairs (bp) to ensure meaningful calculations of major and minor groove features. If an uploaded structure does not meet any of these requirements, then the user is notified via an error message and structures available in the PDB that do not meet these requirements will not be available in the DNAproDB database.
Of the 3886 DNA–protein complexes currently available in the PDB, ∼90% fall within the specified total molecular weight. Of these remaining structures, ∼15% contain multiple double helical regions, 9% contain no helical region, <1% contains fewer than five base pairs, 2% contain too many non-standard residues or residues with many missing atoms and 5% could not be processed for miscellaneous reasons. The resulting total number remaining thus corresponds to the 2441 structures we currently provide in the DNAproDB database.
Structure pre-processing
The first pre-processing step is to generate coordinates of the biological assembly via symmetry operations, which must be included with an uploaded structure file. Any new chains generated from these symmetry operations are assigned unique identifiers, which will appear in the structure reports (see ‘Structure Analysis’ section) and output of the processing pipeline. For any chain that is generated from a symmetry operation, its parent chain in the asymmetric unit will be clearly identified. In the case of uploaded structures in PDB file format, the provided coordinates must already be those of the relevant biological assembly. Structure files are parsed using the PDB module of the Biopython package (24), which is also used throughout the pipeline.
Residues or nucleotides that are missing more than 50% of their heavy atoms (excluding terminal oxygens) are removed by default during pre-processing because some of the incorporated programs used in the feature extraction stage of the pipeline can produce errors if a residue or nucleotide is missing heavy atoms. We do provide, however, an option for the user to add missing heavy atoms to an uploaded structure using the program PDB2PQR (25,26). Hydrogen atoms are added to the structure using Reduce (27) of the MolProbity software suite (28).
We currently support 19 commonly occurring chemically modified nucleotides and protein residues, including 5-methylcytosine (Supplementary Table S1). Any non-standard nucleotide or residue that is currently not supported is removed from the structure before further processing.
Feature extraction
In the feature extraction stage, structural features of the complex are extracted using various incorporated programs and libraries. DNA base pairing, shape parameters and conformation are derived from the 3DNA program suite (29) with a 10.0 Å cut-off for helix breaking. The DNA helical axis is calculated with CURVES (30). For each protein chain, DSSP (31) is used to assign a three-state protein secondary structure. Various components of the solvent-accessible surface area (SASA) for individual residues and nucleotides and the buried solvent accessible surface-area (BASA) between individual residues and nucleotides are calculated using the library freeSASA (32), which implements the Lee–Richards algorithm (33) with a solvent radius of 1.4 Å. These features are described in more detail in the Supplementary Data.
Hydrogen bonds are computed by HBPLUS (34) with default parameters. Van der Waals (vdW) interactions are computed using the KDTree module in Biopython (24) with a cut-off distance of 3.9 Å. Nucleotide–residue interaction geometry (stacking, pseudo-pairing or other) is determined using SNAP, a new component of the 3DNA program suite (35). SNAP also serves as a fall-back for calculating hydrogen bonds if HBPLUS cannot process the file. Hydrophobicity scores for each protein residue in the protein surface are computed using the spatial aggregation propensity (SAP) algorithm, as described in (36), with a 5.0 Å cut-off radius. Additional features mentioned in the ‘Data Aggregation’ section and Figure 2 are computed with in-house code.
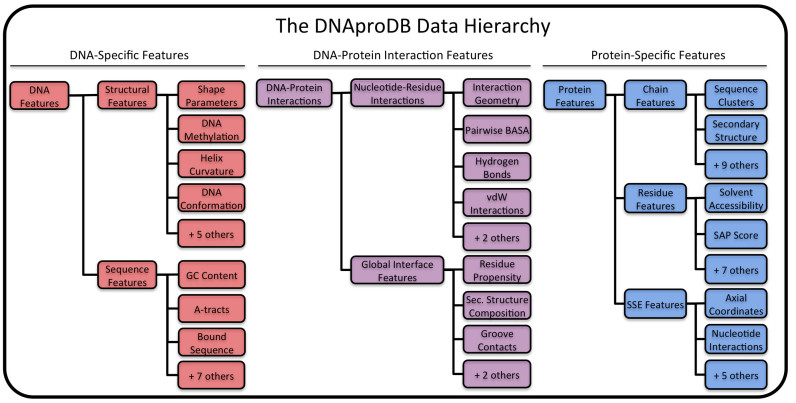
Figure 2.
Illustration of a subset of the data hierarchy used by DNAproDB. Extracted data are categorized into three feature hierarchies: DNA-specific features, protein-specific features and DNA–protein interaction features. Data generated by the processing pipeline are stored as JSON files with a near one-to-one correspondence with the hierarchy. The figure shows a subset of the available features that can easily be retrieved by parsing one of these JSON files. A feature may represent a single value or an array of values, depending on the context. DNAproDB can be searched for structures based on any combination of these features and from any of the three hierarchies.
Data retrieval
In the case of structures retrieved from the PDB, external databases provide additional information that allow for more advanced queries when searching the DNAproDB database. For every protein chain in the complex, the UniProt identifiers, protein names and source organism are retrieved from the UniProt entry (37) for that chain. The RCSB PDB (38) provides BLAST (39) sequence clusters at various sequence similarities for all protein sequences that occur in structures contained in the PDB. For each protein chain in the complex, the pipeline retrieves the representative chain for each sequence cluster the protein chain belongs to. CATH (40) structural classifications are also included for each protein chain in the database.
Data aggregation
In the final stage of the pipeline, features generated in the previous stages are parsed, organized and combined. The data is organized in three main hierarchies: protein-specific features, DNA-specific features and DNA–protein interaction features (Figure 2).
Protein features include information specific to the protein(s) in the complex, and fall under chain features, residue features or secondary structure element (SSE) features. Chain features include basic information about each protein chain (e.g. primary and secondary structure, residue identifiers, parent chain in the asymmetric unit, whether or not it interacts with the DNA in the complex). For structures retrieved from the PDB, additional external database content is stored for each chain (see the ‘Data Retrieval’ section).
Features of individual protein residues in the DNA–protein interface (i.e. amino acids interacting with the DNA) are included under residue features. Residues are judged to be part of the DNA–protein interface if the BASA value of their side-chains is ≥5% of the side-chain SASA in an Ala-X-Ala tripeptide, known as the relative SASA. For each residue, its secondary structure, parent chain, component BASA values, total number of hydrogen bonds and vdW interactions and hydrophobicity score are recorded.
SSEs are determined from the secondary structure of each chain, and features of each SSE are stored under SSE features. Contiguous segments of helix or strand residues are identified and assigned a unique identifier using their chain identifier and order in the chain. For example, the first helix (starting from the N-terminus) that appears in chain A is assigned the ID HA1, and the third strand in chain C is assigned the ID SC3. The same information that is available for residues (excluding hydrophobicity scores) is accumulated from the constituent residues of the SSE. Loop SSEs are treated slightly differently—each loop residue in the interface is considered an SSE of length one and identified according to the residue name, chain and number. The SSE identifiers are used as the default labels in the visualizations discussed in the ‘Structure Analysis’ section. In addition to the accumulated residue properties, SSEs are assigned a set of coordinates in a generalized coordinate system that we refer to as axial coordinates. In this coordinate system, every point in space is referenced with respect to a curve through the space that corresponds to the DNA helix axis. For a complete definition of the coordinate system, see Supplementary Figure S1.
A vector position for each SSE is calculated by computing the weighted vector average of each alpha carbon position of the SSE's constituent residues, where the weights used are the side-chain BASA values for each residue. This vector is then converted to axial coordinates, which are later used to generate the contact maps described in the ‘Structure Analysis’ section.
The second data hierarchy, DNA features, contains information specific to DNA in the complex and falls into structural features, sequence features or nucleotide features. Structural features contain information such as global DNA conformation, local DNA shape parameters, base-pairing information, helical curvature and Cartesian coordinates of the DNA helix axis. Sequence features describe binding-site motifs, sequence length, GC content, presence of A-tracts and other information that can be derived from sequence. Information about each nucleotide (similar to information for protein residues) is provided under nucleotide features.
The third data hierarchy, DNA–protein interaction features, describes interactions between the DNA and protein at two levels of detail. At the most detailed level, individual nucleotide–residue interactions are identified under nucleotide–residue interaction features. For each interaction, the geometry, hydrogen bonds, vdW interactions and BASA value between the residue and nucleotide are given. A nucleotide–residue interaction is determined by the presence of at least one hydrogen bond, one vdW interaction or a BASA value greater than zero. From this list of pairwise interactions, global properties of the interface can be calculated. The overall secondary structure composition of the interface (determined by the BASA), residue propensities and the total BASA, number of hydrogen bonds and number of vdW interactions in each groove by SSE type are recorded under interface features, which describe global features of the DNA–protein interface.
The data produced by the processing pipeline is output as a JSON file that can be parsed by any modern programming language while being human-readable, or can be viewed in-browser through our website. Example JSON files and full explanations of every data item are available on the documentation page at http://dnaprodb.usc.edu/documentation.
WEB SERVER INTERFACE
Database and query functionality
DNAproDB provides a database of DNA–protein complexes that are retrieved from the PDB and meet the requirements outlined in the ‘Structure Requirements’ section. The database is implemented with mongoDB (41) and stores the JSON document produced by the processing pipeline for each DNA–protein complex structure. Users may use the database to search directly for a structure or list of structures by their PDB identifiers or, more powerfully, to search for structures based on any combination of the available features (see the ‘Data Aggregation’ section and Figure 2). By combining different features, users can search for structures based on characteristics of the DNA, protein or DNA–protein interactions. Interaction features can be included in the search at the level of individual nucleotide–residue interactions or at the level of global interface properties.
For example, the user could search for structures where an arginine forms at least one hydrogen bond with a guanine in the major groove of the DNA and with the arginine being located within a helix SSE. Alternatively, the user could simply search for structures where there are any contacts in the major groove with a protein helix. This search could be combined with DNA features, such as constraining the length of the DNA target to between 8 and 20 bp and the DNA conformation to be B-form. An example is a search for structures that have helices bound in the minor groove, no major groove contacts and the DNA is at least 10-bp long. Select structures from the returned results of this search are shown in Table 1 and Supplementary Figure S2 (42–45). The reader can replicate this query and explore the different structures that are returned.
Table 1. Selected results from a search for structures with protein helices in the minor groove, no major groove contacts and a DNA length of at least 10 bp.
| PDB ID | Protein name(s) | Organism | DNA Sequence | DNA Axis Curvature | Interface SSE composition |
|---|---|---|---|---|---|
| 1J46 | Sex-determining region Y protein | Homo sapiens | CCTGCACAAACACC | Curved in-plane | Mainly Helix |
| 1JFI | Dr1-associated corepressor, TATA-box-binding protein | Homo sapiens | TGGCTATAAAAGGGCTC | Curved in-plane | Mainly Strand |
| 2GKD | CalC | Micromonospora echinospora | GCATATGATAG | Linear | Mainly Helix |
| 3U2B | Transcription Factor SOX-4 | Mus musculus | GTCTCTATTGTCCTGG | Curved in-plane | Mainly Helix |
Fifteen structures were returned in total. Summary information about the selected structures is shown, as available from the report pages of these structures. DNA axis curvature describes how the DNA helix axis is curved in 3D space. Most of these structures show DNA that is bent, which allows the minor groove to widen and better accommodate the bulky protein helices. Interface SSE composition describes the overall composition of SSEs at the DNA–protein interface, as measured by BASA. Mainly helix means helix contacts contribute most to the total interface BASA and vice versa for Mainly Strand.
The DNAproDB database provides powerful search capabilities that few other web servers or databases offer. Users can search the database to quickly retrieve data for a DNA–protein complex, discover new structures or generate datasets based on structural criteria. By combining a list of PDB identifiers and structural features, users can filter a list of known structures based on the chosen features.
Structure upload
Users can process a structure using the DNAproDB pipeline and can visualize extracted features from a generated report page for the structure by uploading a structure file of a DNA–protein complex to our server at http://dnaprodb.usc.edu/cgi-bin/upload. Users should verify that their structure meets all the listed requirements on the upload page. Once the file is uploaded, the user is given a private URL to a report page, where the user can download extracted features in JSON format or visualize features of the structure using our interactive visualization tools. User data are stored on the DNAproDB server. However, no other person can access the data unless they know the private URL, which contains a random alphanumeric string that acts as a secure password and cannot be indexed by search engines or guessed.
Structure analysis
DNAproDB generates a report page for every processed structure, whether retrieved from the PDB or uploaded to the server. Report pages provide interactive visualizations for the user that display details about the DNA–protein interface and DNA–protein interactions at different levels of abstraction.
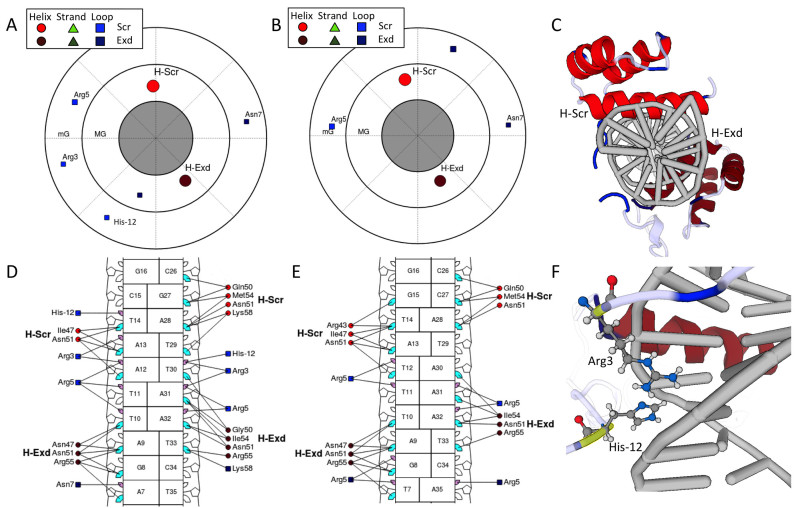
The most abstract and unique visualization is the polar contact map (Figures 3A, B and 4A, C; Supplementary Figures S2 and 3A) in which protein SSEs are plotted in a circular plot that represents a projection onto a series of planes perpendicular to the DNA helix axis. In separate annuli, the major groove, minor groove and backbone contacts are plotted. Each marker corresponds to a specific type of SSE (red circles are helices; green triangles are beta strands; and blue squares are loop residues). Markers are labeled as described in the ‘Data Aggregation’ section. Marker size indicates the total BASA of the SSE (i.e. extent of the contact between the SSE and DNA) in a particular groove. The angular position of the SSEs in the plot corresponds to the axial coordinate ϕ. The radial position within each annulus corresponds to ρ, which measures the distance from each SSE to the DNA helix axis (see Supplementary Figure S1). When looking down the DNA helix axis in the 3D view of the structure (Figure 3C), the angular position of SSEs relative to one another is reflected in the contact map.
Figure 3.
The visualizations from DNAproDB for a heterodimer of the Hox protein Sex combs reduced (Scr) and its cofactor Extradenticle (Exd) bound to two different DNA sequences (PDB IDs: 2R5Z and 2R5Y) (47). Only major groove (MG) and minor groove (mG) contacts are shown. Joshi et al. (47) showed that for this protein complex, Scr loop residues Arg3 and His−12 are important for conferring sequence specificity via shape recognition of the minor groove. In the plots of panels (A and B) and (D and E), the colored markers indicate SSEs. Helices are represented as red circles, beta strands (not present for these structures) are represented as green triangles and loop residues are represented as blue squares. (A) Polar contact map showing major groove (inner circle) and minor groove (outer circle) contacts for the Scr–Exd structure bound to the Scr in vivo site (PDB ID: 2R5Z). The angular distribution of the SSEs in the plot corresponds to the distribution about the DNA helix axis in the structure, as seen in (C). The DNA-binding domains of Scr and Exd are distinguished by applying different color shades. The Scr residues Arg3 and His−12 are seen making contacts in the DNA minor groove. (B) Polar contact map of Scr–Exd bound to a Hox consensus site (PDB ID: 2R5Y). Here, the Scr residues Arg3 and His−12 cannot be seen to make contact with the minor groove, due to differences in the intrinsic shape profile of this DNA sequence as described in (47), which explain the preference for the Scr in vivo site. (C) 3D view looking down the DNA helix in the orientation of the contact maps in (A) and (B). (D) Nucleotide–residue contact map showing individual nucleotide–residue interactions for the preferred binding site. Residues are grouped into SSEs, labeled H-Scr and H-Exd for helices in the DNA-binding domains. Small and large markers on each nucleotide represent the major and minor groove contacts, respectively. Lines joining a residue to nucleotide groove markers indicate interactions in that groove. Filled-in cyan (major groove) and pink (minor groove) markers highlight which nucleotides are contacted by at least one residue in the respective groove. (E) The same visualization as in (D) for the structure of Scr–Exd bound to a Hox consensus site. (F) Scr residues Arg3 and His−12 (highlighted in yellow) are inserted into the minor groove for the preferred binding site. The 3D view of the structures are linked with the contact maps. Clicking on the protein residues in (D) will highlight them in the 3D view (shown in yellow). Hydrogen atoms have been added to the structure as described in the main text.
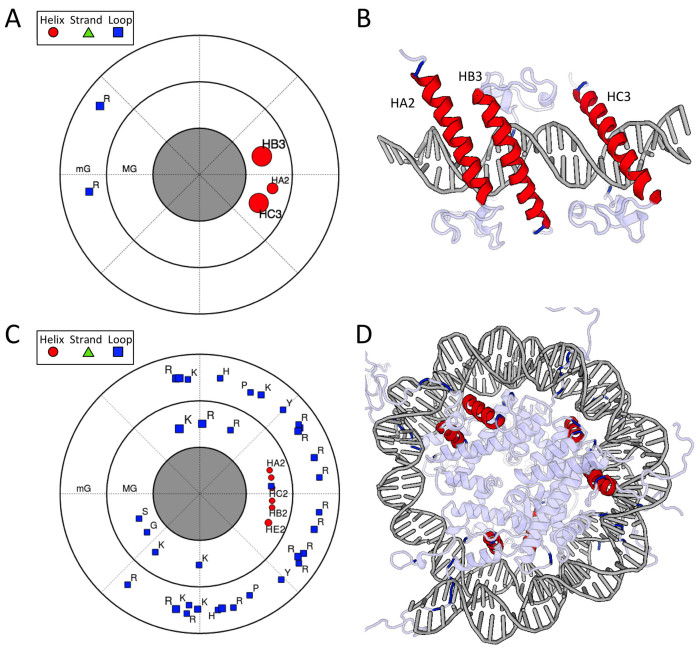
Figure 4.
Visualizations for (A and B) three DNA binding domains of the human Doublesex and Mab-3 Related Transcription factor-1 (DMRT1) with DNA (PDB ID: 4YJ0) (48) and (C and D) a nucleosome (PDB ID: 1KX5) (50). Polar contact maps and the 3D views of the structures are shown. The polar contact map represents the projection of the position of each SSE onto a plane that moves along, and is always perpendicular to, the DNA helix axis. (A) Polar contact map for a complex of three DNA-binding domains of DMRT1 bound to an essentially straight DNA target. Alpha helices bind in the major groove and positively charged arginine residues in loops contact the minor groove (49). The helix contacts cluster in a small region of the plot, which reflects that the helices are positioned on the same face of the DNA with one of the helices (HA2) forming weaker DNA contacts. Arginine residues in loop regions anchor in the minor groove on the opposite face stabilized by DNA shape readout (49). (B) 3D view of three DMRT1 DNA-binding domains with different SSEs that contact the major groove or minor groove, colored according to secondary structure (SSEs that do not make direct contact with the DNA bases are left uncolored). (C) Polar contact map for a nucleosome structure, showing major groove and minor groove contacts. The DNA is wrapped around the histone octamer, so there is no single direction in 3D space that defines the family of planes to which projections are made, as is the case for DNA with a linear helical axis such as in (A). Helices in the map cluster in a narrow region of the major groove (MG), which implies that the helices are each roughly in-phase with the helical pitch of the groove as it winds around the protein. The loop regions of the protein wrap around the DNA and make contacts along the minor groove (mG). In this sense, both protein complexes in this figure have a similar mode of binding despite the formation of very different DNA topologies in the respective structures. (D) 3D view of the nucleosome structure with the same color scheme as in (B). Only SSEs that contact the base pairs are shown in red.
Since the coordinates plotted in the polar contact map are axial, this map also works well for structures where the DNA helix axis has a large degree of curvature (Supplementary Figure S3A), as in a DNA complex with the TATA-box binding protein (Supplementary Figure S3B). This visualization allows the user to visualize what types of SSEs are bound in each groove, how they are distributed around the DNA (i.e. an enveloping or single-sided fashion) and how much contact each SSE makes. The visualization offers a very compact representation that is general enough for any DNA–protein complex for which a DNA helix axis can be defined.
Another option for visualization is the linear contact map (Supplementary Figure S3C), in which the linear DNA sequence is displayed as a ladder of base pairs. SSE contacts to each nucleotide in the DNA are shown, similar to NUCPLOT representations (46). Lines connecting SSEs and nucleotides represent interactions between them. Attached to each nucleotide are markers representing the major groove, minor groove and backbone regions of the DNA; these are used to specify which regions of the DNA are in contact with a specific SSE. Contacts to each DNA strand are shown independently; SSEs on the left side make contact with the first DNA strand, and SSEs on the right side make contact with the second DNA strand. Hence, a DNA-contacting protein domain will often appear twice in the contact map, once for each DNA strand.
In the linear contact map, the axial coordinates ρ and s are plotted (Supplementary Figure S1). ρ (the distance from the helix axis) is plotted along the horizontal axis and s (the distance along the DNA helix axis) is plotted on the vertical axis. The position of the base pairs indicates their position on the DNA helix axis; hence, the distance between adjacent base pairs is roughly equivalent to the corresponding shape parameter rise of those base pairs. As with the polar contact map, the use of axial coordinates allows these plots to be constructed regardless of DNA curvature. For simplicity, DNA is always shown as a straight ladder.
A third visualization of the interface is the nucleotide–residue contact map (Figure 3D and E; Supplementary Figure S3D), whose layout is similar to the linear contact map. This visualization shows individual nucleotide–residue interactions, where the residues are grouped into their corresponding SSEs. Axial coordinates are not used, and the position of each residue and SSE group is optimized for readability.
In all visualizations, the user can hover the mouse cursor over any SSE, residue or interaction indicator line to obtain additional information. Clicking on an SSE or residue marker will highlight that residue and display additional details in the 3D representation of the structure. Thus, the user can explore various interactions in the DNA–protein interface individually at different levels of detail and in manageable steps. The user has the option to customize the visualizations. Different SSE types and specific groove contacts can be turned on or off. The user could decide, for example, to visualize only loop contacts in the minor groove, and turn off helices and strands and all major groove and backbone contacts. Additionally, the user can apply a different color scheme to different chains in the structure; this approach is very useful for distinguishing different domains of a protein complex. Custom labels can be applied to individual SSEs, residues or nucleotides.
An example analysis of two ternary complexes of the Hox protein Sex combs reduced (Scr) and its cofactor Extradenticle (Exd) bound to two DNA targets (PDB IDs: 2R5Z and 2R5Y) (47) using the different types of visualizations including customized labels and color schemes is shown in Figure 3. Figure 4 shows two additional examples where the polar contact map illustrates that proteins can bind DNA in a similar manner despite drastically different topologies of the DNA in these complexes. Figure 4A and B show the quaternary complex of three DNA-binding domains of the Doublesex and Mab-3 Related Transcription factor-1 (DMRT1) with DNA (PDB ID: 4YJ0) (48,49). The alpha helices are arranged in a linear array along an essentially straight DNA target. In Figure 4C and D, DNA wraps around the histone octamer in a nucleosome (PDB ID: 1KX5) (50). The polar contact map illustrates that the protein helices contact the DNA double helix only on one side in both cases.
The report page provides a link where the user can download the data for a structure as a JSON file, or view it in-browser from the report page. All visualizations are constructed from the data available in these JSON files. Therefore, the user may use these data to produce their own visualizations or to extract various structural features of the complex. The report page also provides a link to the RCSB PDB structure summary page for structures retrieved from the DNAproDB database where additional annotations and validation reports for the structure can be obtained.
CONCLUSION
DNAproDB has many search and reporting capabilities for rapid structure analysis of DNA–protein complexes. DNAproDB enables researchers to upload newly determined structures, structures derived from simulation or modeling or to search the DNAproDB database for pre-processed structures and use the developed tools to analyze said structures. To replicate all of the data and visualization capabilities that DNAproDB provides, it would be necessary to install a large suite of software and libraries, each of which comes with its own interface, output format, learning curve and pitfalls.
DNAproDB provides unique, interactive visualizations for each structure, which can be exported and downloaded at high resolution. Each visualization depicts the DNA–protein interface and interactions at different levels of detail and abstraction. The polar contact map (Figures 3A, B and 4A, C; Supplementary Figures S2 and 3A) is useful for visual comparison of a large number of structures simultaneously and provides a compact representation of the DNA–protein interface. The linear contact map (Supplementary Figure S3C) is useful for understanding the extent to which each SSE contacts different nucleotides and how far into the groove or from the backbone the contacts are positioned. The nucleotide–residue contact map (Figure 3D and E; Supplementary Figure S3D) is useful for looking at the interface in detail and understanding the role of each residue. The only existing visualization tool that is widely used and designed to work for DNA–protein structures is NUCPLOT (46). This tool focuses on protein side chain–DNA interactions but neglects secondary structure and has limited options for customization. Visualizations in DNAproDB show secondary structure, allow for customization, display more interaction types and are interactive with the option of exporting a static figure.
DNAproDB currently supports only proteins bound to dsDNA. Our approach, however, is general enough that, in the future, we will be able to include proteins bound to single-stranded DNA and other DNA forms, such as G-quadruplexes and Holliday junctions. Furthermore, because DNAproDB utilizes a non-relational database, new structure- or sequence-based features can be easily integrated into the search and processing capabilities. Users are encouraged to submit feature requests through our contact page at http://dnaprodb.usc.edu/cgi-bin/contact. DNAproDB is open access, and there are no login requirements.
Supplementary Material
ACKNOWLEDGEMENTS
This work was performed in part while H.M.B. was on sabbatical leave from Rutgers University and a Visiting Professor in the Rohs lab. The authors thank Luigi Manna for his assistance in setting up, configuring and maintaining the DNAproDB server at USC and for helpful discussions regarding technical aspects of the project. The authors also thank Maggie Gabanyi for helpful discussions regarding the website usability and Robert Lowe for helpful discussions regarding the website design and layout.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM106056, U01GM103804 to R.R., R01HG003008 to R.R., in part]; USC Bridge Institute Catalyst Grant (to R.R.); Alfred P. Sloan Research Fellowship (to R.R.). Funding for open access charges: USC (to R.R.); National Science Foundation [MCB-1413539 to R.R.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Harrison S.C. A structural taxonomy of DNA-binding domains. Nature. 1991; 353:715–719. [DOI] [PubMed] [Google Scholar]
- 2. Luscombe N.M., Austin S.E., Berman H.M., Thornton J.M.. An overview of the structures of protein-DNA complexes. Genome Biol. 2000; 1, REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garvie C.W., Wolberger C.. Recognition of specific DNA sequences. Mol. Cell. 2001; 8:937–946. [DOI] [PubMed] [Google Scholar]
- 4. Prabakaran P., Siebers J.G., Ahmad S., Gromiha M.M., Singarayan M.G., Sarai A.. Classification of protein-DNA complexes based on structural descriptors. Structure. 2006; 14:1355–1367. [DOI] [PubMed] [Google Scholar]
- 5. Biswas S., Guharoy M., Chakrabarti P.. Dissection, residue conservation, and structural classification of protein-DNA interfaces. Proteins. 2009; 74:643–654. [DOI] [PubMed] [Google Scholar]
- 6. Malhotra S., Sowdhamini R.. Re-visiting protein-centric two-tier classification of existing DNA-protein complexes. BMC Bioinformatics. 2012; 13:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones S., van Heyningen P., Berman H.M., Thornton J.M.. Protein-DNA interactions: a structural analysis. J. Mol. Biol. 1999; 287:877–896. [DOI] [PubMed] [Google Scholar]
- 8. Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S.. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010; 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schneider B., Cerný J., Svozil D., Cech P., Gelly J.C., de Brevern A.G.. Bioinformatic analysis of the protein/DNA interface. Nucleic Acids Res. 2014; 42:3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corona R.I., Guo J.T.. Statistical analysis of structural determinants for protein-DNA-binding specificity. Proteins. 2016; 84:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Locasale J.W., Napoli A.A., Chen S., Berman H.M., Lawson C.L.. Signatures of protein-DNA recognition in free DNA binding sites. J. Mol. Biol. 2009; 386:1054–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hancock S.P., Ghane T., Cascio D., Rohs R., Di Felice R., Johnson R.C.. Control of DNA minor groove width and Fis protein binding by the purine 2-amino group. Nucleic Acids Res. 2013; 41:6750–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dror I., Zhou T., Mandel-Gutfreund Y., Rohs R.. Covariation between homeodomain transcription factors and the shape of their DNA binding sites. Nucleic Acids Res. 2014; 42:430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang X., Dantas Machado A.C., Ding Y., Chen Y., Lu Y., Duan Y., Tham K.W., Chen L., Rohs R., Qin P.Z.. Conformations of p53 response elements in solution deduced using site-directed spin labeling and Monte Carlo sampling. Nucleic Acids Res. 2014; 42:2789–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng Z., Wang Q., Liu Z., Zhang M., Dantas Machado A.C., Chiu T.P., Feng C., Zhang Q., Yu L., Qi L. et al. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 2015; 6:7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Norambuena T., Melo F.. The protein-DNA interface database. BMC Bioinformatics. 2010; 11:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim R., Guo J.T.. PDA: an automatic and comprehensive analysis program for protein-DNA complex structures. BMC Genomics. 2009; 10(Suppl. 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shazman S., Lee H., Socol Y., Mann R.S., Honig B.. OnTheFly: a database of Drosophila melanogaster transcription factors and their binding sites. Nucleic Acids Res. 2014; 42:D167–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer M., Zhang Q.C., Dey F., Chen B.Y., Honig B., Petrey D.. MarkUs: a server to navigate sequence-structure-function space. Nucleic Acids Res. 2011; 39:W357–W361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuang X., Dhroso A., Han J.G., Shyu C.R., Korkin D.. DOMMINO 2.0: integrating structurally resolved protein-, RNA-, and DNA-mediated macromolecular interactions. Database (Oxford). 2016; 2016, bav114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Contreras-Moreira B. 3D-footprint: a database for the structural analysis of protein-DNA complexes. Nucleic Acids Res. 2010; 38:D91–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Westbrook J.D., Fitzgerald P.M.D.. Bourne PE, Gu JE. The PDB format, mmCIF formats, and other data formats. Structural Bioinformatics. 2009; 2nd edn, Hoboken, NJ: John Wiley & Sons, Inc; 271–291. [Google Scholar]
- 24. Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A.. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004; 32:W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A.. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007; 35:W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Word J.M., Lovell S.C., Richardson J.S., Richardson D.C.. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999; 285:1735–1747. [DOI] [PubMed] [Google Scholar]
- 28. Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu X.J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lavery R., Sklenar H.. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989; 6:655–667. [DOI] [PubMed] [Google Scholar]
- 31. Kabsch W., Sander C.. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983; 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 32. Mitternacht S. FreeSASA: an open source C library for solvent accessible surface area calculations. F1000Res. 2016; 5:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee B., Richards F.M.. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971; 55:379–400. [DOI] [PubMed] [Google Scholar]
- 34. McDonald I.K., Thornton J.M.. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994; 238:777–793. [DOI] [PubMed] [Google Scholar]
- 35. Lu X.J., Olson W.K.. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008; 3:1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chennamsetty N., Voynov V., Kayser V., Helk B., Trout B.L.. Prediction of aggregation prone regions of therapeutic proteins. J. Phys. Chem. B. 2010; 114:6614–6624. [DOI] [PubMed] [Google Scholar]
- 37. The UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 40. Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G. et al. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015; 43:D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Plugge E., Membrey P., Hawkins T.. The Definitive Guide to MongoDB: The NoSQL Database for Cloud and Desktop Computing. 2010; New York, NY: Springer-Verlag New York Inc. [Google Scholar]
- 42. Murphy E.C., Zhurkin V.B., Louis J.M., Cornilescu G., Clore G.M.. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 2001; 312:481–499. [DOI] [PubMed] [Google Scholar]
- 43. Kamada K., Shu F., Chen H., Malik S., Stelzer G., Roeder R.G., Meisterernst M., Burley S.K.. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell. 2001; 106:71–81. [DOI] [PubMed] [Google Scholar]
- 44. Singh S., Hager M.H., Zhang C., Griffith B.R., Lee M.S., Hallenga K., Markley J.L., Thorson J.S.. Structural insight into the self-sacrifice mechanism of enediyne resistance. ACS Chem. Biol. 2006; 1:451–460. [DOI] [PubMed] [Google Scholar]
- 45. Jauch R., Ng C.K., Narasimhan K., Kolatkar P.R.. The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem. J. 2012; 443:39–47. [DOI] [PubMed] [Google Scholar]
- 46. Luscombe N.M., Laskowski R.A., Thornton J.M.. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997; 25:4940–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A., Crickmore M.A., Jacob V., Aggarwal A.K., Honig B., Mann R.S.. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007; 131:530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murphy M.W., Lee J.K., Rojo S., Gearhart M.D., Kurahashi K., Banerjee S., Loeuille G.A., Bashamboo A., McElreavey K., Zarkower D. et al. An ancient protein-DNA interaction underlying metazoan sex determination. Nat. Struct. Mol. Biol. 2015; 22:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rohs R., Dantas Machado A.C., Yang L.. Exposing the secrets of sex determination. Nat. Struct. Mol. Biol. 2015; 22:437–438. [DOI] [PubMed] [Google Scholar]
- 50. Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J.. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002; 319:1097–1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.