Abstract
Since 2009 the EMBL-EBI provides free and unrestricted access to several bioinformatics tools via the user's browser as well as programmatically via Web Services APIs. Programmatic access to these tools, which is fundamental to bioinformatics, is increasingly important as more high-throughput data is generated, e.g. from proteomics and metagenomic experiments. Access is available using both the SOAP and RESTful approaches and their usage is reviewed regularly in order to ensure that the best, supported tools are available to all users. We present here an update describing the latest enhancement to the Job Dispatcher APIs as well as the governance under it.
INTRODUCTION
The EMBL-EBI Job Dispatcher (1–3) framework provides an interface between High Performance Compute clusters and command-line applications. It integrates tools and generates uniform interfaces that are used to generate Web, SOAP and RESTful APIs. It also produces statistics in a common format for each tool and makes it possible to analyze detailed usage with common analytic tools. At present, tools include sequence similarity search services (https://www.ebi.ac.uk/Tools/sss/) such as BLAST (4), FASTA (5) and PSI-Search (6), multiple sequence alignment tools (https://www.ebi.ac.uk/Tools/msa/) such as Clustal Omega (7), MAFFT (8) and KAlign (9), and other sequence analysis tools (https://www.ebi.ac.uk/Tools/pfa/) such as InterProScan5 (10). The use of sequence similarity search tools comprises 45 000 distinct sequences libraries from ENA (11), Ensembl Genomes (12), UniProt (13), InterPro (14) and Pfam (15). These contain sequences from whole genomes and complete proteomes, gene sequence submissions, transcripts, reference proteomes, amplicons, metagenomes, metatranscriptomes and assemblies from metagenomic studies, sequences from patents and specialized collections, such as sequence from immunological studies.
During 2016, usage totaled 152 million jobs. Usage is from the academic and industry scientists and is supplemented by training and support activities in collaboration with the EMBL-EBI training program (16).
THE TOOLS FRAMEWORK
The Job Dispatcher framework consists of: a tools configuration module; a cluster scheduling interface that communicates with the queuing system; and results management and rendering modules that take care of coordinating how results are displayed. The framework is developed using the Java JAX-WS APIs for creating XML-based SOAP and RESTful Web Services. Extensive validation routines are built into the web service to ensure that the correct types of data and parameter values are sent to the tools. All outputs are examined in order to verify tool execution, detect errors and produce human readable reports. Visual representations of tool results are provided to help the user understand the job output both interactively using web browser or programmatically using the web services clients. These clients, written in C#, Java, Perl, Python, PHP and Ruby, are available for many tools that can be used directly from the command line as part of a workflow or pipeline, or as a template for integrating tool functionality into complex applications. Work is in progress to add Common Workflow Language (CWL) (https://github.com/common-workflow-language/common-workflow-language) descriptions which will allow the clients to be further integrated into workflow management systems that support CWL, such as Taverna (http://www.taverna.org.uk/), Arvados (https://arvados.org/) and Galaxy (https://galaxyproject.org/). Tools such as HMMER3, with in-memory database support are in the pipeline, as well as new modern compute resources that scale better with current usage. Due to popular demand, a complete collection of clients written in Python is in the making that will support JSON technologies and allow users to interface results with analytical suites, such as the R package. Already, the Job Dispatcher framework provides a high level of interoperability by allowing users to specify output formats such as XML, JSON, CSV and TSV. Furthermore, these significantly ease the effort of integrating tool functionality into third-party portals. Contextually, the framework is equivalent to provisioning ‘software as a service’ and in the context of bioinformatics, this fits well with the mission of EMBL-EBI.
NEW ANALYSIS TOOLS AND DATABASES
New tools include: HMMER3 (17), that uses probabilistic models called hidden Markov models for searching sequence databases for sequence homologs; Simple_Phylogeny, which replaces ClustalW2_Phylogeny and that provides access to phylogenetic tree generation methods; PredComp (1), that compares a set of predicted annotations against actual automated annotations existing in UniProt-TrEMBL and generates a comprehensive graphical report and PSI-Search2 (18), an improved version of PSI-Search, that can reduce the frequency of false-positive alignments more than 20-fold compared with psiblast. A complete list of currently supported categories and tools is shown in Table 1. ChEMBL (19) and MEROPS (20), ENA Barcode, Geospatial and non-coding sequences (11), have been added to the sequence similarity search libraries. Importantly, new UniProt Reference Proteomes (13) and Enzyme Centric (21) libraries are also now available.
Table 1. Tool services available in the Job Dispatcher framework (2017).
| Category | Tool |
|---|---|
| EMBOSS Programs (https://www.ebi.ac.uk/Tools/emboss/) | needle, stretcher, water, matcher, transeq, sixpack, backtranseq, backtranambig, pepinfo, pepstats, pepwindow, cpgplot, newcpgreport, isochore and seqret |
| Multiple Sequence Alignment (https://www.ebi.ac.uk/Tools/msa/) | clustal omega, kalign, mafft, mafft_addseq, muscle, mview, tcoffee and prank |
| Pairwise Sequence Alignment (https://www.ebi.ac.uk/Tools/psa/) | needle, stretcher, water, matcher, lalign, wise2dba, genewise and promoterwise |
| Phylogeny Analysis (https://www.ebi.ac.uk/Tools/phylogeny/) | simple_phylogeny and raxml_epa |
| Protein Functional Analysis (https://www.ebi.ac.uk/Tools/pfa/) | interproscan5, pfamscan, phobius, pratt, prosite scan and radar |
| RNA Analysis (https://www.ebi.ac.uk/Tools/rna/) | infernal_cmscan and mapmi |
| Sequence Format Conversion (https://www.ebi.ac.uk/Tools/sfc/) | seqret and mview |
| Sequence Operation (https://www.ebi.ac.uk/Tools/so/) | seqcksum |
| Sequence Similarity Search (https://www.ebi.ac.uk/Tools/sss/) | ncbiblast+, fasta, ggsearch, glsearch, psiblast, psisearch, psisearch2 and ssearch |
| Sequence Statistics (https://www.ebi.ac.uk/Tools/seqstats/) | pepinfo, pepstats, pepwindow, saps, cpgplot, newcpgplot and isochore |
| Sequence Translation (https://www.ebi.ac.uk/Tools/st/) | transeq, sixpack, backtranseq and backtranambig |
TOOLS AND DATABASE RETIREMENTS
Workflow tools such as ps_scan (22), InterProScan (23) and FingerPrintScan (24) have been retired, although some remain part of the InterProScan5 tool. ClustalW2 (25), WU-Blast (26), MaxSprout (27), DaliLite (28), DBClustal (29) and ReadSeq (30) have also been removed.
TOOLS GOVERNANCE
EMBL-EBI is proud to provide free access to data and analytical tools. There are many variables that need to be taken into account when deciding to provide access to tools and databases. These range from operational requirements to the relative size of the user community of a tool. Importantly, the scientific suitability of a particular tool to produce up-to-date and relevant results need to be considered. In order to manage the process, a governance model has been set up that comprises expert users of bioinformatics tools, developers, infrastructure managers and usability specialists. It is also important that the users’ opinions count and these are obtained through annual surveys (please see: http://www.ebi.ac.uk/about/our-impact). The governance model requires detailed analysis of usage statistics. This includes the use of storage, CPU, memory, number of runs, provenance, interface used (www, SOAP or REST), as well as availability of support for enhancing or fixing bugs, publications and current training.
TOOLS USAGE
The top 10 tools, by job number alone are: InterProScan5, which, as a workflow, is currently running 19 protein domain and structural domain detection methods. This is followed by the BLAST+ programs, which give access to ∼45 000 libraries of sequences from ENA, UniProt and EnsemblGenomes. Clustal Omega and Muscle are the most popular multiple sequence alignment methods. water and needle from the EMBOSS suite give access to local and global pairwise alignments methods. seqret is very popular for sequence reformatting, pfamscan for searching Pfam HMMs and Phobius (31) for predicting transmembrane regions and signal peptides. Finally, simple_phylogeny, which is used for generating phylogenetic trees using Neighbor-Joining (32) and UPGMA (33) methods.
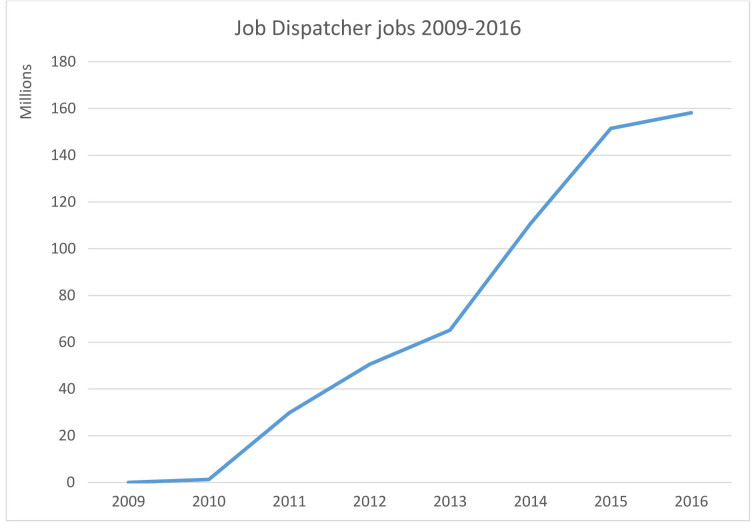
About 56% of all usage occurs using the RESTful APIs, while 26 and 18% are using the SOAP and www interfaces, respectively. Users come from all over of the world, but predominantly from: Germany with 36%; USA with 28%; Japan with 10%; UK with 6%; France with 5%; China and India with 4%; Portugal, Spain and South Korea with 2%. Uptake by users has been steady since 2009 as can be seen in Figure 1, which shows job submissions to the Job Dispatcher framework during 2009–2016.
Figure 1.
Job Dispatcher jobs 2009–2016.
DISCUSSION
Providing robust and reliable access to bioinformatics tools is one important focus of this framework since 2009. However, these tools represent the workhorses of modern bioinformatics and the continuous improvement of the framework ensures the APIs interoperate as easily as possible with workflow management systems. Having a governance model ensures that resources are available to run the tools and meet user demand in a measured and optimal way, and that the acquisition of results and data from EMBL-EBI is consistent, uniform and importantly, as up-to-date as possible. Support and training are important efforts in understanding usage, and users are also encouraged to provide feedback via https://www.ebi.ac.uk/support.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the following: Simone Badoer, Andrea Cristofori and Philip Lewis for web administration support. We also would like to thank all EMBL-EBI teams involved in providing sequence data.
FUNDING
European Molecular Biology Laboratory (EMBL). Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R.. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015; 43:W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lopez R., Cowley A., Li W., McWilliam H.. Using EMBL-EBI services via web interface and programmatically via web services. Curr. Protoc. Bioinformatics. 2014; 48:3.12.1–3.12.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R.. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013; 41:W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pearson W.R., Lipman D.J.. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W., McWilliam H., Goujon M., Cowley A., Lopez R., Pearson W.R.. PSI-Search: iterative HOE-reduced profile SSEARCH searching. Bioinformatics. 2012; 28:1650–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J.J.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lassmann T., Frings O., Sonnhammer E.L.. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009; 37:858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Toribio A.L., Alako B., Amid C., Cerdeño-Tarrága A., Clarke L., Cleland I., Fairley S., Gibson R., Goodgame N., Ten Hoopen P. et al. European nucleotide archive in 2016. Nucleic Acids Res. 2017; 45:D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kersey P.J., Allen J.E., Armean I., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Falin L.J., Grabmueller C. et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016; 44:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.Y., Dosztányi Z., El-Gebali S., Fraser M. et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Welch L., Brooksbank C., Schwartz R., Morgan S.L., Gaeta B., Kilpatrick A.M., Mietchen D., Moore B.L., Mulder N., Pauley M. et al. Applying, evaluating and refining bioinformatics core competencies (An update from the curriculum task force of ISCB's education committee). PLoS Comput. Biol. 2016; 12:e1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R.. HMMER web server: 2015 update. Nucleic Acids Res. 2015; 43:w30–w38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pearson W.R., Li W., Lopez R.. Query-seeded iterative sequence similarity searching improves selectivity 5–20-fold. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Krüger F.A., Light Y., Mak L., McGlinchey S. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014; 42:D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rawlings N.D., Barrett A.J., Finn R.. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016; 44:d343–d350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pundir S., Onwubiko J., Zaru R., Rosanoff S., Antunes R., Bingley M., Watkins X., O’Donovan C., Martin M.J.. An update on the Enzyme Portal: an integrative approach for exploring enzyme knowledge. Protein Eng. Des. Sel. 2017; 30:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I.. New and continuing developments at PROSITE. Nucleic Acids Res. 2013; 41:D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R.. InterProScan: protein domains identifier. Nucleic Acids Res. 2005; 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Attwood T.K., Blythe M.J., Flower D.R., Gaulton A., Mabey J.E., Maudling N., McGregor L., Mitchell A.L., Moulton G., Paine K., Scordis P.. PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res. 2002; 30:239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 26. Lopez R., Silventoinen V., Robinson S., Kibria A., Gish W.. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 2003; 31:3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holm L., Sander C.. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J. Mol. Biol. 1991; 218:183–194. [DOI] [PubMed] [Google Scholar]
- 28. Holm L., Park J.. DaliLite workbench for protein structure comparison. Bioinformatics (Oxford, England). 2000; 16:566–567. [DOI] [PubMed] [Google Scholar]
- 29. Thompson J.D., Plewniak F., Thierry J., Poch O.. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 2000; 28:2919–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gilbert D. Sequence file format conversion with command-line readseq. Curr. Protoc. Bioinformatics. 2003; Appendix 1:Appendix 1E. [DOI] [PubMed] [Google Scholar]
- 31. Käll L., Krogh A., Sonnhammer E.L.. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004; 338:1027–1036. [DOI] [PubMed] [Google Scholar]
- 32. Saitou N., Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 33. Sneath P.H.A., Sokal R.R.. Numerical Taxonomy. 1973; San Francisco, CA: WH Freemanand Co; 230–234. [Google Scholar]