Abstract
Background
We aimed to investigate the potential of standard hematologic and serum biochemical parameters to provide an independent and substantial contribution to the prediction of survival in patients with newly diagnosed brain metastases (BM).
Methods
Hemoglobin, white blood cell count, platelet count, serum albumin, creatinine, lactate dehydrogenase (LDH), and C-reactive protein (CRP) were assessed at diagnosis of BM in a discovery cohort of 1200 cancer patients. A multivariable Cox regression model was used to derive the LabBM score. The LabBM score was externally validated in an independent cohort consisting of 366 patients.
Results
Hemoglobin below lower limit of normal (<LLN; hazard ratio [HR] 1.28; P = .001), platelet count <LLN (HR: 1.36; P = .013), albumin <LLN (HR: 1.19; P = .038), LDH above upper limit of normal (>ULN; HR: 1.51; P < .001), and CRP >ULN (HR: 1.52; P < .001) were associated with survival in a multivariable Cox regression model and were included in the calculation of the LabBM score. Multivariable analysis including the LabBM score and graded prognostic assessment class revealed an independent and significant association of the LabBM score with overall survival (OS) (HR: 1.42; 95% CI: 1.29–1.57; P < .001). The strong and independent association of LabBM score (HR: 1.93; 95% CI: 1.54–2.42) with OS prognosis was confirmed in the validation cohort.
Conclusion
Standard clinical blood parameters, combined in the easy-to-calculate LabBM score, provide strong and independent prognostic information in patients with BM. The LabBM score is an objective, inexpensive, and reproducible tool to plan clinical management strategies in BM patients and to improve patient selection and stratification for clinical trials.
Keywords: albumin, brain metastases, CRP, hemoglobin, laboratory parameters, lactate dehydrogenase
Importance of the study
We introduce the easy-to-calculate LabBM score, which is based on standard clinical blood parameters and provides strong and independent association with OS, irrespective of established prognostic factors in patients with newly diagnosed BM. The LabBM score is an objective, inexpensive, and reproducible tool to plan clinical management strategies in BM patients and to improve patient selection and stratification for clinical trials.
Brain metastases (BM) are a frequent complication occurring in up to 40% of cancer patients and are associated with high morbidity and mortality. Treatment modalities used for BM include neurosurgical resection, radiation therapy (radiosurgery and whole brain radiation therapy), chemotherapy, and increasingly also novel systemic drugs such monoclonal antibodies and tyrosine kinase inhibitors.1,2
So far, BM are in general incurable and median overall survival (OS) times are in the range of a few months.3 However, survival times are highly variable, with some patients succumbing to disease within a few weeks and others achieving longer-term survival of several months or even years. Several BM-specific prognostic scores, such as the recursive partitioning assessment score, the graded prognostic assessment (GPA) score, and the diagnosis-specific GPA, have been established to facilitate estimation of patient outcomes for clinical decision making and use in clinical trials.3 These scores are based on clinical characteristics such as patient age, KPS, status of the extracranial disease, number of BM, and primary tumor type.3 Although the use of BM-specific prognostic scores provides valuable information for patient management and has been widely adopted, especially in the context of clinical trials, the prediction of survival times is inaccurate and needs improvement.4 Laboratory parameters routinely assessed in clinical practice have been shown to correlate with patient outcome in several diseases, including cancer.5–8 Therefore, we hypothesized that standard hematologic and serum biochemical parameters could be valuable for prediction of survival in BM patients. We tested and confirmed our hypothesis in a large and well-defined discovery cohort of 1200 patients treated for newly diagnosed BM at the Medical University of Vienna and an independent validation cohort of 366 patients treated at the University Hospital Zurich. We provide an easy-to-calculate score based on standard clinical blood values that may be useful for survival prediction of BM patients in the clinical setting and in clinical trials (“LabBM score”).
Methods
Patients
The discovery cohort encompassed patients treated for newly diagnosed BM from solid extracranial cancers at the Medical University of Vienna between 1990 and 2013. The independent validation cohort included patients treated for newly diagnosed BM from solid extracranial cancers at the University Hospital Zurich between 2004 and 2014. The discovery and the validation cohort were treated by independent multidisciplinary teams according to good clinical practice guidelines.
Clinical data including information on the primary tumor, clinical course, and survival times were retrieved by chart review. GPA was calculated according to previously published clinical characteristics.3,9 The ethics committee of the Medical University of Vienna (Vote 078/2004) and Zurich (Vote KEK-ZH-Nr. 2015-0559) approved the study.
Analyses of Laboratory Parameters
Hemoglobin level (g/dL), platelet count (G/L), white blood cell count (WBC, G/L), serum albumin (g/L), serum creatinine (mg/dL), serum lactate dehydrogenase (LDH; U/I), and serum C-reactive protein (CRP; mg/dL) were analyzed as part of the routine clinical assessment in the local department of laboratory medicine. We retrieved for this study only blood values that were analyzed within 14 days before or after the diagnosis of BM. Local standard cutoff parameters were used for definition of lower limit of normal (LLN), within normal range (NR), and upper limit of normal (ULN; Supplementary Tables 1 and 2).
Statistical Analysis
OS was defined as time in months from diagnosis of BM to death or date of last follow-up. Primary tumor types with a frequency <5 were combined in the group “other primary tumor.” Laboratory parameters were classified into dummy variables (<LLN vs NR vs >ULN) according to the established local standard clinical cutoff values (Supplementary Tables 1 and 2). Then a univariable survival analysis was carried out for all parameters using Kaplan–Meier curves and 2-sided log rank tests. All laboratory parameters showing a statistically significant association (P < .05) with survival prognosis in univariable analysis were included in the multivariable analysis.10 Dummy variables were defined as <LLN vs not <LLN and >ULN vs not >ULN as appropriate for the specific laboratory parameter. Laboratory parameters with statistically significant association with survival in the multivariable model were included in the LabBM score. The regression coefficient B was used to calculate the LabBM score. In order to obtain an easy-to-use score, the regression coefficient B was multiplied by 2 and rounded, resulting in values between 0.5 and 1.0. Thus, 0 points were given for laboratory values within the NR and, depending on the parameter, 0.5 to 1.0 points for values out of the normal range (LLN or ULN). The LabBM score was calculated for each patient in the discovery cohort. Based on this score, 3 LabBM score groups, each containing one third of patients, were defined in the discovery cohort, to give prognostic groups useful for clinical prognostic assessment and clinical trial planning. Patients with LabBM score 0–1 were defined to belong to the low LabBM score group, 1.5–2 to the medium LabBM score group, and 2.5–3.5 to the high LabBM score group. Therefore, the higher the LabBM score group, the more pathological laboratory parameters were present in the individual patient. For further statistical analysis, the LabBM score groups (low, medium, high) were used.
Association of the LabBM score with survival was again analyzed in a univariable analysis (log rank test), as well as in a multivariable analysis (Cox regression model) including the LabBM score in addition to the established clinical prognosis score GPA. The Harrell’s C Index was calculated to investigate the prognostic accuracy of the LabBM score.11 Then the LabBM score was calculated for the patients in the independent validation cohort and analyzed for association with survival. Again, association of the LabBM score group was investigated in a univariable analysis (log rank test) as well as in a multivariable analysis (Cox regression model) including the LabBM score group and the GPA class (entered as a categorical variable) as the most frequently applied prognostic assessment.9 To evaluate the added value of the LabBM score groups, Harrell’s C index was calculated for both LabBM score and GPA class. A 2-tailed P-value of <.05 was considered significant. The study was conducted according to the TRIPOD statement guidelines.12
Results
Patients’ Characteristics
The discovery cohort consisted of 1200 patients and the validation cohort of 366 patients, all with newly diagnosed BM from a histologically proven extracranial solid cancer. Table 1 lists further patients’ characteristics, including distribution of the investigated laboratory parameters. Due to the retrospective nature of this project, not all blood parameters were available in all patients. A complete set of all investigated laboratory parameters was available in 811/1200 (67.6%) patients in the discovery and 177/366 (48.4%) patients in the validation cohort. Albumin and LDH were most commonly missing, while all other parameters were available in the vast majority of cases (Supplementary Table 3). No difference in survival according to the availability of a complete set of all investigated laboratory parameters was observed in the discovery cohort (7 mo vs 6 mo; P = .355; log rank test). In the validation cohort, patients with missing laboratory parameters had better survival than patients with a complete set of all investigated parameters (13 mo vs 7 mo; P = .015; log rank test).
Table 1.
Patients’ characteristics
| Characteristic | Discovery Cohort (n = 1200) | Validation Cohort (n = 366) | P-value | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Median age at diagnosis of BM, y (range) |
59 (24–89) |
61 (29–105) |
0.464 | ||
| Sex | .005 | ||||
| Male | 569 | 47.4 | 204 | 55.7 | |
| Female | 631 | 52.6 | 162 | 44.3 | |
| Primary tumor | |||||
| Lung cancer | 472 | 39.3 | 157 | 42.9 | |
| Breast cancer | 265 | 22.1 | 44 | 12.0 | |
| Melanoma | 163 | 13.6 | 66 | 18.0 | <.001 |
| Renal cell carcinoma | 121 | 10.1 | 13 | 3.6 | |
| Colorectal cancer | 101 | 8.4 | 21 | 5.7 | |
| Cancer of unknown primary Others |
20 58 |
1.7 4.8 |
12 53 |
3.3 14.5 |
|
| Median hemoglobin (g/dL) | |||||
| Male | 13.60 | 13.80 | |||
| Male range | 5.54–19.40 | 7.10–19.0 | |||
| Female | 12.60 | 13.00 | |||
| Female range | 3.50–21.10 | 7.00–16.20 | |||
| Hemoglobin (g/dL) | <.001 | ||||
| <LLN | 499 | 41.6 | 122 | 33.3 | |
| NR | 692 | 57.7 | 232 | 63.4 | |
| >ULN | 9 | 0.8 | 12 | 3.3 | |
| Median platelet count (G/L) range |
250 6–936 |
273 6–1107 |
|||
| Platelet count (G/L) | .005 | ||||
| <LLN | 117 | 9.8 | 21 | 5.7 | |
| NR | 901 | 75.1 | 304 | 83.1 | |
| >ULN | 182 | 15.2 | 41 | 11.2 | |
| Median WBC (G/L) range |
8.7 0.77–82.10 |
9.3 1.7–79.1 |
|||
| WBC (G/L) | .001 | ||||
| <LLN | 58 | 4.8 | 3 | 0.8 | |
| NR | 660 | 55.0 | 189 | 52.1 | |
| >ULN | 482 | 40.2 | 171 | 47.1 | |
| Median albumin (g/L) range |
38.9 22.40–64.10 |
39.0 21.0–60.0 |
|||
| Albumin (g/L) | <.001 | ||||
| <LLN | 239 | 27.3 | 134 | 52.9 | |
| NR | 635 | 72.7 | 124 | 48.1 | |
| Median creatinine (mg/dL) | |||||
| Male | 0.99 | 0.93 | |||
| Male range | 0.38–3.56 | 0.31–3.04 | |||
| Female | 0.84 | 0.76 | |||
| Female range | 0.42–5.60 | 0.27–1.38 | |||
| Creatinine (mg/dL) | .411 | ||||
| <LLN | 32 | 2.7 | 10 | 3.0 | |
| NR | 795 | 68.3 | 236 | 71.7 | |
| >ULN | 337 | 29.0 | 83 | 25.2 | |
| Median LDH (U/I) range |
212.0 9.0–3804.0 |
392.0 147.0–4763.0 |
|||
| LDH (U/I) | <.001 | ||||
| <LLN | ‒ | ‒ | 16 | 5.9 | |
| NR | 666 | 64.5 | 178 | 66.2 | |
| >ULN | 366 | 35.5 | 75 | 27.9 | |
| Median CRP (mg/dL) range |
0.59 <0.01–165.00 |
0.30 <0.01–23.60 |
|||
| CRP (mg/dL) | <.001 | ||||
| NR | 539 | 48.5 | 226 | 61.7 | |
| >ULN | 573 | 51.5 | 140 | 38.3 | |
| GPA at diagnosis of BM | |||||
| 3.5–4 | 101 | 8.4 | 6 | 1.6 | <.001 |
| 3 | 139 | 11.6 | 32 | 8.7 | |
| 1.5–2.5 | 724 | 60.3 | 209 | 57.1 | |
| 0–1 | 236 | 19.7 | 119 | 32.5 | |
| First line treatment for BM | <.001 | ||||
| Stereotactic surgery | 437 | 36.4 | 8 | 2.2 | |
| Chemotherapy | 6 | 0.5 | 11 | 3.0 | |
| Surgery | 539 | 44.9 | 305 | 83.3 | |
| Whole brain radiation therapy | 202 | 16.8 | 35 | 9.6 | |
| Best supportive care | 16 | 1.3 | 7 | 1.9 | |
| Alive at last follow-up | <.001 | ||||
| Yes | 56 | 4.7 | 59 | 16.1 | |
| No | 1144 | 95.3 | 307 | 83.9 | |
| Median OS from diagnosis of BM, mo (range) |
7 (0–207) |
10 (0–106) |
<.001 | ||
Prognostic Impact of Laboratory Parameters in the Discovery Cohort
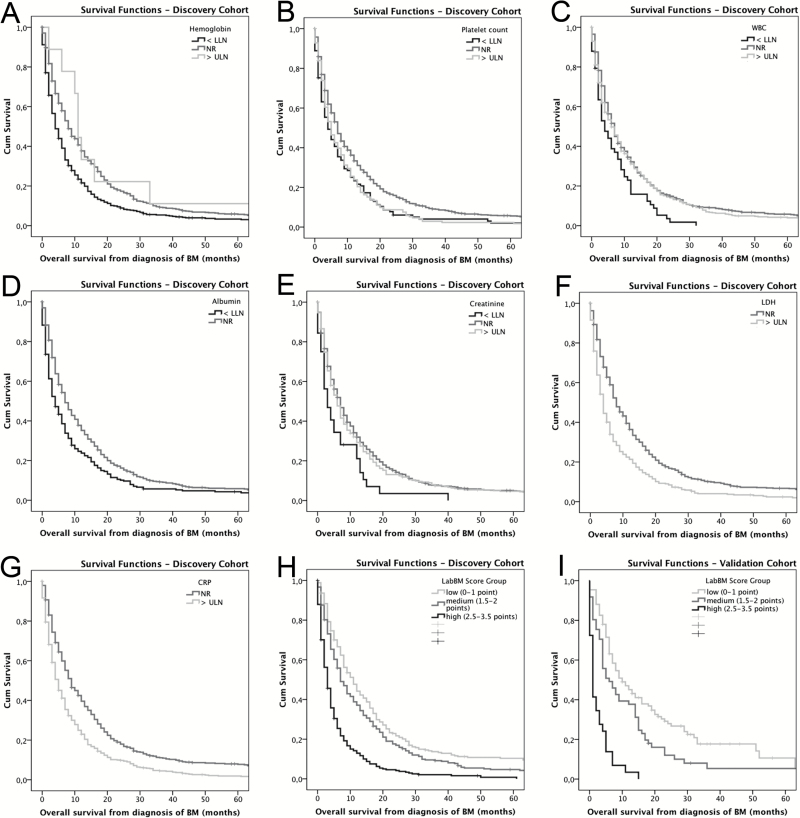
Hemoglobin (P < .001; log rank test), platelet count (P < .001; log rank test), WBC (P = .005; log rank test), albumin (P < .001; log rank test), creatinine (P = .018; log rank test), LDH (P < .001; log rank test), and CRP (P < .001; log rank test) showed an association with survival on univariable analysis (Table 2, Fig. 1A–G).
Table 2.
Overall survival from diagnosis of BM in the discovery cohort according to laboratory parameters at univariate and multivariable analyses
| Univariate Analysis (log rank test) | Multivariable Analysis (Cox regression model) | |||||
|---|---|---|---|---|---|---|
| Median OS, mo | P-value | HR | 95% CI | B | P-value | |
| Hemoglobin | ||||||
| <LLN* | 4 | <.001 | 1.280 | 1.100–1.491 | 0.247 | .001 |
| NR | 8 | |||||
| >ULN | 11 | 0.927 | 0.413–2.084 | -0.076 | .855 | |
| Platelet count | ||||||
| <LLN* | 4 | <.001 | 1.365 | 1.068–1.744 | 0.311 | .013 |
| NR | 7 | |||||
| >ULN | 5 | 1.199 | 0.970–1.481 | 0.181 | .093 | |
| WBC | ||||||
| <LLN | 4 | .005 | 1.217 | 0.845–1.751 | 0.196 | .292 |
| NR | 7 | |||||
| >ULN | 6 | 1.084 | 0.931–1.262 | 0.081 | .299 | |
| Albumin | ||||||
| <LLN* | 4 | <.001 | 1.191 | 1.010–1.405 | 0.175 | .038 |
| NR | 7 | |||||
| Creatinine | ||||||
| <LLN | 3 | .018 | 1.261 | 0.748–2.123 | 0.232 | .384 |
| NR | 7 | |||||
| >ULN | 6 | 1.044 | 0.891–1.222 | 0.043 | .596 | |
| LDH | ||||||
| NR | 8 | <.001 | ||||
| >ULN* | 4 | 1.515 | 1.303–1.760 | 0.415 | <.001 | |
| CRP | ||||||
| NR | 9 | <.001 | ||||
| >ULN* | 5 | 1.525 | 1.312–1.774 | 0.422 | <.001 | |
Parameters marked with * were included in LabBM score calculation.
Fig. 1.
Overall survival according to laboratory parameter in the discovery cohort. (A) Hemoglobin. (B) Platelet count. (C) White blood cell count (WBC) (D) Albumin. (E) Creatinine. (F) Lactate dehydrogenase (LDH). (G) C-reactive protein (CRP). (H) Survival according to LabBM score group in the discovery cohort. (I) Validation cohort.
Development of the LabBM Score
All laboratory parameters were entered in a Cox regression model for multivariable analysis and score development. Here, hemoglobin <LLN (hazard ratio [HR]: 1.280; P = .001), platelet count <LLN (HR: 1.365; P = .013), albumin <LLN (HR: 1.191; P = .038), LDH >ULN (HR: 1.515; P < .001), and CRP >ULN (HR: 1.525; P < .001) showed an association with survival and were included in the further development of the LabBM score. Table 2 gives further details of the survival prognosis according to laboratory values in the discovery cohort. Next, the LabBM score was formulated as indicated in the Methods section (Table 3). Based on the laboratory parameters, the LabBM score was calculated for 815/1200 (67.9%) patients in the discovery cohort, resulting in a score between 0 and 3.5 (Table 4). In 385/1200 (32.1%) patients, calculation of the LabBM score was not possible due to missing values (Supplementary Table 3). Importantly, Harrell’s C index of the LabBM score model was 0.6386 compared with 0.6465 if using all laboratory markers, showing that the information lost by using the easy-to-use LabBM score compared with the exact algorithm is minimal. In the discovery cohort, survival of patients with missing LabBM score did not differ from survival of patients with available LabBM score (7 mo vs 7 mo; P = .266; log rank test).
Table 3.
The LabBM score is calculated as follows
| 0 points | 0.5 point | 1.0 point | |
|---|---|---|---|
| Hemoglobin | NR OR >ULN | <LLN | |
| Platelet count | NR OR >ULN | <LLN | |
| Albumin | NR | <LLN | |
| LDH | NR | >ULN | |
| CRP | NR | >ULN |
Add points according to laboratory parameters to obtain the LabBM score. LabBM score groups: 0–1 (low LabBM score group); 1.5–2 (medium LabBM score group); 2.5–3.5 (high LabBM score group).
Table 4.
LabBM score in the discovery and validation cohort
| Discovery Cohort (n = 815) | Validation Cohort (n = 199) | |||
|---|---|---|---|---|
| n | % | n | % | |
| Median LabBM score (range) | 1 (0–3.5) | 1 (0–3.5) | ||
| LabBM score group | ||||
| Low (0–1 point) | 268 | 32.9 | 109 | 54.8 |
| Medium (1.5–2 points) | 299 | 36.7 | 61 | 30.7 |
| High (2.5–3.5 points) | 248 | 30.4 | 29 | 14.6 |
| Median OS according to LabBM score (mo) | ||||
| Low (0–1 point) | 11 | 10 | ||
| Medium (1.5–2 points) | 7 | 6 | ||
| High (2.5–3.5 points) | 3 | 1 | ||
| HR | 1.579 | 1.985 | ||
| 95% CI | 1.435–1.738 | 1.588–2.483 | ||
Median LabBM score was 1 (range 0–3.5). LabBM score group 0–1 comprised 268/815 (32.9%) patients; LabBM score group 1.5–2 comprised 299/815 (36.7%) patients, and LabBM score group 2.5–3.5 comprised 248/815 (30.4%) patients (Table 4).
The LabBM score group showed a significant association with OS from diagnosis of BM in the discovery cohort. Patients in the low LabBM score group (0–1 points) had a median OS of 11 months; patients in the medium LabBM score group (1.5–2 points) of 7 months, and patients in the high LabBM score group (2.5–3.5 points) of 3 months (P < .001; log rank test; Table 4; Fig. 1H). Accordingly, the LabBM score group showed an HR of 1.579 (95% CI: 1.435–1.738; P < .001; Cox regression model).
The GPA class presented with a statistically significant association with survival prognosis in the discovery cohort (HR: 1.563; 95% CI: 1.445–1.690; P < .001; Cox regression model). To check whether the LabBM score contains information in addition to the GPA class, both variables were entered in a multivariable analysis. Here, the GPA class (HR: 1.506; 95% CI: 1.370–1.654; P < .001; Cox regression model) as well as the LabBM score group (HR: 1.428; 95% CI: 1.296–1.573; P < .001; Cox regression model) showed an independent association with OS. The LabBM score group presented with an independent statistically significant association with survival prognosis (HR: 1.447; 95% CI: 1.312–1.597; P < .001; Cox regression model) when entered with the individual data of the GPA, that is, age (HR: 0.833; 95% CI: 0.692–1.002; P = .053; Cox regression model), KPS (HR: 0.404; 95% CI: 0.326–0.500; P < .001; Cox regression model), number of BM (HR: 0.623; 95% CI: 0.518–0.749; P < .001; Cox regression model), and presence of extracranial metastases (HR: 0.812; 95% CI: 0.694–0.951; P = .010; Cox regression model). To address the added value of the LabBM score compared with the existing and established GPA score Harrell’s C index was calculated for both scores. Here, the GPA class showed a Harrell’s C index of 0.619, indicating a gain of 24% in prognostic accuracy compared with a null model with Harrell’s C index of 0.5. The LabBM score group resulted in a Harrell’s C index of 0.6386 and therefore the prognostic accuracy increased by 28% compared with the null model and by 4% compared with the model including only the GPA. Importantly, a combination score defined as the sum of GPA class and LabBM score showed a Harrell’s C index of 0.680 and thereby an increase of prognostic accuracy by 36.1% compared with the null model, and by 12.1% compared with GPA only (Supplementary Table 5).
As the primary tumor type and applied BM therapy are additional important prognostic parameters, a multivariable analysis including the LabBM score group, the GPA class, the primary tumor type, and the applied therapy was calculated. As expected, the GPA class (P < .001), the primary tumor type (P < .001), as well as the applied therapy (P < .001) showed associations with OS. In addition, the LabBM score group was significantly and independently associated with OS (HR: 1.490; 95% 1.249–1.645; P < .001; Cox regression model).
Validation of the LabBM score
External validation was performed in an independent validation cohort, consisting of patients treated at the University Hospital Zurich. Calculation of the LabBM score was possible in 199/366 (54.4%) patients, while 167/366 (45.6%) patients were excluded from the analysis due to missing laboratory parameters. Survival prognosis in patients with available laboratory parameters for LabBM score calculation was inferior to that in patients with missing LabBM score (7 mo vs 13 mo; P = .010; log rank test). The GPA class was statistically significantly associated with survival prognosis in the validation cohort (HR: 1.477; 95% CI: 1.228–1.777; P < .001; Cox regression model).
Here, the LabBM score groups confirmed the association with survival on univariable analysis. In the validation cohort, patients in the low LabBM score group (0–1 points) had a median OS of 10 months, patients in the medium LabBM score group (1.5–2 points) of 6 months, and patients in the high LabBM score group (2.5–3.5 points) of 1 month (P < .001; log rank test; Table 4; Fig. 1I). In line with the results from the discovery cohort, the LabBM score group had an HR of of 1.985 (95% CI: 1.588–2.483; P < .001; Cox regression model). Again, the independent association of the LabBM score group (HR: 1.932; 95% CI: 1.542–2.420; P < .001; Cox regression model) was retained at the multivariable analysis, including the GPA class (HR: 1.249; 95% CI: 0.978–1.595; P = .075; Cox regression model).
Discussion
BM are an increasing challenge in general oncology and their prevalence is likely to increase. The prognosis of BM is highly variable. Here we report that standard laboratory blood parameters, combined in the LabBM score, have a robust and independent prognostic value in patients with newly diagnosed BM. We identified low hemoglobin levels, low platelet counts, low albumin levels, high LDH levels, and high CRP as adverse prognostic factors. Importantly, all the parameters are routinely tested in cancer patients and display as surrogate parameters important information on the bone marrow reserve, liver function, tumor cell turnover, and infection. Several of these parameters have previously been reported as prognostically relevant in patients with advanced cancer as well as nonmalignant disorders.5,7,8,13–16 The causes of laboratory anomalies may be manifold in cancer patients and may include previous applied therapies, paraneoplastic factors, effects of chronic disease, bleedings, malnutrition, toxicities of prior or concurrent therapies, and others. Low hemoglobin and platelet counts may be surrogate parameters of impaired bone marrow reserve; low albumin levels may indicate malnutrition; and high LDH and CRP levels may be associated with high tumor load or underlying infections.15,17 We did not intend to analyze the specific cause of abnormal blood values in individual cases, but view the LabBM score rather as a general indicator of disease activity and the patient’s biological constitution at the time of BM diagnosis. Of note, we investigated in our study only blood parameters measured at diagnosis of brain metastases and not during the disease course. Future research may evaluate the prognostic impact of blood value changes over time.
We consider the LabBM score as easy to apply in clinical practice, because it is based on routinely investigated parameters. In addition, the LabBM score is based on objectively measurable parameters, as blood values are in general assessed in specialized, certified, and quality controlled laboratories and according to strict standard operating procedures. Other prognostic scores used for BM patients are based on more subjective criteria. The physician-assessed clinical performance score (eg, KPS), which is prone to some interobserver variability, is a core parameter of the GPA.18 Importantly, the prognostic value of the LabBM score was independent of the established prognostic GPA score as well as the histologic tumor type and the applied therapy, thus indicating that consideration of blood values in addition to clinical parameters has added value for estimation of survival probabilities in BM patients.
Despite the large investigated patient cohort of 1566 patients overall, the opportunity to investigate 2 separate cohorts, and the availability of high-quality and detailed clinical data, our study has limitations. Due to the retrospective nature of this project, not all blood parameters were available in all patients. A complete set of all laboratory parameters of interest was available in 811/1200 (67.6%) patients in the discovery and 177/366 (48.4%) patients of the validation cohort. Albumin and LDH were missing in 28% and 17% of patients, respectively, while all other parameters were available in the vast majority of cases. Patients with missing values had a longer survival compared with patients with the full set of investigated laboratory parameters in the validation cohort. Although this finding might be a chance association, it might also be hypothesized that patients in a general good health status might be less likely to receive a full set of laboratory investigation compared with patients in an impaired health status. However, we decided to include all available patients in the calculation and validation of the LabBM score to exclude any kind of inclusion bias. For optimal application of the LabBM score in the clinical setting, standardized measurement of all 5 relevant parameters should be ensured. The 2 investigated cohorts were treated at 2 different centers and therefore resemble the standard real life cohorts at these particular centers. Although the cohorts differ in some clinical characteristics, the LabBM score resulted in comparable results in both cohorts, as an independent association with estimated survival was shown in both. Therefore, these data suggest that although our cohorts presented with some clinical differences, the LabBM score is applicable to real life cohorts across centers. Although the large sample size and the utilization of 2 cohorts provide large statistical power and external validation of our results, the retrospective nature of our data needs to be acknowledged as a limitation, and makes prospective validation of the LabBM score desirable.
In conclusion, the LabBM score provides an objective, inexpensive, and easily reproducible tool to estimate survival of patients with newly diagnosed BM. The LabBM score has an independent association with OS prognosis, irrespective of other established prognostic factors like the GPA class, and adds substantial prognostic accuracy. In the future, the LabBM score may help to plan clinical management strategies in BM patients or to improve patient selection and stratification for clinical trials.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
The costs for this project were covered by the research budget of the Medical University of Vienna.
Conflict of interest statement. The authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
We thank Mela Medjedovic, Aleksandra Angelovski, Lisa Füreder, Benjamin Prascher, Hanna Plazer, Pedro Gomes, and Marion Anderl for assistance in data collection and Harald Heinzl for critical discussion.
References
- 1. Jenkinson MD, Haylock B, Shenoy A, et al. Management of cerebral metastasis: evidence-based approach for surgery, stereotactic radiosurgery and radiotherapy. Eur J Cancer. 2011;47(5):649–655. [DOI] [PubMed] [Google Scholar]
- 2. Arvold ND, Lee EQ, Mehta MP, et al. Updates in the management of brain metastases. Neuro Oncol. 2016;18(8):1043–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sperduto PW, Kased N, Roberge D, et al. Summary report on the graded prognostic assessment: an accurate and facile diagnosis-specific tool to estimate survival for patients with brain metastases. J Clin Oncol. 2012;30(4):419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kondziolka D, Parry PV, Lunsford LD, et al. The accuracy of predicting survival in individual patients with cancer. J Neurosurg. 2014;120(1):24–30. [DOI] [PubMed] [Google Scholar]
- 5. Danner BC, Didilis VN, Wiemeyer S, et al. Long-term survival is linked to serum LDH and partly to tumour LDH-5 in NSCLC. Anticancer Res. 2010;30(4):1347–1351. [PubMed] [Google Scholar]
- 6. Forrest LM, McMillan DC, McArdle CS, et al. Comparison of an inflammation-based prognostic score (GPS) with performance status (ECOG) in patients receiving platinum-based chemotherapy for inoperable non-small-cell lung cancer. Br J Cancer. 2004;90(9):1704–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ulas A, Turkoz FP, Silay K, et al. A laboratory prognostic index model for patients with advanced non-small cell lung cancer. PLoS One. 2014;9(12):e114471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nieder C, Dalhaug A. A new prognostic score derived from phase I study participants with advanced solid tumours is also valid in patients with brain metastasis. Anticancer Res. 2010;30(3):977–979. [PubMed] [Google Scholar]
- 9. Sperduto PW, Berkey B, Gaspar LE, et al. A new prognostic index and comparison to three other indices for patients with brain metastases: an analysis of 1960 patients in the RTOG database. Int J Radiat Oncol Biol Phys. 2008;70(2):510–514. [DOI] [PubMed] [Google Scholar]
- 10. Bursac Z, Gauss CH, Williams DK, et al. Purposeful selection of variables in logistic regression. Source Code Biol Med. 2008;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uno H, Cai T, Pencina MJ, et al. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30(10):1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162(1):55–63. [DOI] [PubMed] [Google Scholar]
- 13. Eigentler TK, Figl A, Krex D, et al. ; Dermatologic Cooperative Oncology Group and the National Interdisciplinary Working Group on Melanoma. Number of metastases, serum lactate dehydrogenase level, and type of treatment are prognostic factors in patients with brain metastases of malignant melanoma. Cancer. 2011;117(8):1697–1703. [DOI] [PubMed] [Google Scholar]
- 14. Lagerwaard FJ, Levendag PC, Nowak PJ, et al. Identification of prognostic factors in patients with brain metastases: a review of 1292 patients. Int J Radiat Oncol Biol Phys. 1999;43(4):795–803. [DOI] [PubMed] [Google Scholar]
- 15. Walenta S, Mueller-Klieser WF. Lactate: mirror and motor of tumor malignancy. Semin Radiat Oncol. 2004;14(3):267–274. [DOI] [PubMed] [Google Scholar]
- 16. Asegaonkar SB, Asegaonkar BN, Takalkar UV, et al. C-reactive protein and breast cancer: new insights from old molecule. Int J Breast Cancer. 2015;2015:145647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arrieta O, Michel Ortega RM, Villanueva-Rodríguez G, et al. Association of nutritional status and serum albumin levels with development of toxicity in patients with advanced non-small cell lung cancer treated with paclitaxel-cisplatin chemotherapy: a prospective study. BMC Cancer. 2010;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor AE, Olver IN, Sivanthan T, et al. Observer error in grading performance status in cancer patients. Support Care Cancer. 1999;7(5):332–335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.