Abstract
The advent of polypharmacology paradigm in drug discovery calls for novel chemoinformatic tools for analyzing compounds’ multi-targeting activities. Such tools should provide an intuitive representation of the chemical space through capturing and visualizing underlying patterns of compound similarities linked to their polypharmacological effects. Most of the existing compound-centric chemoinformatics tools lack interactive options and user interfaces that are critical for the real-time needs of chemical biologists carrying out compound screening experiments. Toward that end, we introduce C-SPADE, an open-source exploratory web-tool for interactive analysis and visualization of drug profiling assays (biochemical, cell-based or cell-free) using compound-centric similarity clustering. C-SPADE allows the users to visually map the chemical diversity of a screening panel, explore investigational compounds in terms of their similarity to the screening panel, perform polypharmacological analyses and guide drug-target interaction predictions. C-SPADE requires only the raw drug profiling data as input, and it automatically retrieves the structural information and constructs the compound clusters in real-time, thereby reducing the time required for manual analysis in drug development or repurposing applications. The web-tool provides a customizable visual workspace that can either be downloaded as figure or Newick tree file or shared as a hyperlink with other users. C-SPADE is freely available at http://cspade.fimm.fi/.
INTRODUCTION
Drug discovery has witnessed a paradigm shift from the traditional one drug-one target view to the multi-target approach, creating a need for novel computational tools that could aid in polypharmacological studies. Polypharmacology refers to the ability of a drug molecule to interact with multiple targets simultaneously (1,2). Such promiscuous drug-target interactions may lead to both therapeutic effects and off-target side effects. Recent efforts in drug discovery have made use of polypharmacology as a means of repurposing or repositioning approved drugs for novel disease targets (3), which in-turn may significantly reduce both the economic cost and time invested in drug development process (4,5). Polypharmacology can also help to design selectively promiscuous drugs, such as multi-kinase anti-cancer agents (2,6). Furthermore, polypharmacology is being used to identify and explain the off-target activities of drugs that result in harmful side-effects, thereby assisting in compound optimization applications (7). The existing computational approaches for investigating polypharmacological effects and predicting drug-target interactions can be broadly categorized as target-centric (8,9) or compound-centric (10,11). The underlying hypothesis in the target-centric approaches is that structurally similar targets are expected to have similar selectivity properties, and hence are likely to bind the same compound. Therefore, this approach is useful for target-based drug discovery approach, although it is applicable only to targets whose structural information is available. The compound or chemo-centric approach is based on the principle that structurally similar compounds tend to bind to the same targets; therefore, this approach is best-suited for identifying compound analogs, hence supporting the phenotype-based and polypharmacology paradigm of drug discovery.
Chemoinformatic web-applications have been developed for target-centric visualization of broad spectrum activity of compounds against well-studied protein families such as kinases; e.g. Kinome Render (12), TREEspot and KinMap (13). For the compound-centric analyses, similar easy-to-use web-tools are not available, although the computational protocol to estimate compound similarities has been detailed by Vilar et al. (14). Most of the existing tools to analyze drug screening assays through clustering and visualization of compounds and their corresponding bioactivity measurements have been implemented as stand-alone tools requiring local installation, e.g. Scaffold Hunter (15), ChemTreeMap (16), Mona 2 (17) and Data warrior (18) or as a browser-based tool, e.g. ChemMineTools (19). However, most tools assume the users to have chemoinformatics and database management skills (MySQL), and either require the structure of the compounds or their PubChem compound identifiers as a primary input (20). Furthermore, even though the existing tools can address a broad range of questions arising in drug discovery applications, they often lack an interactive interface and other user options required by a chemical biologist for real-time sharing, visualization and interpretation of bioactivity data from drug screening assays. To address these limitations, we have capitalized on recent developments in server management and Data Object Model (DOM) frameworks, and implemented a fully-automated, open-source web-based application, named Compound SPecific bioActivity DEndrogram (C-SPADE), which enables biologists with little to no informatics skills to interactively visualize, annotate and investigate the relationships between the compounds’ structural similarities and phenotypic responses through compound-centric bioactivity clustering.
MATERIALS AND METHODS
Implementation
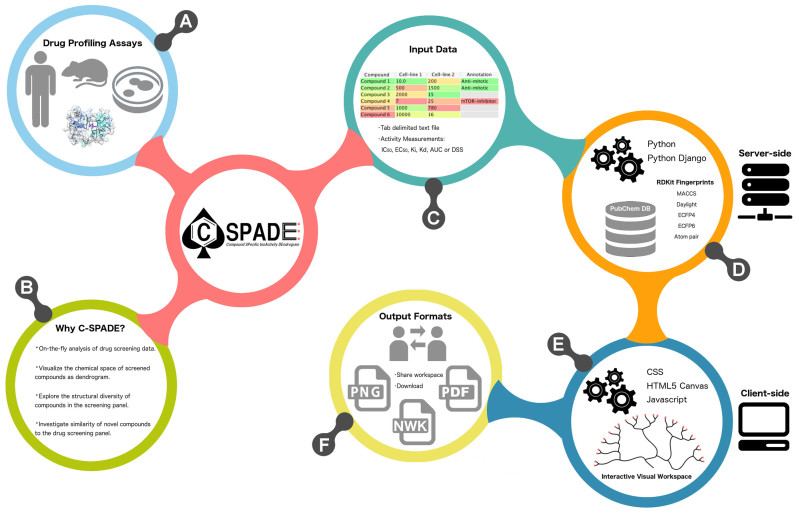
C-SPADE web-application adapts a client-server model (Figure 1), in which session management is maintained through Python Django (version 1.9), and the server-side computation is implemented using Python (version 2.7). The compound names in the input screening data are automatically queried against the PubChem database (21) to retrieve structural description as SMILES. The user can also directly upload the SMILES information of the compounds, which is useful especially when multiple custom structures are used as input. The input data is later displayed on the Data Preview page. C-SPADE utilizes the RDKit python module (version 2016.09.1) to calculate various compound fingerprints (FPs) and their similarities. With the MACCS and Daylight FPs, the structural similarity is calculated using Tanimoto similarity (22), whereas with Atom-pair and Morgan (ECFP-like) FPs using Dice similarity (23) coefficient. The Scipy module (version 0.13.2) then constructs an agglomerative hierarchical cluster using the compound similarities, where the linkage distance between clusters are estimated using the Ward minimum variance method. The resulting compound dendrogram is annotated with bioactivity values and saved as a JavaScript Object Notation (JSON) file for client input. The client-side uses DOM implemented through D3 JavaScript library (http://d3js.org/), HTML5 Canvas and CSS for the interactive analysis and visualization of the compound similarity dendrogram. C-SPADE has been checked for compatibility with all standard modern browsers (Mozilla Firefox, Google Chrome and Safari) and operating systems (Windows, MacOSX and various Linux distributions).
Figure 1.
An infographic illustration of C-SPADE web-tool and its functionalities. (A) Various types of biological drug screening assays (biochemical, cell-based, cell-free or target-based assays) can be analyzed. (B) The key functionalities implemented in C-SPADE. (C) The input consists of a tab-delimited text file of drug screening data, with compound names and bioactivity values, where a wide range of activity measurements are supported. (D) On the server-side, the compound name is used to retrieve the compound structural information from the PubChem database and various compound features are calculated using the RDKit package. (E) The client-side options provide an interactive interface to visualize and customize the compound clusters. (F) The output visualization from C-SPADE can be either shared with collaborators or downloaded as publication-quality images or as a Newick tree file.
Session management
C-SPADE enables the analysis of multiple inputs simultaneously and displays each submission in a separate row on the My Projects page, providing links to Data Preview page and Visualization page as icons that are color-coded based on the status of data processing. The link to Visualization page is not available until the user invokes it from the Data Preview page, which serves as a check to ensure that the user has verified the automatically-retrieved information. The user can either bookmark the web address or save the workspace to enable saving and processing the results at a later time. Each user session is managed anonymously and has an expiration period of 10 days, providing the user sufficient time to analyze the output. C-SPADE visualization has been designed for low-throughput studies; a profiling data of ∼500 compounds takes less than 6 min to process and visualize. Larger datasets (>600 compounds), although computationally more intensive and time consuming, can still be processed, and the user can retrieve the compound similarity dendrogram as a Newick tree file for further analyses.
Input format
C-SPADE currently accepts a tab-delimited text file (.txt) of a preprocessed drug screening data as input. The input file should include the following columns: Compound (required), the unique names of the compounds used in the screen; Smiles (optional), the compounds whose SMILES information is provided will be directly used by C-SPADE to calculate their structural features; otherwise, the compound names will be queried against the PubChem database to retrieve the SMILES; one or more screening assays (optional) (i.e. protein targets, cell lines, patient samples, etc.), each assay in a separate column providing numerical bioactivity values (e.g., IC50, EC50, Ki, Kd, area under the dose response curve (AUC) or drug sensitivity score (DSS) (24)); Annotation (optional), additional annotations of the compounds (e.g. compound class, compound properties, etc.), if available.
Compound selection
The Data Preview page allows the user to edit, curate and select a subset of data for visualization. Compounds whose SMILES are either provided by user or retrieved from PubChem are automatically selected for the visualization. The PubChem compound identifiers (CID) in the Data Preview page are hyperlinked to the PubChem database, giving the user the possibility to check the retrieved compound's information. The user can visualize the chemical diversity of a subset of compounds from the screening panel by selecting the compounds using the check boxes located at the beginning of each row in the table (note: a minimum of 10 compounds are required to generate the clustering dendrogram). Through the Add Compounds option, C-SPADE facilitates on-the-fly similarity investigation, where the similarity of one or more investigational molecules to the drugs in the screening panel can be visualized. Selecting this option adds a new row in the table, where the user is expected to provide the name of the compound and its structural information as SMILES. These compounds can be clustered and visualized with the selected compounds in the table using the Visualize option.
Cluster visualization
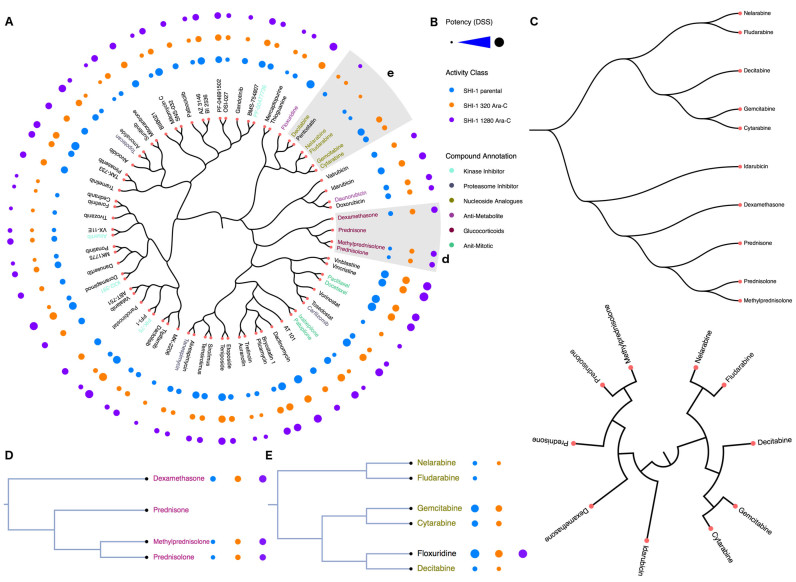
Similar to a traditional phylogenetic tree, the main visual interface in C-SPADE displays the chemical space of compounds used in the screen as a hierarchically-clustered dendrogram, where each compound forms the node in the tree annotated with their respective bioactivity values as bubbles (Figure 2A). Currently, C-SPADE uses molecular fingerprints as features to measure the compound similarity; the closer two compounds are in the tree, the higher is their chemical similarity in terms of their similarity coefficient. The bioactivity values from individual screens of each compound are categorized into five potency classes using log-transformed IC50, EC50, Ki, Kd values (≤1 nM, ≤10 nM, ≤100 nM, ≤1 uM, ≤10 uM) and displayed in the dendrogram. With the summary measurements, such as AUC and DSS, the actual bioactivity values are used and represented as circular annotations in the dendrogram. Visual key for the bioactivity values, activity classes, and compound annotations are shown in the legend of the dendrogram to interpret the output visualization (Figure 2B).
Figure 2.
(A) Compound-centric bioactivity dendrogram visualization of the example drug screening data (25). The drug panel contained a total of 250 compounds screened across three cytarabine-resistant SHI-1 cell-line variants (Parental, 320 Ara-C and 1280 Ara-C) to identify drugs that could counteract cytarabine resistance. A subset of 75 compounds were selected and their chemical space was visualized using the C-SPADE web-tool. The nodes are the compounds and the distance between the nodes represents the degree of their structural similarity based on the selected ECFP4 fingerprints. The bioactivity measurements (DSS) of each compound are represented as circles, where the radius of the circle is proportional to the bioactivity level. Inset: the distinct sub-clusters of glucocorticoids (d) and nucleoside analogues (e). (B) The provided bioactivities, activity classes and compound annotations, if available, are color-coded and displayed in the legend. (C) The types of dendrogram layouts and styles that are currently available in C-SPADE. The glucocorticoids (D) showed an increased sensitivity in the cytarabine resistant cell-lines, whereas the nucleoside analogues (E) showed a co-resistance pattern in the cytarabine resistant SHI-1 cells. In this example, C-SPADE enables the user to (i) map the chemical space of the compounds screened, (ii) explain the sub-clustering patterns that one commonly encounters in such drug-screening data analysis, and (iii) investigate and predict structurally similar compounds that cluster into a desired activity cluster prior to a future compound screening application.
Interactive options
C-SPADE serves as an exploratory tool and provides a highly interactive and customizable visual workspace. For instance, the user has the options to upload multiple datasets, and in real-time change the molecular features for calculating the compound similarity clusters. The visual workspace that displays the compound dendrogram provides several options in the sidebar to interactively customize the visualization. The layout of the displayed dendrogram can be altered between a tree and radial layout (Figure 2C). Features corresponding to the branches and nodes of the tree, such as thickness, radius and colors, can also be dynamically changed. The font sizes of compound labels and annotations are adjustable. Color coding of different assay classes and compound annotations can be interactively changed, and these changes are simultaneously updated in the figure legend. In addition to the traditional zoom and pan functions, C-SPADE provides also other toolbar options, such as a search bar to search and highlight a compound in the tree and rotate option to rotate a radial tree to any given angle. Hovering the mouse pointer over a bioactivity bubble or a compound name displays a tooltip that shows the input bioactivity value or structure of the compound, respectively. The Save Workspace option saves the last performed changes by the user in the visual workspace, hence providing the most updated version of the visualization when revisited.
Output formats
Once customized, the compound clustering can be downloaded either as Portable Network Graphics image (.png) or as a Portable Document Format file (.pdf). The web-tool enables background transparency and maintains a high degree of spatial resolution for these outputs files, thereby generating publication-ready figures. The user can also choose to download the compound similarity dendrogram in a Newick file format (.nwk) to post-process the hierarchical tree object independent of the C-SPADE. By using the Share option, the user can share the hyperlink to the visual workspace, providing the possibility for collaborators to visualize and customize the results interactively.
Documentation and example data
To enable easy start, we have provided an extensive documentation in the help section that explains all the features of C-SPADE in detail. For testing the web-tool we also provide an example data in the form a preprocessed subset of bioactivity data from a cell-based drug screening assay by Malani et al. (25). This example dataset contains 75 compounds screened across three cell-types (Figure 2A), using DSS as the bioactivity measure, and with a subset of compounds annotated by the inhibitor type (Figure 2D and E) (e.g. glucocorticoids (Figure 2D), nucleoside analogue (Figure 2E), anti-metabolite, etc.). For templates of the input data, the user can download this example data in various formats. We have also implemented a quick tutorial section to help the users to quickly go through C-SPADE's functionalities (available at http://cspade.fimm.fi/).
CONCLUSION AND FUTURE WORK
We have implemented C-SPADE, a secure web-based application that enables the users to interactively map and explore drug screening data. Through constructing and visualizing compound-specific dendrograms, C-SPADE, provides a timely solution to the critical need for an efficient and easy-to-use compound-centric visualization tool, hence facilitating chemical biology researchers in various phenotypic screening and polypharmacology-based drug discovery applications. By merely requiring the compound names (and no structural information), C-SPADE serves as a one click tool, thereby significantly reducing the time needed from drug screening to data analysis and interpretation. The web-tool was designed mainly for the needs of biologist; to understand the diversity of the screening panel, to explore investigational compounds’ similarity to the screening panel, and for polypharmacological explorations. However, C-SPADE may also aid chemo-informaticians to draw conclusions related to drug-target interaction predictions. When compared with the traditional heat-map visualization of drug screening data, C-SPADE, through its compound-specific clustering, provides a novel perspective to analyze drug screening data.
Like any other browser-based application, C-SPADE is limited by the size of the data that can be processed and visualized. The current version of C-SPADE uses standard fingerprint measurements from SMILES as compound features, and one future improvement will be to incorporate 2D and 3D molecular descriptor information in feature calculation. The next version of C-SPADE will incorporate also sub-cluster analysis and implement the Maximum Common Substructure (26) algorithm to aid users in compound-optimization studies.
ACKNOWLEDGEMENTS
We thank our collaborators and colleagues at FIMM for beta-testing of the C-SPADE tool. We thank Disha Malani and her colleagues for providing and explaining the example drug screening data. We thank Jing Tang, Daniel Abankwa, Sarang Talwelkar, Mahesh Tambe and Prson Gautam for their constructive comments when developing the tool. We thank Krister Wennerberg for critically reading the manuscript.
Footnotes
Present address: Gopal Peddinti, VTT Technical Research Centre of Finland, Espoo, Finland.
FUNDING
Academy of Finland [269862, 272437, 295504 to T.A.; 265966 to G.P.]; Cancer Society of Finland. Funding for open access charge: Academy of Finland.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hopkins A.L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008; 4:682–690. [DOI] [PubMed] [Google Scholar]
- 2. Paolini G.V., Shapland R.H., van Hoorn W.P., Mason J.S., Hopkins A.L.. Global mapping of pharmacological space. Nat. Biotechnol. 2006; 24:805–815. [DOI] [PubMed] [Google Scholar]
- 3. Lin H., Sassano M.F., Roth B.L., Shoichet B.K.. A pharmacological organization of G protein-coupled receptors. Nat. Methods. 2013; 10:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chong C.R., Sullivan D.J. Jr. New uses for old drugs. Nature. 2007; 448:645–646. [DOI] [PubMed] [Google Scholar]
- 5. Li J., Zheng S., Chen B., Butte A.J., Swamidass S.J., Lu Z.. A survey of current trends in computational drug repositioning. Brief. Bioinf. 2016; 17:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peters J.U. Polypharmacology—foe or friend. J. Med. Chem. 2013; 56:8955–8971. [DOI] [PubMed] [Google Scholar]
- 7. Wermuth C.G. Selective optimization of side activities: the SOSA approach. Drug Discov. Today. 2006; 11:160–164. [DOI] [PubMed] [Google Scholar]
- 8. Cheng F., Liu C., Jiang J., Lu W., Li W., Liu G., Zhou W., Huang J., Tang Y.. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol. 2012; 8:e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lavecchia A., Di Giovanni C.. Virtual screening strategies in drug discovery: a critical review. Curr. Med. Chem. 2013; 20:2839–2860. [DOI] [PubMed] [Google Scholar]
- 10. Liu X., Xu Y., Li S., Wang Y., Peng J., Luo C., Luo X., Zheng M., Chen K., Jiang H.. In Silico target fishing: addressing a ‘Big Data’ problem by ligand-based similarity rankings with data fusion. J. Cheminf. 2014; 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mervin L.H., Afzal A.M., Drakakis G., Lewis R., Engkvist O., Bender A.. Target prediction utilising negative bioactivity data covering large chemical space. J. Cheminf. 2015; 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chartier M., Chenard T., Barker J., Najmanovich R.. Kinome Render: a stand-alone and web-accessible tool to annotate the human protein kinome tree. PeerJ. 2013; 1:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eid S., Turk S., Volkamer A., Rippmann F., Fulle S.. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinf. 2017; 18:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vilar S., Uriarte E., Santana L., Lorberbaum T., Hripcsak G., Friedman C., Tatonetti N.P.. Similarity-based modeling in large-scale prediction of drug-drug interactions. Nat. Protoc. 2014; 9:2147–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wetzel S., Klein K., Renner S., Rauh D., Oprea T.I., Mutzel P., Waldmann H.. Interactive exploration of chemical space with Scaffold Hunter. Nat. Chem. Biol. 2009; 5:581–583. [DOI] [PubMed] [Google Scholar]
- 16. Lu J., Carlson H.A.. ChemTreeMap: an interactive map of biochemical similarity in molecular datasets. Bioinformatics. 2016; 32:3584–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hilbig M., Rarey M.. MONA 2: a light cheminformatics platform for interactive compound library processing. J. Chem. Inf. Model. 2015; 55:2071–2078. [DOI] [PubMed] [Google Scholar]
- 18. Sander T., Freyss J., von Korff M., Rufener C.. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015; 55:460–473. [DOI] [PubMed] [Google Scholar]
- 19. Backman T.W., Cao Y., Girke T.. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011; 39:W486–W491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Humbeck L., Koch O.. What can we learn from bioactivity data? Chemoinformatics tools and applications in chemical biology research. ACS Chem. Biol. 2017; 12:23–35. [DOI] [PubMed] [Google Scholar]
- 21. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bajusz D., Racz A., Heberger K.. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations?. J. Cheminf. 2015; 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sørensen T. A method of establishing groups of equal amplitudes in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol. Skr. – K. Dan. Vidensk. Selsk. 1948; 5:1–34. [Google Scholar]
- 24. Yadav B., Pemovska T., Szwajda A., Kulesskiy E., Kontro M., Karjalainen R., Majumder M.M., Malani D., Murumagi A., Knowles J. et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. 2014; 4:5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malani D., Murumagi A., Yadav B., Kontro M., Eldfors S., Kumar A., Karjalainen R., Majumder M.M., Ojamies P., Pemovska T. et al. Enhanced sensitivity to glucocorticoids in cytarabine-resistant AML. Leukemia. 2016; doi:10.1038/leu.2016.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gardiner E.J., Gillet V.J., Willett P., Cosgrove D.A.. Representing clusters using a maximum common edge substructure algorithm applied to reduced graphs and molecular graphs. J. Chem. Inf. Model. 2007; 47:354–366. [DOI] [PubMed] [Google Scholar]