Abstract
Pathway analysis is widely used in omics studies. Pathway-based data integration and visualization is a critical component of the analysis. To address this need, we recently developed a novel R package called Pathview. Pathview maps, integrates and renders a large variety of biological data onto molecular pathway graphs. Here we developed the Pathview Web server, as to make pathway visualization and data integration accessible to all scientists, including those without the special computing skills or resources. Pathview Web features an intuitive graphical web interface and a user centered design. The server not only expands the core functions of Pathview, but also provides many useful features not available in the offline R package. Importantly, the server presents a comprehensive workflow for both regular and integrated pathway analysis of multiple omics data. In addition, the server also provides a RESTful API for programmatic access and conveniently integration in third-party software or workflows. Pathview Web is openly and freely accessible at https://pathview.uncc.edu/.
INTRODUCTION
Pathway-based analysis is a powerful strategy widely used in omics studies. A wide range of databases and resources have been built (KEGG (1), Reactome (2), Wikipathways (3), MetaCyc (4), PANTHER (5), Pathway Commons (6) etc.) and numerous statistical methods and tools (generally applicable gene-set enrichment (GAGE) (7), GSEA (8), SPIA (9) etc.) developed for pathway analysis. Visualization is a critical component of data analysis in generally and especially so for pathway-based analysis. In fact, pathways themselves are primarily defined and presented as graphs in major pathway databases mentioned above. A number of options/tools exist for pathway visualization (10–15), each with its unique focus and strength. These tools have been extensively reviewed and compared in recent literature (10,16,17).
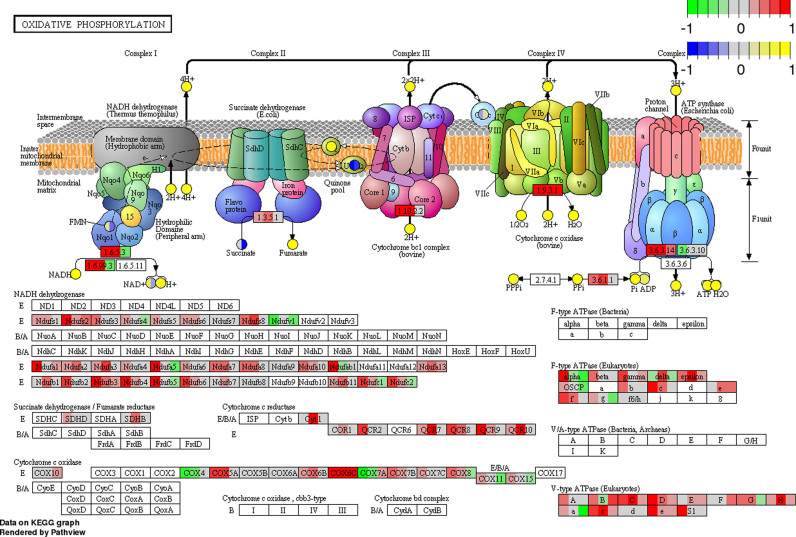
As part of these efforts, we developed an R package called Pathview (10), recently. Pathview maps, integrates and renders a large variety of biological data on molecular and generates pathway graphs with publication quality (as demonstrated by Supplementary Figure S1). Pathview has three important features (10): (i) intuitive and biologically relevant pathway visualization. It adheres to human readable pathway definitions and layouts like KEGG with all visual elements for data presentation (Supplementary Figure S1); (ii) strong data mapping and integration capacity with a wide range of data types, formats, over 3000 species and dozens of molecular IDs (Supplementary Figure S1); (iii) high interoperability, easy fit into pathway analysis pipelines with different analysis tools (as shown in Example 4 online). Detailed description of Pathview and comparison to other tools were covered previously (10). Pathview quickly became a major tool in pathway visualization with tens of thousands of downloads per year (statistics online at http://bioconductor.org/packages/stats/bioc/pathview/).
Despite the usefulness of Pathview, important technical issues remain unsolved. First of all, the software package is only directly accessible by a small minority of scientists with sufficient R programming skills. In general, pathway-based data visualization and integration is demanding for computing resources and also requires a deep understanding of graphics and analytics. A graphical web application would lower the technical threshold greatly for the potential users. Currently, several online tools offer intuitive and useful functions for pathway and data visualization, including MAPMAN (18), VANTED (19), iPath (15), Pathway Projector (12), G-language Pathway Visualization Web Application (20) and Escher (21). In addition, KEGG also provides its own data mapping and visualization web services, i.e. KEGG WebLinks (22) and KEGG Mapper (23) (details in Supplementary Note 1). However, more comprehensive data visualization function is needed as to support a wide range of reference pathways, species, data types and user options. Frequently we need to combine or compare multiple data types, samples or experiments in a study. There has not been a web tool for such integrated visualization and analysis based on pathways.
To address all these needs, here we developed the Pathview Web server. Pathview Web features an intuitive graphical web interface and a user centered design (Figure 1). We also provide a comprehensive online help system and multiple quick-start example analyses. Even with no special computing training and resources, users may still accomplish pathway-based data visualization and integration independently.
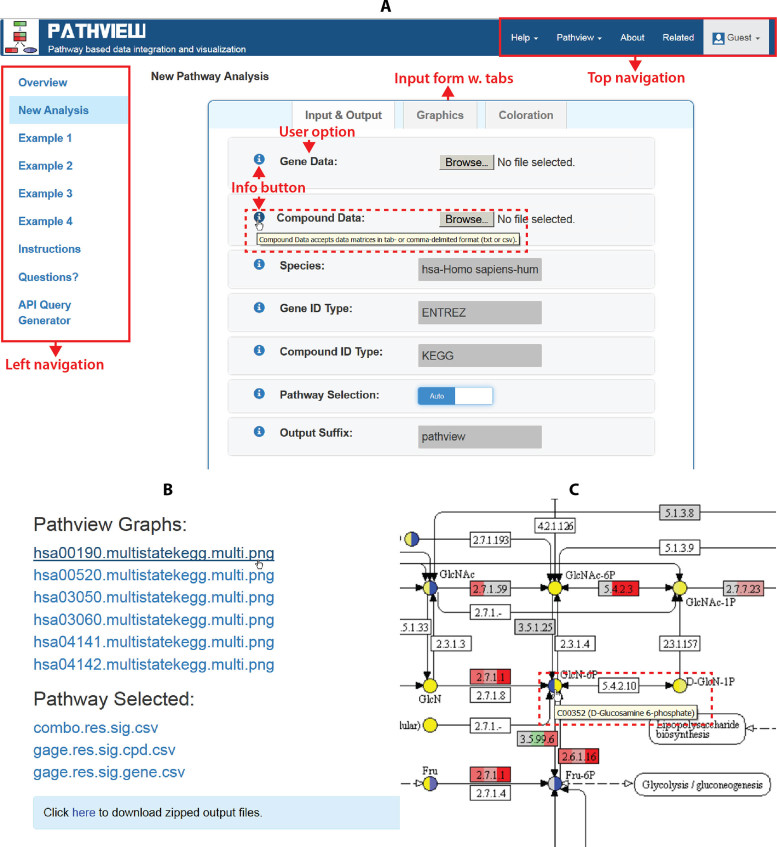
Figure 1.
The Pathview Web server GUI. (A) The main analysis page with the user input web form; (B) the main result page with lists of outputs; (C) an example of interactive pathview graph viewed in browser. Important features are annotated by red tags and texts. Red dashed boxes mark the interactive features with hyperlinks and hover info.
Pathview Web not only expands the core functions of Pathview package, but also provides many unique and useful features. We deployed a full pathway analysis workflow on the server, supporting multiple types of omics data and their integrated analysis. Unlike the R package, Pathview Web is constantly improved in its function and updated in pathway data. In addition to the interactive GUI, the server also provides a RESTful API (Application Program Interface). Developers or expert users can conveniently integrate the web service into their software or workflows.
FUNCTIONS AND FEATURES
Expansion on Pathview core functions
Pathview Web server makes all core functions of Pathview package (10) easily accessible, i.e. (i) pathway visualization, (ii) data mapping and integration and (iii) (interoperability with different) integrated pathway analysis workflows. Supplementary Figure S1 demonstrates the main function. It is advisable to try the example analyses online to see exactly what Pathview and the server can do. These example analyses are straightforward and run in ∼10–30 s each. In addition to user-friendliness, the server expands these functions substantially (Table 1).
Table 1. Comparison of the three versions of Pathview.
| Version | Package | Web | API |
|---|---|---|---|
| Interface | R | GUI | Bash shell |
| Access | open | open and registered | open |
| Host | bioconductor.org/packages/pathview/ | pathview.uncc.edu | pathview.uncc.edu |
| Visualization | KEGG view (png files), Graphviz view (pdf files) | same as R version, hyperlinked graphs | same as R version |
| Data mapping | >3000 KEGG species + Orthology, 12 gene ID, 21 compound ID | same as R version | same as R version |
| Data integration | any mappable data, arbitrary number of conditions/samples | same as R version | same as R version |
| Analysis (Interoperability) | any workflow through R, notably GAGE/Pathview workflow | integrated workflow on server | any workflow through shell script, integrated workflow on server |
| Analysis history | no | yes | no |
| Data sharing | no | yes | no |
| Installation | with R/Bioconductor | not needed | single bash script |
| Update release | every 6 months with Bioconductor | constant (software); every month (data) | constant (software); every month (data) |
| User support | support.bioconductor.org, bioinformatics forums, email | on server, email | on server, email |
Both Web and API versions are part of the web server.
For pathway visualization, the result graphs are interactive and hyperlinked when browsed online (Figure 1C). Similar features were implemented in KEGG website for function (gene, compound or connected pathways) annotation. But they are designated as user data centered features on Pathview Web. All nodes (molecules, genes/proteins/enzymes and metabolites/compounds) with user data mapped are colored and highlighted on the graphs. Importantly, these colored nodes are annotated with hover information, which indicates specifically which molecules from the user data are mapped there. The unmapped entries associated with the node will not show. In addition, these nodes are clickable and hyperlinked to reference pages in KEGG, which is convenient for results interpretation. Blank nodes indicate no user data mapped and are not clickable.
The data mapping and integration function become interactive too (Figure 1A). The full lists of species, gene and compound ID types and reference pathway IDs are available in drop-down boxes with autocompletion and spelling check. In addition, pathway IDs are presented in a listbox for click-selection. Notably, these options are interconnected in the web interface (Figure 1A). The lists of gene ID types and pathway IDs automatically switch corresponding to the species selected. On the other hand, compound IDs are not species specific. These features not only expand the data mapping/integration function of Pathview, but also reduce the otherwise complicated and error-prone steps into a few clicks.
Pathview is highly interoperable in that it easily fits into different pathway analysis workflows with different data types or tools. The web server capitalizes on this feature and provides a generic and integrated pathway analysis workflow (Figure 2B and Pathway Selection option in Figure 1A). This workflow is widely useful for different types of gene data and compound data (Figure 2B). Importantly, it can also be used for combined analysis of both gene data and compound data (Figure 3 and Table 2, Example 4 online). While the combined analysis has been implemented previously (24,25), Pathview Web server makes such analysis possible for a wide range of species and data types. Like Pathview itself, the pathway analysis workflow works for both numeric (e.g. abundance or expression levels) and categorical (e.g. gene or compound ID list) data. For numeric data, GAGE (7) is used due to its high versatility. For categorical data, over representation analysis is implemented using hypergeometric test. When both gene data and compound data are present, pathway analysis is done on each dataset separately first, then the results are combined into more robust global statistics/P-values through meta-analysis (Figure 2B). Note that the meta-analysis allows integration across numeric and categorical data.
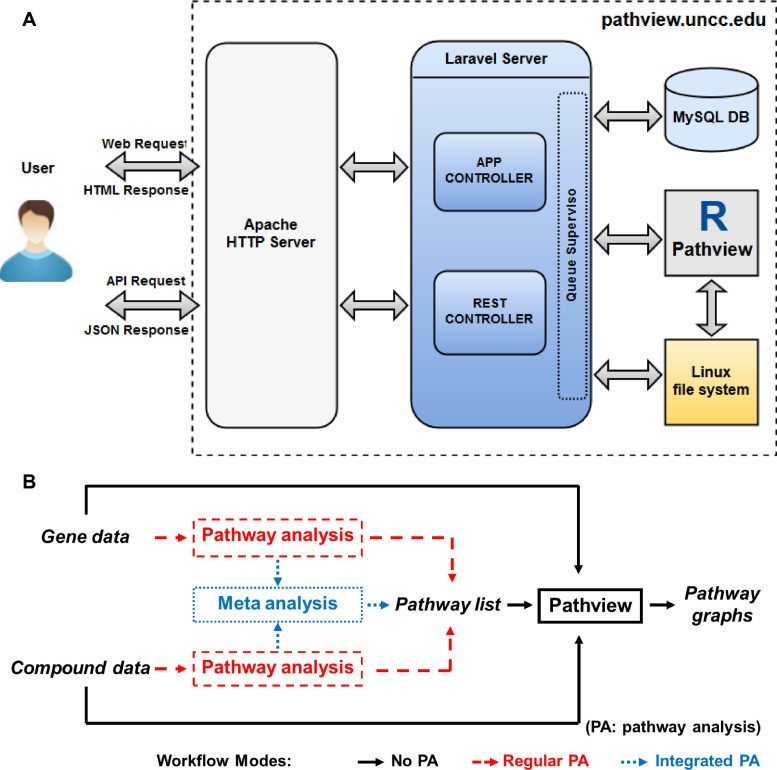
Figure 2.
The design of Pathview Web server: (A) architecture, (B) workflow and different modes. Note the workflow works with either gene data or compound data (No PA, Regular PA) or both together (No PA, Integrated PA). In the pathway analysis step, generally applicable gene-set enrichment (GAGE) is used for numeric data and over representation analysis for categorical data. In meta analysis step, P-values from regular pathway analysis are summarized into global P-values and pathways are then selected based on the latter.
Figure 3.
Integrated pathway analysis and visualization of both gene expression and metabolomics data (Example 4 online). Shown here is the KEGG pathway hsa00190 Oxidative phosphorylation. The analysis statistics is shown in Table 2.
Table 2. Results of integrated pathway analysis of both gene expression and metabolomics data (Example 4 online).
| Pathway | stat.gene | size.gene | stat.cpd | size.cpd | p.gene | p.cpd | p.val | q.val |
|---|---|---|---|---|---|---|---|---|
| hsa04141 Protein processing in endoplasmic reticulum | 3.79 | 144 | NA | 1 | 1.07E-10 | NA | 1.07E-10 | 2.17E-08 |
| hsa00190 Oxidative phosphorylation | 2.76 | 97 | 1.71 | 16 | 2.74E-06 | 1.99E-02 | 9.67E-07 | 9.77E-05 |
| hsa04142 Lysosome | 2.77 | 110 | NA | 4 | 2.28E-06 | NA | 2.28E-06 | 1.53E-04 |
| hsa03050 Proteasome | 2.68 | 39 | NA | NA | 6.66E-06 | NA | 6.66E-06 | 3.36E-04 |
| hsa00520 Amino sugar and nucleotide sugar metabolism | 1.52 | 41 | 2.68 | 83 | 9.29E-03 | 1.88E-04 | 2.48E-05 | 1.00E-03 |
| hsa03060 Protein export | 2.46 | 18 | NA | NA | 6.14E-05 | NA | 6.14E-05 | 2.07E-03 |
| hsa04510 Focal adhesion | −2.30 | 192 | NA | 2 | 8.43E-05 | NA | 8.43E-05 | 2.43E-03 |
| hsa04060 Cytokine-cytokine receptor interaction | −2.17 | 226 | NA | NA | 1.92E-04 | NA | 1.92E-04 | 4.84E-03 |
| hsa04080 Neuroactive ligand-receptor interaction | −1.49 | 242 | 2.20 | 53 | 1.01E-02 | 2.21E-03 | 2.61E-04 | 5.86E-03 |
| hsa04145 Phagosome | 2.07 | 131 | NA | 1 | 3.99E-04 | NA | 3.99E-04 | 8.06E-03 |
Example visualization is shown in Figure 3. Columns include test statistics for gene and compound data (2 and 4) and P-values (6–7), gene set sizes (3 and 5) and P- and Q-values for combined analysis (8–9)
Online server only features
In addition to the Pathview core functions, the web server provides extra features and a unique experience that are not available to the offline R package users (Table 1).
Pathview Web is constantly updated and improved in its interfaces and functions. The server synchronizes monthly with KEGG source databases through their REST API. The user always have access to the latest, most complete and accurate pathway graphs and data. In addition, the update processes run as part of web servers background routines and do not affect users. In opposite, R package users receive major updates in both pathway source data and software function every 6 months with Bioconductor release cycle.
The web server provides free registered user accounts (although registration is not required). All analysis history is saved with these accounts, including input data, analysis settings and results. Users can review, replicate and even share their analyses to other users easily. This feature enables collaborative research and reproducible science. In the meantime, all unauthorized access of user data are blocked.
The web server provides an important channel for user support and engagement for the Pathview project. Users can make comments and suggestions, or ask for help in the designated page. The server collects usage data, which help us better understand users needs. We also share these insights or measurement about the project with the users. For example, the summary usage statistics of both the sever and package are shown as real-time graphs in the frontpage. These user engagements and statistics would lead to better informed development.
The web server also serves as the homepage for the whole Pathview project. This centralized online resource include multiple supportive pages covering project description, documentation, references, tutorials, news, contacts and related links.
SYSTEM DESIGN AND IMPLEMENTATION
Input
The web form in the analysis page (GUI) collects all user inputs (Figure 1A). The most important user inputs and the only options with no default are the user data to be visualized or analyzed. There are two major categories of input data: (i) gene data cover any data that map to unique gene IDs including genes, transcripts, genomic loci, proteins, enzymes and their attributes; (ii) compound data cover any data that map to unique compound IDs including compounds, metabolites, drugs, small molecules and their attributes. Such generic definitions of gene and compound data allow Pathview to work with a wide range of data types. In addition, the input data may include just a few entries/molecules up to the full omic profiling, i.e. the entire genome, transcriptome, proteome or metabolome. They can be either matrices or vectors, both absolute and relative molecular abundance (gene expression etc). Two most commonly used data file formats are supported: tab-delimited text (txt) or comma-separated values (csv). Other essential user options include species, gene and compound ID types, the method for pathway selection (manual or auto) or/and the target pathway IDs (if manual selection). All user options are self-explanatory and fully described online with examples.
Output
The main results/outputs are pathway graphs with user data mapped (Figures 1C and 3). Pathview generates graphs in two styles: either the native KEGG view (Example 1 online) or the Graphviz view (Example 2 online). The former renders user data on native KEGG pathway graphs (raster images) and is more interpretable with abundant context and meta-data. The latter layouts pathway graph using Graphviz engine (vector images) and provides better control over node/edge attributes and graph topology. In the browser, native KEGG graphs are interactive with hover and hyperlinked annotations (Figure 1C).
When automatic pathway selection is chosen, pathway analysis will be done before data visualization on selected pathways (Figure 3 and Table 2, Example 4 online). The pathway analysis statistics will be returned and all analysis results will be included in a zipped folder for download (Figure 1B).
Help
We also provide a comprehensive help system with Pathview Web (Figure 1A). The help system has four components: (i) a centralized help pages, including documentation of all user options, input and output description; (ii) multiple example analyses with input data pre-loaded and options pre-set, plus dedicated tutorials; and (iii) the help button next to each user option in the web form, which directly links to its help page (Figure 1A); (iv) extra user support channels through emails and the question page.
Architecture
Figure 2A is the block diagram for the Pathview Web server architecture. We used the LAMP (Linux, Apache, MySQL and PHP) stack as the basic development environment. The central components include the Apache HTTP server and the PHP application server using Laravel 5 Framework. The GUI or web form was written in HTML and JavaScript, which accepts and validate the user inputs. The Apache server acts as the gateway between the user interface and the application server, i.e. routing the user requests to application server and returning the responses to the user. The application server coordinates the back-end processes. It interacts with MySQL database to read and write user or analysis related data and meta-data. It starts an R session with Pathview for each submitted analysis. The bulk computing or the actual data analysis is by Pathview in R. The Pathview process has been previously described (10) and fully documented (26). All analysis results and log files are written to the Linux file system. The application server then reads these files and creates a response for the user request.
Queue
The Apache server provides default request queuing mechanisms, which handles multiple user requests in parallel. Pathview server GUI can still be overloaded based on the user number estimated from R package. To ensure fair and efficient access, we implement a request queue explicitly through Laravel Queue Supervisor. We set up a fixed number of threads/workers on to serve the user requests. The number of analysis requests a user (or IP address) can submit is unlimited, but only a limited number of them enter the execution queue while others will be cached until some earlier analysis is completed.
API
Pathview Web server can also be accessed programmatically through a RESTful API. Users just need to download a single Bash shell script. No installation is needed and minimal local computing resource is required. For users with moderate internet access, the API is usually faster than GUI and R versions. All user options in the GUI are implemented in API. API users can directly use the integrated pathway analysis workflow (Figure 2B) on the server or they can freely assemble their own analysis workflow.
The API has a dedicated help system with four components (Figure 1A): (i) like for GUI, the server provides a centralized help page for API, which includes a tutorial and user option documentation. (ii) All example analyses have the API version too. (iii) To help new users with less programming experience, we set up a unique API query generator. Users can click through the same GUI web form and assemble error-free API query statements interactively. The result page returns both the full API statement with all options and a shorter version skipping default values. (iv) In addition to online help, the API script also features a detailed command-line help page like other shell programs.
Pathview Web API is RESTful as it adheres to the REST(Representational State Transfer) architecture. Pathview Web API and GUI have different front ends (command line versus web page) and response formats (JSON versus HTML), but they share the same backend (Figure 2A).
EXAMPLES AND USE CASES
To help users get started, we devised four example analyses (left navigation in Figure 1A). In these examples, we used real data or simulated data. These example data analyses are made easily accessible from the sticky left navigation column. The example analysis interfaces look identical to the regular analysis, except with input data pre-loaded and options pre-set for the intended analysis. All users need to do is to click and submit the analysis. To help them learn from these example analyses, all input data files can be clicked, viewed or downloaded. Users can easily explore and customize all example analyses by modifying the input options.
The four example analyses represent the most representative use cases: (i) KEGG view of multiple-sample data, (ii) Graphviz view of multiple-sample data, (iii) visualization of data with ID mapping and (iv) integrated pathway analysis with both gene and compound data. While Example analyses 1–3 covers the two primary functions of Pathview package: visualization, data mapping and integration (10). Pathview also has a third core feature: highly interoperable and easy fit into different pathway analysis workflows (10). Example 4 covers the comprehensive workflow (Figure 2B) including pathway analysis, data integration and visualization, i.e. built on all above-mentioned core features. It demonstrates a study including both high throughput gene expression and metabolomics profiling. The user options of this analysis are similar to other examples, except that Pathway Selection option is set to auto instead of manual. In this case, pathway analysis is done as to determine the target pathways for downstream visualization and interpretation. Example 4 is even more sophisticated with both gene and compound data included. Therefore, pathway analysis is done on these data separately first, followed by data integration.
As an example of integrated pathway analysis workflow (Figure 2B), Example 4 returns: (i) separate pathway analysis results for gene data and compound data (Figure 1B); (ii) meta-analysis results that integrates both gene data and compound data (Table 2 and Figure 1B); (iii) Pathview graphs that map and integrate gene data and compound data into the selected pathways from the pathway and meta-analysis above (Figure 3). Apparently, both gene data (expression) and compound data (metabolomic) results contribute to the meta-analysis, although the former gets more weight. These results make sense biologically and suggest that multiple metabolic processes are greatly upregulated in breast cancer (ductal carcinoma in situ) samples versus normal controls. These findings are mostly novel and not reported in the original study (27). This example demonstrates the power of pathway analytics and visualization via Pathview Web server.
DISCUSSION
We have presented Pathview Web, a server for pathway-based visualization and data integration. Pathview Web provides both graphical online portal (GUI) and programmatic access (API) to Pathview. It extends the static package into an interactive bioinformatics server. With no special training or computing resources, users now have full access to high quality and novel solutions for pathway-based data visualization, integration and analytics. These solutions support a wide range of omics data and their integration. Pathview Web could further increase the impact and user base of the Pathview project substantially.
Being an integral part of the Pathview project, the web server will grow with the project too. For example, we have been funded for a major expansion of the core functions of Pathview package (NSF ABI-1565030). Once mature, these expanded functions will be incorporated into the server.
AVAILABILITY
The Pathview Web server is freely and openly available at https://pathview.uncc.edu/.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Jinbo Xu at Toyota Technological Institute at Chicago for his comments on the manuscript, also thank the Department of Bioinformatics and Genomics and the Bioinformatics Service Division (BiSD) at UNC Charlotte for the extra support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [ABI-1565030 to W.L.]; UNC Charlotte CCI Faculty Innovation Award [2015 to W.L.]. Funding for open access charge: National Science Foundation [ABI-1565030 to W.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999; 27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Croft D., O’Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B. et al. . Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011; 39:D691–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kelder T., van Iersel M.P., Hanspers K., Kutmon M., Conklin B.R., Evelo C.T., Pico A.R.. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012; 40:D1301–D1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caspi R., Altman T., Billington R., Dreher K., Foerster H., Fulcher C.A., Holland T.A., Keseler I.M., Kothari A., Kubo A. et al. . The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014; 42:D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mi H., Muruganujan A., Thomas P.D.. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013; 41:D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C.. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011; 39:D685–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J.. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009; 10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tarca A.L., Draghici S., Khatri P., Hassan S.S., Mittal P., Kim J.S., Kim C.J., Kusanovic J.P., Romero R.. A novel signaling pathway impact analysis. Bioinformatics. 2009; 25:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo W., Brouwer C.. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013; 29:1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu Z., Chang Y.C., Wang Y., Huang C.L., Liu Y., Tian F., Granger B., Delisi C.. VisANT 4.0: Integrative network platform to connect genes, drugs, diseases and therapies. Nucleic Acids Res. 2013; 41:W225–W231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kono N., Arakawa K., Ogawa R., Kido N., Oshita K., Ikegami K., Tamaki S., Tomita M.. Pathway projector: web-based zoomable pathway browser using KEGG atlas and Google Maps API. PLoS One. 2009; 4:e7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kutmon M., van Iersel M.P., Bohler A., Kelder T., Nunes N., Pico A.R., Evelo C.T.. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput. Biol. 2015; 11:e1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T.. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011; 27:431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada T., Letunic I., Okuda S., Kanehisa M., Bork P.. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011; 39:W412–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kramer F., Bayerlova M., Beissbarth T.. R-based software for the integration of pathway data into bioinformatic algorithms. Biology. 2014; 3:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villaveces J.M., Koti P., Habermann B.H.. Tools for visualization and analysis of molecular networks, pathways, and -omics data. Adv. App. Bioinformatics Chem. 2015; 8:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M.. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004; 37:914–939. [DOI] [PubMed] [Google Scholar]
- 19. Junker B.H., Klukas C., Schreiber F.. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006; 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arakawa K., Kono N., Yamada Y., Mori H., Tomita M.. KEGG-based pathway visualization tool for complex omics data. In silico Biol. 2005; 5:419–423. [PubMed] [Google Scholar]
- 21. King Z.A., Drager A., Ebrahim A., Sonnenschein N., Lewis N.E., Palsson B.O.. Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput. Biol. 2015; 11:e1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M.. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012; 40:D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamburov A., Cavill R., Ebbels T.M., Herwig R., Keun H.C.. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011; 27:2917–2918. [DOI] [PubMed] [Google Scholar]
- 25. Xia J., Sinelnikov I.V., Han B., Wishart D.S.. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015; 43:W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo W. Pathview package: a tool set for pathway based data integration and visualization. 2013. [DOI] [PMC free article] [PubMed]
- 27. Emery L.A., Tripathi A., King C., Kavanah M., Mendez J., Stone M.D., de las Morenas A., Sebastiani P., Rosenberg C.L.. Early dysregulation of cell adhesion and extracellular matrix pathways in breast cancer progression. Am. J. Pathol. 2009; 175:1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.