Abstract
Background
Inhibitors of fibroblast growth factor receptors (FGFRs) have recently arisen as a promising treatment option for patients with FGFR alterations. Gene fusions involving FGFR3 and transforming acidic coiled-coil protein 3 (TACC3) have been detected in diffuse gliomas and other malignancies, and fusion-positive cases have responded well to FGFR inhibition. As high FGFR3 expression has been detected in fusion-positive tumors, we sought to determine the clinical significance of FGFR3 protein expression level as well as its potential for indicating FGFR3 fusions.
Methods
We performed FGFR3 immunohistochemistry on tissue microarrays containing 676 grades II–IV astrocytomas and 116 grades II–III oligodendroglial tumor specimens. Fifty-one cases were further analyzed using targeted sequencing.
Results
Moderate to strong FGFR3 staining was detected in gliomas of all grades, was more common in females, and was associated with poor survival in diffuse astrocytomas. Targeted sequencing identified FGFR3-TACC3 fusions and an FGFR3-CAMK2A fusion in 10 of 15 strongly stained cases, whereas no fusions were found in 36 negatively to moderately stained cases. Fusion-positive cases were predominantly female and negative for IDH and EGFR/PDGFRA/MET alterations. These and moderately stained cases show lower MIB-1 proliferation index than negatively to weakly stained cases. Furthermore, stronger FGFR3 expression was commonly observed in malignant tissue regions of lower cellularity in fusion-negative cases. Importantly, subregional negative FGFR3 staining was also observed in a few fusion-positive cases.
Conclusions
Strong FGFR3 protein expression is indicative of FGFR3 fusions and may serve as a clinically applicable predictive marker for treatment regimens based on FGFR inhibitors.
Keywords: biomarker, gene fusion, glioblastoma, targeted sequencing
Importance of the study
FGFR3 gene fusions, initially discovered in glioblastoma, have since been reported in a wide spectrum of other malignancies. FGFR3 fusion-positive tumors have responded well to FGFR inhibitors in ongoing clinical trials. We demonstrate that FGFR3 staining is negative in most diffuse gliomas, whereas strong FGFR3 staining is observed in all fusion-positive cases, which facilitates stratification of FGFR3 fusion-positive gliomas. FGFR3 immunostaining does not rely on prior knowledge about fusion breakpoints or partners and allows evaluation of intratumoral heterogeneity by revealing heterogeneous expression levels and FGFR3-negative subclones. Fusions were only detected in aggressive isocitrate dehydrogenase wild-type tumors and were more common in female patients. Interestingly, moderate to strong FGFR3 staining was associated with a lower MIB-1 proliferation index without any prognostic benefit. The obtained information is relevant when FGFR3 fusions are targeted for therapeutic purposes and for better understanding of FGFR3 fusion-driven tumor development and progression.
Glioma pathology has recently experienced great advancements as sequencing studies by the working group of The Cancer Genome Atlas1,2 and others3,4 have identified new molecular criteria for the diagnosis and stratification of gliomas. The World Health Organization recently published new definition criteria for glioma diagnosis.5 Diffuse gliomas can be currently stratified into 3 main categories based on isocitrate dehydrogenase (IDH) mutation status and the presence of 1p/19q codeletion.1–4 IDH wild-type tumors, which are present in all diffuse glioma grades, have usually the poorest survival rates and are generally less responsive to treatment,6 highlighting the need for better treatment options for these patients. Since the discovery of recurrent fibroblast growth factor receptor (FGFR) gene fusions in glioblastoma (GBM),7,8 targeted treatment regimens using FGFR inhibitors have arisen as a promising option for glioma patients with FGFR alterations. Of those, oncogenic FGFR3 fusions and various FGFR1 alterations have been detected in brain tumors.7–14 In addition, FGFR fusions and other FGFR alterations have been detected in several extracranial malignancies.10,15–20 FGFR3 is most commonly fused to the transforming acidic coiled-coil protein 3 gene (TACC3), but other fusion partners also exist, such as recurrent FGFR3–BAIAP2L1 fusions in bladder cancer.18 Several FGFR inhibitors are currently being tested in clinical trials for different cancer types,21 and FGFR3 fusion-positive cells and tumors show the best treatment responses.18,22 Responses to FGFR inhibitor treatment have also been reported in GBM.9,22 Tumors that carry FGFR3 fusions or other responsive FGFR alterations represent a minority of cases in these malignancies, which emphasizes the need for efficient patient stratification tools, as also stated by others.23
We have previously shown that FGFR3 protein expression is suppressed by miR-99a, one of the most abundantly expressed miRNAs in gliomas and normal brain tissue.8 High fusion protein levels have been reported in GBM,8,9 most likely because the miR-99a binding site is removed by genomic rearrangements that generate FGFR3 fusions.8 The miR-99a–mediated suppression of FGFR3 may explain why activating FGFR3 mutations are not observed in diffuse gliomas. The relationship between FGFR3 immunostaining intensity and fusion status or FGFR3 expression levels in fusion-negative cases has not been systematically analyzed before. We wanted to determine whether strong FGFR3 staining, detected using an antibody that recognizes an epitope present in all reported gene fusions, could be used as a marker for FGFR3 gene fusion. We therefore used immunohistochemistry (IHC) to detect FGFR3 in 791 diffuse glioma cases and associated FGFR3 expression with patients’ clinical features. Genetic alterations in FGFR3 and other glioma-associated genes were identified by targeted sequencing of selected cases. Our results demonstrate that strong FGFR3 staining characterizes FGFR3 fusion-positive diffuse gliomas. FGFR3 fusions involved several fusion partners, junctions, and breakpoints, rendering them difficult to detect in a comprehensive manner with PCR-based methods in a diagnostic context.
Materials and Methods
Patient Tissue Samples
The study design was approved by the ethical committee of Tampere University Hospital and the National Authority for Medicolegal Affairs in Finland. Diffuse glioma samples had been obtained from tumor surgery patients at Tampere University Hospital between 1983 and 2009 and diagnosed according to the World Health Organization (WHO) 2007 classification for this study. We used 676 diffuse astrocytoma samples and 116 oligodendroglial tumors for FGFR3 detection (Table 1). Cause-specific survival association of grades II–IV diffuse astrocytomas was analyzed using primary tumors (n = 533) (Table 1, Supplementary Table 1). The samples were fixed in formaldehyde (buffered with 4% phosphate), embedded in paraffin, and processed for tissue microarray (TMA).24
Table 1.
Patient demographic and clinical characteristics in the astrocytic and oligodendroglial tumor cohorts*
| Astrocytoma | Grade II | Grade III | Grade IV | All |
|---|---|---|---|---|
| No. of patients | 114 | 59 | 503 | 676 |
| Sex (n) | ||||
| Male | 82 | 38 | 287 | 407 |
| Female | 32 | 21 | 216 | 269 |
| Age, y | ||||
| Median (mean ± SD) | 46 (46 ± 16) | 50 (52 ± 17) | 65 (65 ± 13) | |
| Minimum | 12 | 23 | 4 | |
| Maximum | 83 | 84 | 90 | |
| Follow-up | ||||
| No. of survivors at the end of follow-up | 25 | 6 | 9 | |
| Follow-up duration for survivors, mo (median [mean ± SD]) | 92 (108 ± 77) | 141 (142 ± 80) | 59 (98 ± 69) | |
| 5-y survival rate, % | 58 | 32 | 3.4 | |
| Tumors (n) | ||||
| Primary | 81 | 41 | 414 | |
| Second recidive | 25 | 13 | 69 | |
| Third recidive | 8 | 4 | 17 | |
| Fourth-sixth recidive | 0 | 1 | 2 | |
| Oligodendroglial tumor | O II | O III | OA II | OA III |
| No. of patients | 38 | 33 | 23 | 22 |
| Sex (n) | ||||
| Male | 19 | 17 | 12 | 8 |
| Female | 19 | 16 | 11 | 14 |
| Age, y | ||||
| Median (mean ± SD) | 45 (45 ± 12) | 57 (57 ± 10) | 52 (46 ± 17) | 42 (46 ± 15) |
| Minimum | 26 | 38 | 5 | 25 |
| Maximum | 69 | 72 | 64 | 74 |
| Follow-up | ||||
| No of survivors at the end of follow-up | 25 | 9 | 9 | 4 |
| Follow-up duration for survivors, mo (median [mean ± SD]) | 83 (106 ± 64) | 96 (66 ± 50) | 94 (161 ± 121) | 203 (221 ± 114) |
| 5-y survival rate, % | 79 | 43 | 84 | 50 |
| Tumors (n) | ||||
| Primary | 34 | 21 | 19 | 18 |
| Second recidive | 4 | 10 | 4 | 4 |
| Third recidive | 0 | 1 | 0 | 0 |
| Fourth-sixth recidive | 0 | 1 | 0 | 0 |
Abbreviations: O, oligodendroglioma, OA, oligoastrocytoma.
*Patient age and follow-up information were calculated using primary cases. Follow-up times are shown in months (mo).
In the 2016 WHO classification, IDH mutation and 1p/19q codeletion are used as additional criteria for more detailed diffuse glioma diagnosis. In our cohort, 113 astrocytomas (20%, 562 analyzed) and 79 oligodendroglial tumors (81%, 97 analyzed) were positive for IDH p.R132H. The 1p/19q codeletion was detected in 28 (90%, 31 analyzed) of IDH p.R132H positive-staining oligodendroglial tumors using fluorescence in situ hybridization.25 Our cohort thus includes 96 IDH-mutant diffuse astrocytomas, 40 IDH-mutant GBMs, and 28 oligodendrogliomas (IDH-mutant and 1p/19q codeleted) according to the WHO 2016 criteria. The remaining tumors with inconclusive genetic testing are classified as diffuse astrocytoma, not otherwise specified (NOS) (80 tumors), oligodendroglioma, NOS (48 tumors), oligoastrocytoma, NOS (37 tumors), and glioblastoma, NOS (463 tumors).
Immunohistochemistry Staining
We stained 24 TMA slides using mouse monoclonal FGFR3 antibody (1:600 dilution; #sc-13121, Santa Cruz Biotechnology) together with the Dako EnVision+ kit (K4006) and antibody diluent S0809. Antigen retrieval was performed in sodium citrate buffer (pH 6.0), and immunoperoxidase reactions were visualized with 3,3'-diaminobenzidine using hematoxylin for counterstaining. IDH p.R132H, MIB-1, and p53 staining have been reported previously.26,27 An experienced neuropathologist (H.H.) and an experienced cell biologist (K.G.) independently scored FGFR3 IHC staining intensities as 0 (no staining), 1 (weak), 2 (moderate), or 3 (strong staining). Consensus interpretation was used in the case of discrepant interpretation. MIB-1 staining on TMAs was used for the proliferation analysis.
For antibody blocking, either peptide corresponding to full-length epitope (amino acids 25–124 of FGFR3 of human origin) (Biocenter Finland Protein Service) or peptide corresponding to amino acids 40–64 (GPEPGQQEQLVFGSGDAVELSCPPP) (GenScript) efficiently blocked antibody staining. Antibody was incubated (30 min room temperature) with or without the excess blocking peptide (5 times the antibody concentration in ng/µL units) in antibody diluent before adding the mixture onto slides.
Statistical Methods
Statistical analyses were performed using R or IBM SPSS statistics 21.0 software for Windows. For tables larger than 2 × 2, the P-values of Fisher’s exact test were calculated using Monte Carlo simulation with 2.5*107replicates.
Targeted Sequencing
Fifty-one samples with staining data from whole-mount tissue samples were analyzed using pulldown-based targeted sequencing. These included 15 strongly stained, 22 moderately stained, 6 weakly stained, and 8 negatively stained samples. Most samples were formalin fixed and paraffin embedded (FFPE). The turXTRAC FFPE DNA kit (Covaris) or AllPrep DNA/RNA Mini Kit (Qiagen) was used for DNA isolation. The QIAamp DNA Mini Kit (Qiagen) was used for 4 freshly frozen GBM samples that were available. Target regions were enriched for sequencing from 1 μg of extracted DNA using the Sureselect XT Target enrichment system and custom-designed RNA probes (Supplementary Table 2). The sequencing libraries were prepared according to the kit (200 ng of DNA samples) with a shorter DNA-shearing protocol (220 sec) and were sequenced with an Illumina MiSeq instrument. Reads were aligned against the GRCh37 human reference genome using Bowtie-2.2.4.28
Somatic Mutation Calling and Fusion Detection
Somatic mutations were called by searching for variants with an alternate allele fraction of at least 10%, and at least 5 supporting reads. Additionally, the allele fraction was required to be 20 times higher than the background error rate (ie, the average allele fraction across all nontumor samples). Variants with a population frequency of 0.5% or above in the 1000 Genomes or ESP6500 database were filtered out. Protein-level outcomes were predicted using ANNOVAR.29 To discover chromosomal rearrangements for fusion detection, unaligned reads from each sample were split into two 30 bp anchors (one from both ends) that were aligned to the hg38 genome using Bowtie-1.1.2. Discordant anchor pairs were grouped by position, and groups with 8 or more supporting reads were flagged as rearrangement candidates and manually curated using IGV and BLAT.
Copy Number Analysis
Read counts were calculated using Bedtools-2.25.0.30 At least 25% of a read was required to be within the targeted capture region for the read to be counted. For each sample, we calculated coverage log ratios at all captured regions against a median reference derived from gender-matched blood samples from healthy patients (2 for each gender). We then corrected for guanine-cytosine content bias in coverage log ratios using a Theil–Sen estimator (after observing that guanine-cytosine content and coverage log ratio bias had a linear relationship in this cohort) (Supplementary Figure 1). Finally, we forced the median log ratio of control baits in infrequently copy number altered chromosomes 5, 8, 11, and 18 to zero by subtracting the median from all log ratios. Copy numbers were called by visual inspection of violin plots that visualize coverage log ratios for each target gene.
Computational Analysis of FGFR3 IHC Staining
FGFR3 staining intensities were computationally analyzed from whole-mount FFPE tissue samples of cases that were used for targeted sequencing. One to 3 representative regions of interest (size 1.4–52 mm2, average 9.7 mm2) were selected from scanned tissue images. Red versus blue ratio, which was used as a readout of staining intensity, was calculated for each pixel in the regions of interest. To remove background and nuclear hematoxylin staining, only pixels with a red/blue ratio ≥1.00 were included in the analysis. The mean of red/blue ratios was calculated from these pixel locations.
Results
Moderate to Strong FGFR3 Staining Associated with Poor Survival and Absence of IDH1 p.R132H Mutation
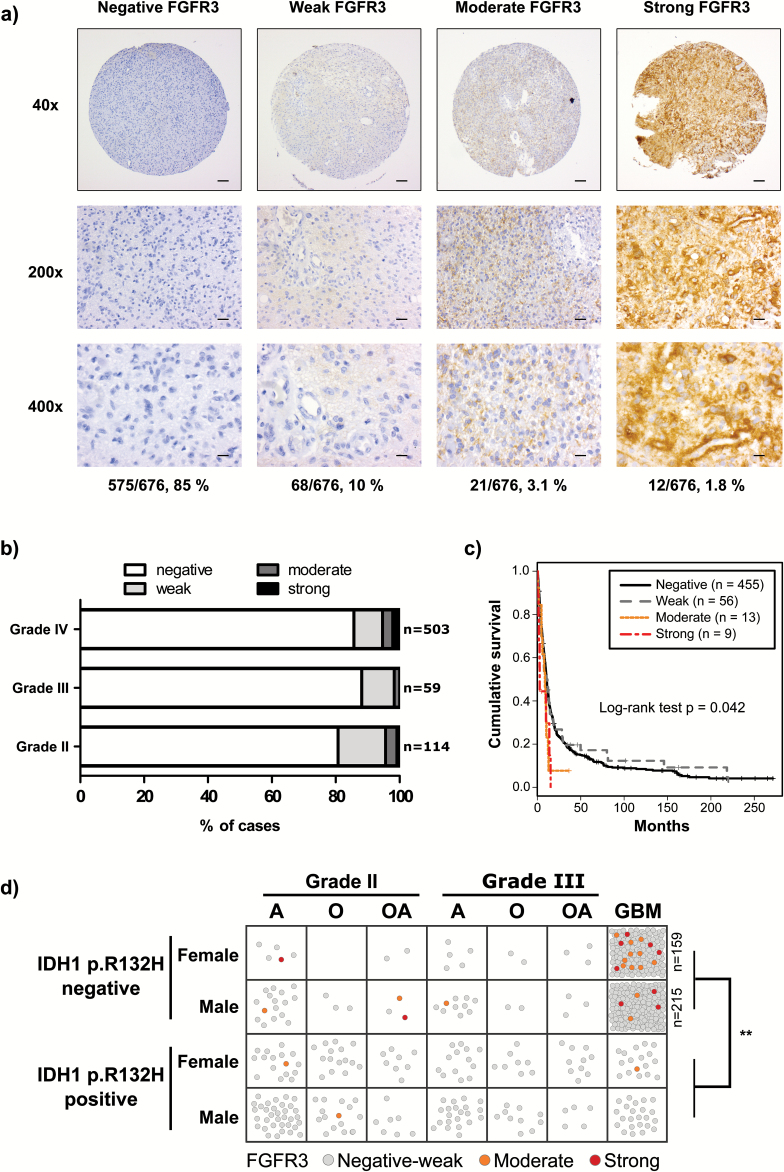
We used an antibody that targets amino acids 25–124 in the FGFR3 N-terminus to perform IHC staining on 676 diffuse astrocytomas and 116 grades II–III oligodendroglial tumors (Table 1). Moderate to strong FGFR3 staining was generally diffuse and cytosolic with occasional nuclear staining, whereas membrane-restricted staining was observed in some of the more weakly stained cases (Fig. 1A, Supplementary Figure 2). Negatively stained blood vessels provided an internal control for antibody specificity (Supplementary Figure 3a). Normal brain tissue was generally negative, with the exception of the cerebellar and cerebral molecular layers, where weak to moderate staining was observed (Supplementary Figure 3B–C). Among astrocytomas, 575 (85%) cases were fully negative for FGFR3. Among samples with FGFR3 immunoreactivity, 68 (10%) showed weak, 21 (3.1%) moderate, and 12 (1.8%) strong positivity. The proportion of moderately to strongly stained cases was within the range of previous estimates of FGFR3 fusion frequency.1,2,7,8 FGFR3 staining was not associated with tumor grade (Fig. 1B). Among 116 oligodendroglial tumors, only 2 cases (1.7%) exhibited moderate and 1 case (0.9%) strong FGFR3 staining; all 3 cases were grade II (Supplementary Figure 4).
Fig. 1.
Moderate or strong FGFR3 staining was associated with worse survival and lack of IDH1 p.R132H mutation. (A) Representative staining images. Proportion of samples for each staining category in grades II–IV astrocytomas is indicated below the representative images. Scale bars: 100 µm in 40×, 20 µm in 200×, and 10 µm in 400× images. (B) FGFR3 staining distribution in grades II–IV astrocytomas was not associated with tumor grade (P = .523, Fisher’s exact test). (C) Moderate or strong FGFR3 staining was associated with poor cause-specific survival (n = 533, P = .042, log-rank test). (D) Moderate to strong FGFR3 staining was more common in IDH1 p.R132H staining-negative cases. Each sample is marked with a circle and colored according to FGFR3 staining. IDH1 p.R132H-negative GBM samples included 159 females and 215 males; thus, all the negatively to weakly stained cases are not visualized separately. **P < .01, Fisher’s exact test. A: astrocytoma, O: oligodendroglioma, OA: oligoastrocytoma.
An association analysis of astrocytic tumors (Suppl ementary Figure 5) revealed that moderate to strong FGFR3 staining was more common in females (8.55%) than in males (2.46%) (P < .001, Fisher’s exact test) and less common in samples with aberrant p53 expression (P < .010, Fisher’s exact test) (Supplementary Figure 6). In the prognostic analysis, moderate and strong FGFR3 staining was associated with a significantly shorter cause-specific survival duration than weak and negative staining (P = .0417, n = 533, log-rank test) (Fig. 1C). Although the association remained significant after adjustment for grade, proliferation, and IDH1 p.R132H mutation status (P = .0422, Cox proportional hazards model) (Supplementary Table 3), no significant associations were observed when analysis was restricted to grade IV GBM (P = .203, n = 414, log-rank test) or cases lacking IDH1 p.R132H mutation (P = .119, n = 370 for grades II–IV astrocytomas and P = .525, n = 343 for GBMs, log-rank test) (Supplementary Figure 7), suggesting that the reduced survival of moderately to strongly stained cases is at least partly explained by their IDH1 mutation status and high proportion of glioblastoma tumors. Indeed, moderate to strong FGFR3 staining was only observed in patients who lacked IDH1 p.R132H mutation, and all except one of them suffered from GBM in the survival analysis (Supplementary Figure 8). In the whole diffuse glioma cohort, we found only 3 IDH1 p.R132H–positive cases with moderate and none with strong FGFR3 staining (P = .0063, Fisher’s exact test, for the IDH1 p.R132H staining association) (Fig. 2D, Supplementary Figure 9). The immunostaining results strongly coincide with previously reported data on FGFR3 fusion-positive tumors (Supplementary Figure 10),1,2,8,9 supporting the use of IHC for FGFR3 fusion detection.
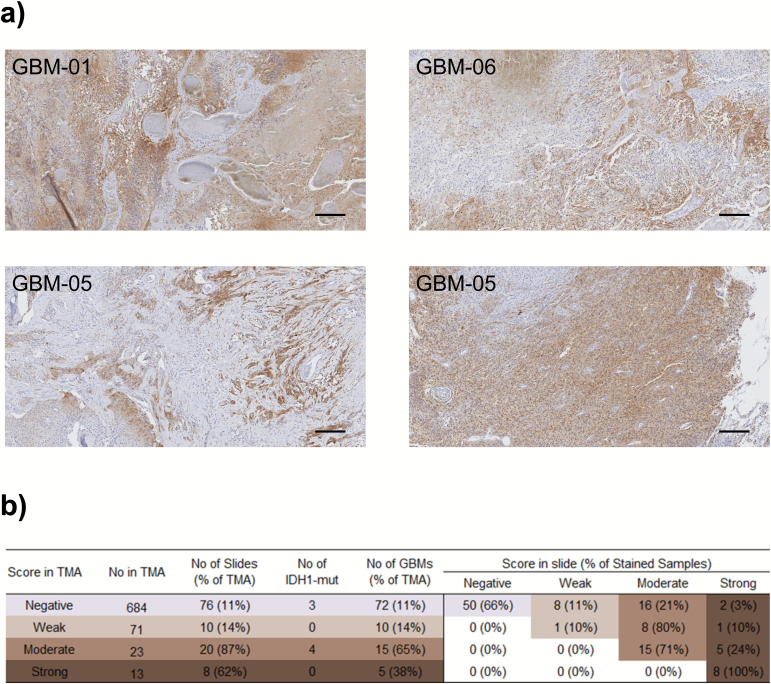
Fig. 2.
FGFR3 staining is heterogeneous. (A) Representative images (50× magnification) of heterogeneously stained areas in 3 cases with strong FGFR3 staining. In sample GBM-05, the staining pattern clearly differed between tumor sites. Scale bar 50 µm. (B) Summary of FGFR3 IHC staining data from whole-mount tissue slides. Most samples (65% of all) retain their original FGFR3 staining score evaluated from TMA data, but especially moderate scoring was observed in many samples (21% of all) with negative to weak staining on TMA. Numbers of TMA samples, whole-mount tissue samples used for staining, IDH1 p.R132H staining-positive samples, and GBM samples are marked on the left-hand side. On the right, shown are the number of whole-mount tissue samples and their proportion of all the stained slides with the specified staining score on TMAs.
FGFR3 Staining Is Spatially Heterogeneous
Next, we performed FGFR3 staining on all the available whole-mount tissue samples with moderate to strong FGFR3 staining on TMA. In addition, a subset (11%) of GBM samples with negative to weak staining on TMA were analyzed using whole-mount tissue sections. Heterogeneous FGFR3 staining of malignant tissue was generally observed (Fig. 2A). Strong staining was subregional, and staining intensities typically varied in a continuous fashion. Most samples (65%, n = 114) had the same FGFR3 staining score on both the TMA and the whole-mount tissue slide (Fig. 2B), but higher staining intensities were observed in one third of the samples. It is thus better to perform FGFR3 staining for diagnostic purposes using whole-mount tissue slides, as they will be more representative than biopsies or TMAs.
Cases with Strong FGFR3 Staining Harbor FGFR3 Fusion Rearrangements
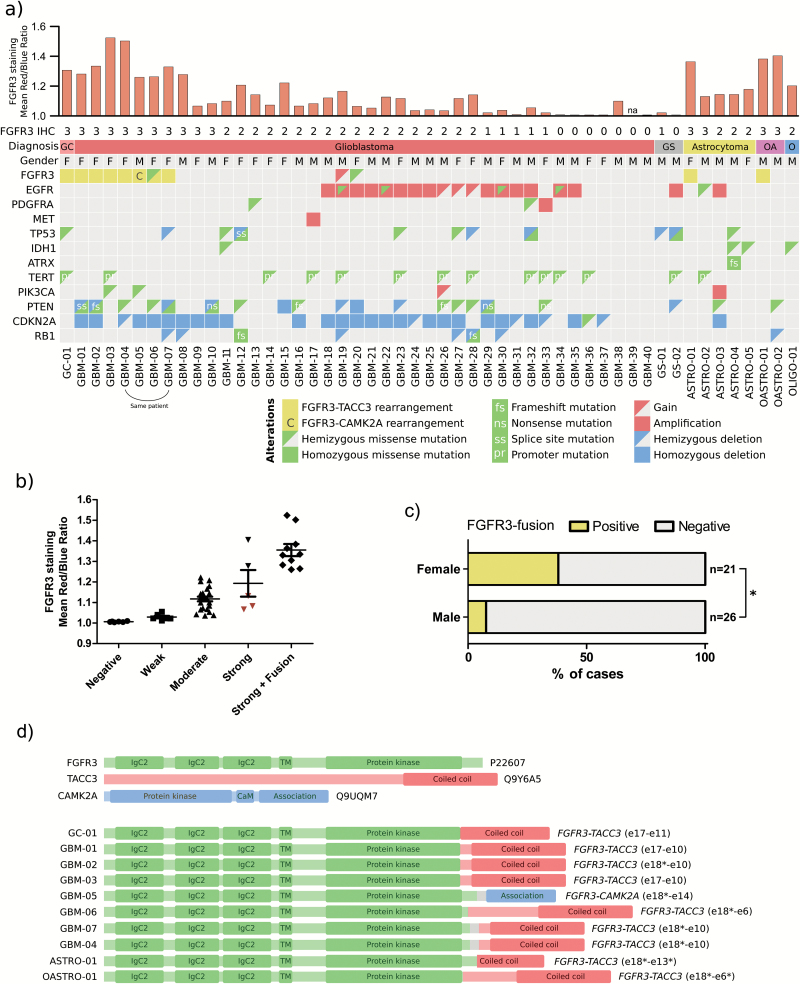
To detect genomic breakpoints in cases with positive FGFR3 staining, we selected 51 diffuse glioma samples (including 43 GBMs, 5 grade II astrocytomas, 2 grade II oligoastrocytomas, and 1 grade II oligodendroglioma according to initial diagnosis) for targeted sequencing. We analyzed all the patients with moderate to strong FGFR3 staining in whole-mount tissue slides and with enough tumor material for sequencing, but the cohort included also negatively to weakly stained cases. In addition to FGFR3 and FGFR1, the sequencing panel targeted genes with reported alterations in diffuse gliomas, such as IDH1, IDH2, TP53, ATRX, CIC, FUBP1, CDKN2A, and RB1. FGFR3 rearrangements that generated gene fusions were detected in 10 cases (Fig. 3A, Supplementary Table 4), including 9 FGFR3-TACC3 fusions and 1 FGFR3-CAMK2A fusion. In addition, case GBM-19 harbored an FGFR3 copy number gain (Fig. 3A) and a rearrangement that joined the WHSC1 gene, located downstream of FGFR3, to TACC3 (data not shown). The DNA breakpoints and resulting FGFR3 fusion junctions were distinct in each tumor (Fig. 3C, Supplementary Figure 11), but the coiled-coil domain of TACC3 was conserved in FGFR3-TACC3 fusions. CAMK2A codes for a subunit of calcium/calmodulin-dependent protein kinase II (CAMK2).31,32 The C-terminal self-association domain of CAMK2A is joined with nearly the full-length FGFR3 protein.31,32 This is also the case for the fusion of FGFR3 and embryonic lethal abnormal vision (ELAV)–like RNA binding protein 3 (ELAVL3) previously reported in oligoastrocytoma1,33 (Supplementary Figure 10). It is thus likely that ELAVL3 and CAMK2A facilitate spontaneous fusion protein oligomerization and drive the oncogenic activity of FGFR3 fusion through increased kinase activity, as suggested for previously reported FGFR3 fusions.18,19
Fig. 3.
FGFR3 fusion-positive cases showed strong FGFR3 staining. Most were female and lacked alterations in IDH, EGFR, and PDGFRA genes. (A) FGFR3 rearrangements and associated alterations were detected by targeted sequencing. FGFR3 staining scores (3: strong, 2: moderate, 1: weak, 0: negative staining) on whole-mount tissue slides and computational analyzed staining intensities are marked above the aberration matrix. The cases had been initially diagnosed as GBMs (one giant cell glioblastoma [GC] and 2 gliosarcomas [GS]), grade II astrocytomas (A), grade II oligoastrocytomas (OA), and a grade II oligodendroglioma (O). M: male, F: female. (B) Computationally analyzed FGFR3 staining intensity is significantly higher in fusion-positive cases than in negatively to moderately stained fusion-negative cases (P < .0001, Kruskal–Wallis test). Cases with focal strong FGFR3 staining are marked in red. (C) FGFR3 fusions were more commonly observed in females than in males (*P < .05, Fisher’s exact test). (D) All FGFR3 fusion partners carried dimerization domains. Predicted fusion protein structures are shown in the figure. Intronic regions that become part of the translated fusion protein are marked with gray. *In-frame fusion junction is located inside the marked exon.
Most positive cases were GBMs, but FGFR3-TACC3 fusion rearrangements were also found in one grade II tumor with astrocytic morphology and one grade II tumor with oligoastrocytic morphology. Both cases were IDH wild-type in targeted sequencing, and thus represent “diffuse astrocytoma, IDH wild-type” in the WHO 2016 classification.5 This is also the case for the IDH wild-type grade II tumors ASTRO-02 and OASTRO-02, which showed strong FGFR3 staining but did not carry detectable FGFR3 fusions.
All FGFR3-TACC3 and FGFR3-CAMK2A fusion-positive samples were strongly FGFR3 positive on IHC (staining sensitivity 100% in sequencing cohort) (Fig. 3A, Supplementary Figure 12). Five cases (out of 41, staining specificity 88% in the sequencing cohort) lacked intergenic FGFR3 rearrangements but showed strong FGFR3 staining in subregions of whole tissue sections (Fig. 3A, Supplementary Figure 12). Computationally analyzed staining intensities were also significantly higher in fusion-positive cases than in negatively to moderately stained fusion-negative cases (P < .0001, Kruskal–Wallis test) (Fig. 3A–B). Fusion-negative cases that show only focal strong FGFR3 staining (GBM-09, GBM-10, and ASTRO-02) resemble more moderately stained cases in the computational analysis (Fig. 3A–B).
FGFR3 Fusion-Positive Cases Are Predominantly Female and Mutually Exclusive with EGFR/PDGFRA/MET Alterations
All cases with FGFR3 fusions were IDH wild-type, which is concordant with the results of previous reports.7, 9 In addition, FGFR3 fusion events were mutually exclusive with amplification or mutation of epidermal growth factor receptor (EGFR)/platelet derived growth factor receptor alpha (PDGFRA)/MET (Fig. 3A, Supplementary Tables 4–5). Mutual exclusivity between EGFR amplification and FGFR3-TACC3 fusions has been previously reported by us and others.8,9
Nearly all patients (7 of 8) with FGFR3-TACC3 fusions were female, the sole exception being a male patient with grade II oligoastrocytoma. Alternative FGFR3-CAMK2A and FGFR3-ELAVL31 fusions were observed in male patients. A higher FGFR3 fusion frequency in females (P = .028, Fisher’s exact test) (Fig. 3B) is consistent with sex-associated FGFR3 staining distribution in our IHC cohort (Supplementary Figure 6a). In all previously reported cohorts, 14 females and 11 males carried FGFR3 fusions.1,2,8,9,13,34 After combining our data and all IDH wild-type cases with sex information in previous reports,1,2,8,13,34 3.1% of males (8 of 255) and 8.4% of females (14 of 152) were FGFR3 fusion positive (P = .027, Fisher’s exact test), suggesting a higher incidence among females.
Predominant FGFR3 Staining Typically Observed in Fusion-Positive Tumors
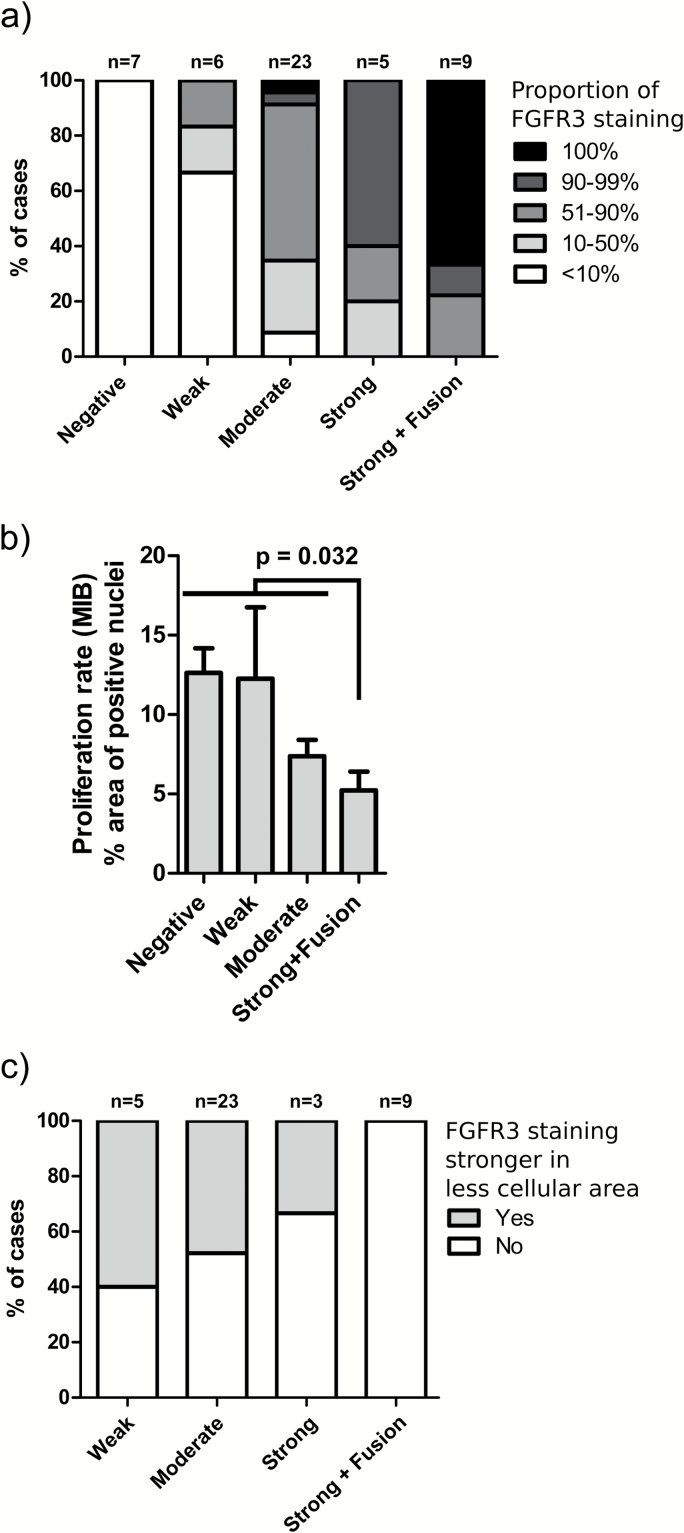
As negative FGFR3 staining was commonly observed in whole-mount tissue slide samples, we determined the proportion of positively (weakly to strongly) stained malignant cells from the tissue slide samples. In 6 of 9 fusion-positive patients, all malignant cells were FGFR3 positive (Fig. 4B). Moderate to strong staining was observed in 4 cases and weak to strong staining in 2 cases. Furthermore, both GBM-04 and GBM-07 samples, derived from different surgeries of one patient, had a small negatively stained tumor region (<10% of malignant cells). There was a significant proportion of negatively stained malignant cells in 2 additional fusion-positive patients (GBM-02 and GBM-05). These cases harbored an FGFR3-CAMK2A fusion and an FGFR3-TACC3 fusion, in which the breakpoint was in the 3' untranslated region (UTR) of FGFR3 but produced fusion protein, as the splicing acceptor site had been deleted during the rearrangement (Supplementary Figure 13). The proportion of positively stained malignant cells was higher in fusion-positive than fusion-negative samples (P = 1.2 × 10–8, Fisher’s exact test) (Fig. 4A), suggesting that positive staining in the whole malignant tissue further supports the presence of FGFR3 fusion. The majority of FGFR3-positive malignant cells were moderately to strongly stained in all fusion-positive cases, which is concordant with moderate to strong FGFR3 staining observed in all the fusion-positive cases on TMA. The use of antibody blocking peptide diminished FGFR3 staining very efficiently in both fusion-positive and fusion-negative tumors, as only red blood cell and hemosiderin staining were apparent in samples that were stained by using a blocking peptide (Supplementary Figure 14).
Fig. 4.
(A) The proportion of negatively stained tumor cells was generally lower in fusion-positive cases. P = 1.2 × 10–8, Fisher’s exact test. (B) Tumors with moderate to strong FGFR3 staining showed lower proliferation rate than negatively to weakly stained tumors (P = .0048, Kruskal–Wallis test). FGFR3 fusion-positive cases significantly differed from fusion-negative ones (P = .032, Mann–Whitney test). GBM samples with whole-mount tissue staining of FGFR3 were included in the analysis. Out of strongly stained samples, only fusion-positive cases were included due to low number of fusion-negative ones. The mean proliferation rate observed on TMA and SEM are shown. (C) FGFR3 staining was associated with lower cellularity within malignant regions in whole-mount tissue sections used for targeted sequencing, but this association was not observed in FGFR3 fusion-positive cases (P = .024, Fisher’s exact test).
Higher FGFR3 Staining Was Associated with Lower Proliferation Rate
We next associated FGFR3 staining data with the proportion of MIB-1 immunopositive nuclei, which is commonly used as a cell proliferation index and a key feature of tumor aggressiveness. Surprisingly, astrocytoma cases with moderate to strong FGFR3 staining exhibited lower proliferation rates than negatively or weakly stained cases independently of tumor grade (P < .05, Kruskal–Wallis test) (Supplementary Figure 15a). This is somewhat contradictory to worse prognosis of moderately to strongly stained cases in the same cohort (Fig. 1C). A similar pattern was also observed in whole-mount tissue GBM samples used for FGFR3 staining (P = .0048, Kruskal–Wallis test) (Fig. 4B, Supplementary Figure 15b). Staining was not associated with patient prognosis in this cohort (Supplementary Figure 16). Lower proliferation rates of FGFR3 fusion-positive tumors may have led to underestimation of their aggressiveness when diagnosis was based on WHO 2007 classification. More detailed inspection of intratumoral FGFR3 staining patterns revealed that stronger FGFR3 staining was observed in less cellular tumor areas with higher differentiation state in a large proportion of fusion-negative cases but not in fusion-positive cases (P = .024, Fisher’s exact test) (Fig. 4C, Supplementary Figure 17). In addition, stronger FGFR3 staining was perivascular in a subpopulation of cases (Supplementary Figure 3a), but this was not associated with fusion status.
Discussion
Our results demonstrate that FGFR3 staining, as detected using IHC, is indicative of FGFR3 gene fusion, which can be further confirmed by PCR- or sequencing-based technologies. This method allows efficient patient selection, as most cases are fully negative for FGFR3, and is fully compatible with current clinical practices throughout the world. Even if sequencing-based technologies are used as the primary tool for FGFR3 fusion detection, information on fusion protein expression levels will be valuable when estimating FGFR inhibitor treatment response. FGFR3 alterations will not be informative if the altered protein is not expressed. Actually, FGFR1 expression levels have been shown to predict treatment responses better than genomic FGFR1 alterations in head and neck squamous cell cancers as well as different lung cancers.35,36 The role of FGFR3 staining as an independent predictive marker needs to be evaluated in the future. We identified 2 IDH wild-type cases with strong and widely positive FGFR3 staining but no evidence for intergenic FGFR3 rearrangements (Fig. 3A). It is relevant to determine whether these cases will benefit from treatment with FGFR inhibitors. Reason for strong FGFR3 staining in fusion-negative samples is unknown, but at least the miR-99a gene was not altered in these tumors. One reason might be suppressed miR-99a expression, despite miR-99a being clearly expressed in all the GBMs in the cohort of The Cancer Genome Atlas. Atypical oncogenic rearrangements, such as intragenic rearrangements in FGFR1,12 may also be difficult to detect, which might cause false negatives in sequencing analysis.
Our sequencing cohort does not fully represent the original staining cohort, as we sequenced all available moderately to strongly stained tumors but only part of the negatively to weakly stained tumors. This was done for practical reasons, as moderate FGFR3 fusion recurrence rate requires a large cohort for fusion analysis, but pulldown-based targeted sequencing expenses and workload are not compatible with hundreds of samples. Our approach allowed us to estimate staining sensitivity and specificity only in the sequencing cohort. Sensitivity was 100%, which is consistent with previous studies, where we and others have detected strong FGFR3 staining in all the available fusion-positive cases.8,9 It is thus unlikely that sensitivity would be significantly less in the cohort with full presentation of negatively to weakly stained cases. Furthermore, the same applies also to estimated staining specificity (88% in the sequencing cohort), as exclusion of most of the negatively to weakly stained cases tends to lead to underestimation of staining specificity.
Positive FGFR3 staining was generally heterogeneous. While positive staining was typically less predominant and associated with lower cellularity in fusion-negative tumors, negatively stained malignant regions were also detected in a proportion of fusion-positive cases (Fig. 4A). This is relevant from a treatment perspective, as negative cells are unlikely to respond to targeted treatment, potentially resulting in treatment failures with FGFR inhibitors. Variable expression levels and negative subclones may also explain why partial responses have been observed in clinical trials.9,22 Further characterization of FGFR3-negative cells is important for improved treatment.
In this study, FGFR3 fusions always involved 3' fusion partners with dimerization domains, and no FGFR3 staining was detected in the majority of samples (Fig. 1), which supports our previous finding that loss of 3' UTR and miR-99a mediated suppression is needed for expression and oncogenicity of the fusion protein in diffuse gliomas.8 Although FGFR3 is mostly fused to TACC3, other fusion partners also exist. If PCR-based tools for detecting gene fusions are used as the sole diagnostic method for fusion detection, cases with alternative 3' partners will be missed during diagnosis. On the other hand, FGFR3 staining does not require pre-knowledge of fusion partners and is fully FFPE compatible together with our targeted sequencing approach. It will also be interesting to determine whether the IHC approach is suitable for fusion detection in other malignancies with low endogenous FGFR3 expression, such as in lung squamous cell carcinoma.20
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by grants from the Academy of Finland (grants 259038 to K.G., 269474 to M.N., and 251790 to O.Y.H.), the Emil Aaltonen Foundation (M.A.), the Päivikki ja Sakari Sholberg Foundation (K.G.), the Finnish Funding Agency for Technology and Innovation Finland Distinguished Professor program, Competitive State Research Financing of the Expert Responsibility area of Tampere University Hospital (J.H., H.H., grant 9T042 to M.N.), and funding from the US National Cancer Institute (R01 CA183153 to W.Z.). W.Z. is supported by the endowed Hanes and Willis Family Professorship in Cancer.
Conflict of interest statement. The authors have no conflicts of interest to disclose.
Supplementary Material
Acknowledgments
We highly appreciate Marika Vähä-Jaakkola, Paula Kosonen, Päivi Martikainen, Marja Pirinen, Katja Liljeström, Riina Kylätie, Dr. Leena Latonen, Osku Alanen, Maria Laaksonen, Dr. Eloise Mikkonen, Riitta Koivisto, Dr. Joanna Ilvesaro, and Satu Salo for their help in sample handling. Personnel at Tampere University Hospital and Fimlab laboratories are acknowledged for their contribution to sample collection. We acknowledge the CSC IT Centre for Science, Finland, for computational resources and Ann M. Sutton (Department of Scientific Publications, University of Texas MD Anderson Cancer Center) for editing this manuscript.
References
- 1. Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. [DOI] [PubMed] [Google Scholar]
- 4. Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 5. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. ed. WHO Classification of Tumours of the Central Nervous System WHO/IARC Classification of Tumours, 4th edition revised Geneva, Switzerland: World Health Organization; 2016. [Google Scholar]
- 6. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parker BC, Annala MJ, Cogdell DE, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123(2):855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21(14):3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parker BC, Engels M, Annala M, Zhang W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J Pathol. 2014;232(1):4–15. [DOI] [PubMed] [Google Scholar]
- 11. Rand V, Huang J, Stockwell T, et al. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102(40):14344–14349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Wu G, Miller CP, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parker BC, Zhang W. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin J Cancer. 2013;32(11):594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Capelletti M, Dodge ME, Ercan D, et al. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res. 2014;20(24):6551–6558. [DOI] [PubMed] [Google Scholar]
- 16. Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20(15):4107–4114. [DOI] [PubMed] [Google Scholar]
- 17. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: Analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22(1):259–267. [DOI] [PubMed] [Google Scholar]
- 18. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230(3):270–276. [DOI] [PubMed] [Google Scholar]
- 21. Dienstmann R, Rodon J, Prat A, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25(3):552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tabernero J, Bahleda R, Dienstmann R, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2015;33(30):3401–3408. [DOI] [PubMed] [Google Scholar]
- 23. Lewin J, Siu LL. Development of fibroblast growth factor receptor inhibitors: kissing frogs to find a prince? J Clin Oncol. 2015;33(30):3372–3374. [DOI] [PubMed] [Google Scholar]
- 24. Sallinen SL, Sallinen PK, Haapasalo HK, et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000;60(23):6617–6622. [PubMed] [Google Scholar]
- 25. Järvelä S, Parkkila S, Bragge H, et al. Carbonic anhydrase IX in oligodendroglial brain tumors. BMC Cancer. 2008;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nordfors K, Haapasalo J, Mäkelä K, et al. Twist predicts poor outcome of patients with astrocytic glioma. J Clin Pathol. 2015;68(11):905–912. [DOI] [PubMed] [Google Scholar]
- 27. Haapasalo H, Sallinen S, Sallinen P, et al. Clinicopathological correlation of cell proliferation, apoptosis and p53 in cerebellar pilocytic astrocytomas. Neuropathol Appl Neurobiol. 1999;25(2):134–142. [DOI] [PubMed] [Google Scholar]
- 28. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin CR, Kapiloff MS, Durgerian S, et al. Molecular cloning of a brain-specific calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci U S A. 1987;84(16):5962–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamauchi T. Neuronal Ca2+/calmodulin-dependent protein kinase II–discovery, progress in a quarter of a century, and perspective: implication for learning and memory. Biol Pharm Bull. 2005;28(8):1342–1354. [DOI] [PubMed] [Google Scholar]
- 33. Kasashima K, Sakashita E, Saito K, Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 2002;30(20):4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bao ZS, Chen HM, Yang MY, et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014;24(11):1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Göke F, Franzen A, Hinz TK, et al. FGFR1 expression levels predict BGJ398 sensitivity of FGFR1-dependent head and neck squamous cell cancers. Clin Cancer Res. 2015;21(19):4356–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20(12):3299–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.