Abstract
The fluorescence of 2-aminopurine (2A)-substituted duplexes (contained in the GATC target site) was investigated by titration with T4 Dam DNA-(N6-adenine)-methyltransferase. With an unmethylated target (2A/A duplex) or its methylated derivative (2A/mA duplex), T4 Dam produced up to a 50-fold increase in fluorescence, consistent with 2A being flipped out of the DNA helix. Though neither S-adenosyl-l-homocysteine nor sinefungin had any significant effect, addition of substrate S-adenosyl-l-methionine (AdoMet) sharply reduced the Dam-induced fluorescence with these complexes. In contrast, AdoMet had no effect on the fluorescence increase produced with an 2A/2A double-substituted duplex. Since the 2A/mA duplex cannot be methylated, the AdoMet-induced decrease in fluorescence cannot be due to methylation per se. We propose that T4 Dam alone randomly binds to the asymmetric 2A/A and 2A/mA duplexes, and that AdoMet induces an allosteric T4 Dam conformational change that promotes reorientation of the enzyme to the strand containing the native base. Thus, AdoMet increases enzyme binding-specificity, in addition to serving as the methyl donor. The results of pre-steady-state methylation kinetics are consistent with this model.
INTRODUCTION
Type II DNA methyltransferases (MTases) generally recognize short palindromic sequences and catalyze methyl group transfer from the donor S-adenosyl-l-methionine (AdoMet) to the N6-amino group of an adenine (Ade), or the C5 atom or N4-amino nitrogen of a cytosine (Cyt) in the target sequence (1). Elucidating the mechanism(s) of action of these enzymes still remains an important problem in the area of biological DNA methylation. Furthermore, due to their high specificity and comparatively simple organization, the Type II MTases are excellent subjects for detailed studies on specific protein–DNA interactions.
The catalytic mechanism (2) as well as the three-dimensional structures of the (Cyt-5) MTases have been described (3–6). A most surprising and exciting result is that the Cyt residue to be methylated is flipped out of the DNA helix (5,6). Among the (N6-Ade) and (N4-Cyt) MTases, structures have been reported for the TaqI, PvuII, DpnM and RsrI MTases, respectively (7–10). With the exception of the M·TaqI, co-crystallization with DNA has not been successful for amino-MTases. The recently solved crystal structure of a M·TaqI–DNA–AdoMet analog ternary complex (11) shows flipping of the target Ade. However, M·TaqI and T4 Dam differ dramatically in their kinetic behavior, e.g., M·TaqI has a kmeth that is 1/10 the rate of release of the enzyme from the methylated DNA product (E.Weinhold, Max-Planck Institute for Molecular Physiology, Dortmund, Germany, personal communication), whereas T4 Dam has a kmeth that is 20-fold higher than kcat (and the rate of release of the enzyme from the methylated DNA product) (12). Thus, the rate-limiting step in the methylation reaction differs for the two MTases, and it is reasonable to believe that there will be differences in both the structure and function of their ternary complexes. In the absence of a comparable structure for T4 Dam, we have taken advantage of alternative methodologies (13–20) to study base flipping by this MTase. One of these methods is based on the substitution of a target Ade by 2-aminopurine (2A), which serves as a fluorescent probe (13–18). The fluorescence intensity of 2A incorporated into double-stranded DNA is very low, but sharply increases if the base (or nucleoside) is flipped out into the cavity of the enzyme’s active site. We have used this property to investigate possible flipping in the methylation reaction catalyzed by the T4 Dam (N6-Ade)-MTase, which recognizes the palindromic sequence GATC (21).
MATERIALS AND METHODS
Enzymes and chemicals
[3H-CH3]AdoMet was from Amersham. S-adenosyl-l-homocysteine (AdoHcy) and sinefungin were from Sigma. Unlabeled AdoMet (Sigma) was purified further by chromatography on a C18-reversed-phase column as described previously (22). Synthetic 20mer oligodeoxyribonucleotide duplexes contained 2-aminopurine (A→2A substitution) and N6-methylAde (A→mA substitution) in the GATC target site (underlined) were used as DNA substrates:
2A/A duplex
5′-CAGTTTAGG2ATCCATTTCAC-3′
3′-GTCAAATCCTAGGTAAAGTG-5′
2A/mA duplex (hemimethylated)
5′-CAGTTTAGG2ATCCATTTCAC-3′
3′-GTCAAATCCTmAGGTAAAGTG-5′
2A/2A duplex (double substitution)
5′-CAGTTTAGG2ATCCATTTCAC-3′
3′-GTCAAATCCT2AGGTAAAGTG-5′
G2ATC/GTAC duplex (double mismatch)
5′-CAGTTTAGG2ATCCATTTCAC-3′
3′-GTCAAATCCATGGTAAAGTG-5′
HPLC-purified oligodeoxyribonucleotide 20mers were synthesized at Midland Certified Reagent Co. (Midland, TX). The concentration of each oligodeoxyribonucleotide was determined spectrophotometrically from the molar extinction coefficients of the individual nucleotides and the known sequence. Duplexes were prepared by annealing complementary single-stranded oligodeoxyribonucleotides.
The T4 Dam MTase was purified to homogeneity as previously described (22). Protein concentration was determined by the Bradford method (23). It was in close agreement with one determined spectrophotometrically in 6.0 M guanidinium hydrochloride, 0.02 M phosphate buffer (pH 6.5) using 33 710 M–1cm–1 for the enzyme molar extinction coefficient at 280 nm, which was calculated from the known composition and molar extinction coefficients of individual aromatic amino acid residues (24).
Fluorescence measurements
A dual beam difference Shimadzu RF-520 spectrofluorimeter was used for fluorescence measurements. The fluorescence was excited and detected at wavelengths of 320 nm (with 5 nm slit) and 370 nm (with 10 nm slit), which correspond to the maxima for 2A excitation and fluorescence, respectively. The spectrofluorimeter was adjusted such that 0.0025 (arbitrary) units (a.u.) correspond to 1 nM of free 2A/A duplex. Measurements were carried out at different DNA concentrations in reaction buffer (100 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 0.2 mg/ml bovine serum albumin and 5% glycerol) (22) at 25°C in the presence of increasing concentrations of T4 Dam MTase. A cuvette lacking duplex DNA was run as a control in order to correct for light scattering by the reaction buffer and protein intrinsic fluorescence. An intrinsic fluorescence of the duplex (without T4 Dam added) was also subtracted. In other experiments, T4 Dam was titrated by increasing oligonucleotide duplex concentrations. For these analyses, a control cuvette lacking T4 Dam was included in order to correct for light scattering and intrinsic fluorescence of the duplex.
Glutaraldehyde crosslinking of T4 Dam MTase
T4 Dam MTase (1 µM) samples were incubated with 0.0025% (v/v) glutaraldehyde in 10 mM sodium phosphate (pH 7.8) at 25°C for 40 min. Sodium borohydride was then added to a final concentration of 100 mM and incubation was continued for an additional 10 min. The samples were diluted into Laemmli sample buffer, boiled for 3 min. and analyzed by SDS–15% PAGE. Crosslinking was also carried out in the presence of specific oligonucleotide duplexes. In this case, T4 MTase was pre-incubated with different concentrations of oligonucleotide duplexes (on ice for 20 min) prior to treatment with glutaraldehyde.
Pre-steady-state T4 Dam MTase assay
The microvolume rapid quench instrument ‘KinTek Corp. RQF-3’ was used for pre-steady-state assays of T4 Dam MTase activity. Assay mixtures contained 100 mM Tris–HCl (pH 8.0), 10 mM EDTA, 10 mM DTT and 0.2 mg/ml bovine serum albumin. The feeding syringe containing the enzyme preparation was kept at 0°C to avoid inactivation of the T4 Dam MTase during the experiment. The other syringes, mixers and age-loops were equilibrated to 25°C, the temperature of the enzyme assay. Prior to the start of the next reaction an aliquot of the enzyme preparation was also equilibrated to 25°C in the sample loop. SDS (0.05% w/v) in 25 mM Tris–HCl (pH 8.3) was used as the quench solution. The quenched samples were collected in Eppendorf tubes, evaporated and adjusted to 100 µl; duplicate aliquots were spotted onto DE81 anion-exchange filter papers (Whatman). The 3H-counts were summed and the molar concentration of (3H-CH3)-groups incorporated into DNA was calculated as described earlier (25). Reaction mixtures without T4 Dam added were treated in the same manner for background corrections.
Data analysis
Kinetic parameters were obtained using the program ‘Scientist 2.01’ (MicroMath Inc.) for non-linear regression analysis.
RESULTS
Fluorescence titration of 2A-containing duplexes with T4 Dam: evidence for base flipping
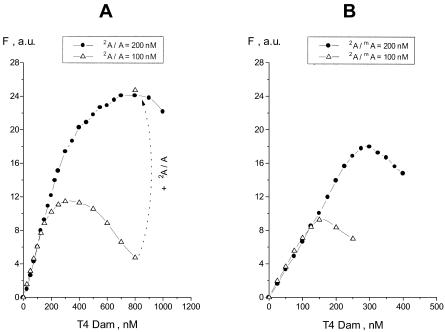
Fluorescence excitation and emission spectra of the 2A-substituted duplex, 2A/A, were determined (data not shown). Based on these spectra, wavelengths of 320 and 370 nm were chosen for excitation and emission, respectively. The 2A fluorescence intensities could be readily observed at a concentration of 1 µM duplex. As shown in Figure 1A, addition of T4 Dam to duplex 2A/A resulted in an increase in 2A fluorescence. The intensity was approximately proportional to the duplex concentration, and exhibited a 50-fold signal increase at saturating enzyme concentration. These results are analogous to those observed with several other MTases (13–18) and consistent with flipping of the 2A residue out of the DNA helix.
Figure 1.
Fluorescence analysis of T4 Dam titration of duplexes 2A/A (A) and 2A/mA (B) in the absence of AdoMet; duplexes were at 100 or 200 nM as indicated. Addition at the last titration point of more duplex (up to 200 nM) to the mixture containing 100 nM of 2A/A resulted in an increase in fluorescence intensity confirming that the fluorescence quenching observed was fully reversible (A). Fluorescence intensity is given in arbitrary units (a.u.), and intrinsic fluorescence of the free duplex was subtracted in this and subsequent figures.
The hemimethylated duplex, 2A/mA, showed an analogous response to T4 Dam compared to the 2A/A duplex (Fig. 1B). However, at a duplex concentration of 200 nM, the fluorescence increase reached a maximum at 300 nM T4 Dam with 2A/mA, compared to 700 nM T4 Dam with 2A/A, and then it decreased at higher enzyme concentrations. This decrease was reversible and not related to enzyme inactivation, because addition of more duplex to the mixture resulted in an increase in fluorescence intensity (Fig. 1A). Therefore, we suggest that the decrease in fluorescence observed at higher enzyme concentrations was due to the formation of altered complexes in which the 2A fluorescence was either quenched within the pocket of the enzyme, or its flipping was inhibited.
Effect of methylation cofactor and inhibitors on fluorescence titration
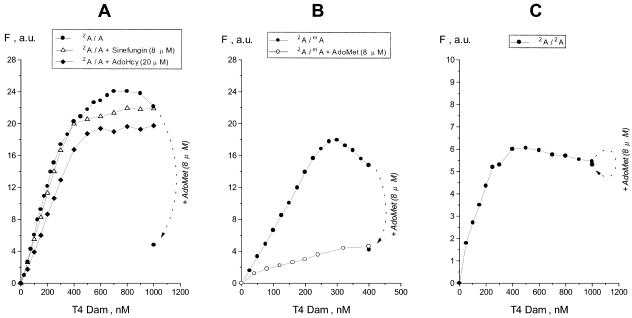
AdoHcy, the product of methyl transfer, is a competitive inhibitor of AdoMet in transmethylation reactions. Presence of AdoHcy did not dramatically reduce the fluorescence during T4 Dam titration of duplex 2A/A (Fig. 2A). Furthermore, another cofactor-analog and competitive inhibitor, sinefungin, had even less of an effect on the character of the titration curve. However, in contrast to the methylation inhibitors, addition of substrate AdoMet led to a sharp reduction in the fluorescence intensity (Fig. 2A). Analogous titrations were carried out with the hemimethylated 2A/mA duplex. Since duplex 2A/mA is unable to serve as a methyl acceptor, an entire T4 Dam titration could be carried out in the presence of AdoMet. As seen in Figure 2B (lower curve) AdoMet reduced the fluorescence response, and the maximum fluorescence attained was similar to that observed when AdoMet was added at the standard titration-end points for the 2A/A and 2A/mA duplexes (compare to Fig. 2A). In contrast, presence of a saturating concentration of either AdoHcy or sinefungin produced only a minor decrease in the maximum level of fluorescence with the 2A/mA duplex (data not shown). Thus, the AdoMet-induced reduction in fluorescence following T4 Dam titration of the 2A/A duplex must occur by a process independent of methylation per se, since it was also observed with the 2A/mA duplex, which is not a substrate for methylation.
Figure 2.
Effect of added AdoMet on fluorescence analysis of T4 Dam titration. Duplexes 2A/A (A), 2A/mA (B) or 2A/2A (C) were at 200 nM; AdoMet was added to a final concentration of 8 µM at the last titration point. For duplex 2A/A (A) titrations are shown in the absence or presence of the reaction inhibitors AdoHcy (20 µM) or sinefungin (8 µM). The lower curve in (B) corresponds to a titration of the 2A/mA duplex in the presence of 8 µM AdoMet.
To investigate the AdoMet effect further, we carried out a T4 Dam titration of a 20mer duplex having 2A substitutions in both strands (2A/2A duplex, Fig. 2C). In contrast to asymmetric 2A/A and 2A/mA duplexes, the doubly substituted 2A/2A duplex possesses a high intrinsic fluorescence, 0.018 a.u./nM versus 0.0025 a.u./nM for 2A/A or 2A/mA duplexes (subtracted from the data points in Fig. 2C). This suggests that the two 2A residues in the recognition site are destabilized in comparison to solitary 2A residues in 2A/A and 2A/mA duplexes. For this reason, interaction of the 2A/2A duplex with T4 Dam resulted in a lower net increase in fluorescence, but the general shape of the titration curve was comparable to those for the 2A/A and 2A/mA duplexes. However, when a saturating concentration of AdoMet was added at the titration end point, there was no decrease in fluorescence. This argues against either an alteration in the environment of the flipped-out base resulting in fluorescence quenching, or a destabilization of the flipped 2A resulting in a decrease in fluorescence. Otherwise, presence of AdoMet should have had the same effect on fluorescence with the 2A/2A duplex as with the 2A/mA and 2A/A duplexes.
Fluorescence titration of T4 Dam with 2A-substituted duplexes
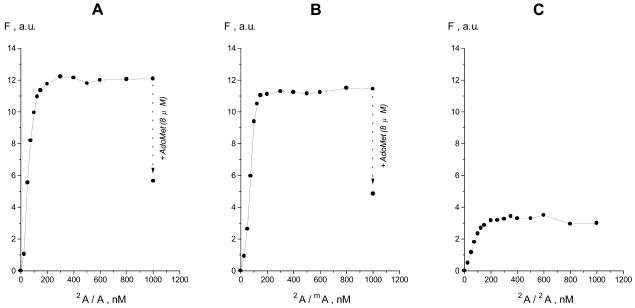
Because significant quenching of 2A fluorescence occurred at high [T4 Dam]/[2A/A] or [T4 Dam]/[2A/mA] ratios (Fig. 1), we used a constant T4 Dam concentration and titrated the enzyme with increasing concentrations of 2A/mA or 2A/A duplex (Fig. 3). Addition of either duplex gave a rapid and large increase in fluorescence, and the two duplexes showed comparable affinities for T4 Dam. However, the characteristics of the curves were not the same as those for the titration of duplex by enzyme (compare with Fig. 1A and B). Firstly, the curves in Figure 3 have sharper increases at the initial stages of the titrations. At equal concentrations (200 nM) of enzyme and duplex 2A/A, the fluorescence intensities were approximately equal and independent of the order of component addition (Figs 1A and 3A). However, after this point the curves differed. That is, the titration of enzyme by duplex quickly reached a plateau in fluorescence (Fig. 3), whereas the fluorescence continued to increase to a 2-fold higher maximum level in the titration of duplex by T4 Dam (Fig. 1A). This suggests that different complexes were formed in the later stages of the two titrations, and that an oligomeric form of T4 Dam might participate in complex formation under conditions of enzyme excess, as suggested earlier from gel shift assays (26).
Figure 3.
Fluorescence titration of T4 Dam (200 nM) with duplex 2A/A (A), 2A/mA (B) or 2A/2A (C). At the last titration point in (A) and (B), cofactor AdoMet was added to a final concentration of 8 µM.
As observed in the T4 Dam titrations of 2A/A and 2A/mA duplexes (Fig. 2A and B), addition of AdoMet produced a substantial drop in fluorescence in both the 2A/A and 2A/mA duplex titrations of T4 Dam (Fig. 3A and B). This effect was reversible for both the 2A/A and 2A/mA duplexes, because subsequent addition of sinefungin, a competitive inhibitor of AdoMet binding, raised the fluorescence close to the original levels (data not shown). Thus, AdoMet-induced reduction in fluorescence was independent of the manner in which the titration was carried out.
Glutaraldehyde crosslinking of T4 Dam
Glutaraldehyde can generate covalent bonds between lysine residues located at the interface between protein subunits (27,28). This crosslinking results in the stabilization of oligomeric forms of the protein, which can be subsequently identified by SDS–PAGE analysis. Therefore, we used glutaraldehyde to investigate the possible presence of oligomeric forms of T4 Dam. In order to approximate the conditions of our fluorescence titration experiments and to reduce chance crosslinking due to the close proximity of proteins at high concentration, we used low concentrations of T4 Dam (1 µM) and glutaraldehyde (0.0025%).
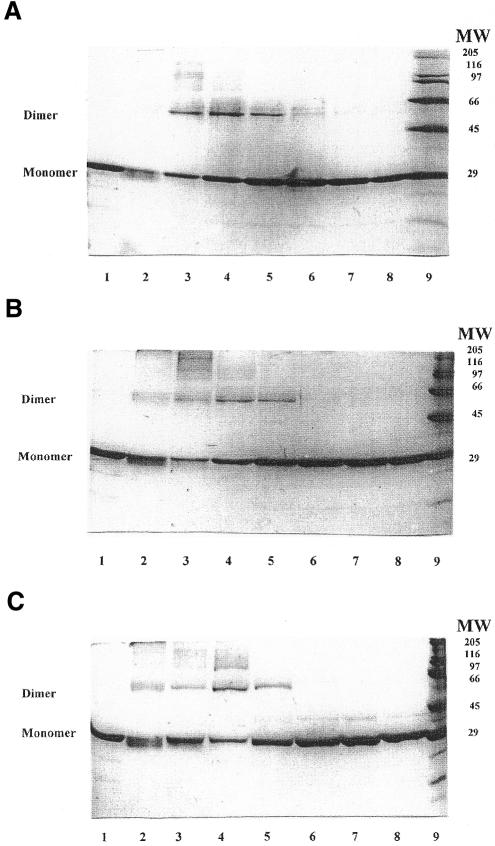
As seen in Figure 4, exposure of the free T4 Dam to glutaraldehyde resulted in the formation of only a small amount of SDS-resistant oligomers (Fig. 4B and C, lanes 2). Addition of either duplex 2A/A or duplex 2A/mA at a high non-stoichiometric ratio [enzyme]/[duplex] of 4.0 significantly enhanced the formation of oligomers, with the dimeric form predominating. However, further addition of either the 2A/A or the 2A/mA duplex led to a progressive reduction in the amount of the oligomers, practically disappearing at a [enzyme]/[duplex] ratio <1.0. Pre-incubation of T4 Dam with AdoMet (at saturating concentration of 5 µM) prior to the addition of the 2A/mA duplex resulted in a similar distribution of T4 Dam forms (Fig. 4C). However, under these conditions loss of more slowly migrating oligomers, including T4 Dam dimers, occurred even when the [enzyme]/[duplex] ratio was >1.0. We conclude that at a high [enzyme]/[duplex] ratio, formation of T4 Dam oligomers (predominantly dimers, but with trimers and larger oligomers also present) occurs, but these revert to monomers as the [enzyme]/[duplex] ratio decreases. These results show that different complexes were indeed present at the titration end points in Figures 1 and 3.
Figure 4.
Glutaraldehyde crosslinking of T4 Dam (1 µM) in the presence of varying concentrations of duplex: (A) duplex 2A/A; (B) duplex 2A/mA. Lane 1, T4 Dam, untreated; lane 2, T4 Dam treated with 0.0025% glutaraldehyde; lanes 3–8, T4 Dam treated in the presence of duplex at concentrations of 0.25, 0.5, 1.0, 2.0, 4.0 and 8.0 µM, respectively; lane 9, molecular weight standards. (C) Crosslinking in the presence of 5 µM AdoMet. Lane 1, T4 Dam, untreated; lane 2, T4 Dam + AdoMet, treated with 0.0025% glutaraldehyde; lanes 3–8, T4 Dam + AdoMet, treated in the presence of the 2A/mA duplex at concentrations of 0.1, 0.25, 0.5, 1.0, 2.0 and 4.0 µM, respectively; lane 9, molecular weight standards.
Methylation kinetics
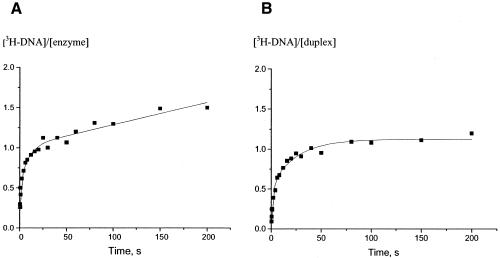
Figure 5A shows the time course of multiple turnover reaction following addition of T4 Dam (saturated by pre-incubation with excess [3H]AdoMet) to duplex 2A/A, where duplex was in excess over enzyme. It is clear that a burst of methylated product was produced, followed by a slower steady-state rate of methylation. This is in agreement with our previous observations in which we obtained a burst of approximately one (i.e., one methyl group transferred per T4 Dam binding) (12). Figure 5B shows the time course of a single turnover reaction following addition of T4 Dam (saturated by pre-incubation with excess [3H]AdoMet), to duplex 2A/A, where enzyme was in excess over duplex. Under these (single turnover) conditions a rapid exponential increase in product formation was observed, which reached a plateau of one, indicating that one methyl group was transferred per bound duplex. Thus, in both types of experiment T4 Dam catalyzed complete methylation of the 2A/A duplex in a single binding event.
Figure 5.
Time course of T4 Dam methylation of duplex 2A/A. T4 Dam and duplex 2A/A concentrations were 0.158 and 1 µM (A) or 2.7 and 0.2 µM (B), respectively. T4 Dam was pre-incubated with 8 µM of [3H]AdoMet in both cases. In (A) the number of CH3 groups transferred per molecule of enzyme bound was plotted (multiple turnover conditions where DNA was in excess); in (B) the number of CH3 groups transferred per bound duplex was plotted (single turnover conditions where enzyme was in excess).
A burst reflects rapid conversion of the initial enzyme–substrate complexes, followed by a relatively slower dissociation of the reaction products and then steady-state cycles of the reaction (29). When the enzyme–AdoMet complex is in excess over the duplex, then the reaction course reflects a single conversion (turnover) of the substrates. If the initial binding is random, then there are two alternative orientations (productive and non-productive) of T4 Dam on the asymmetric DNA duplex. Thus, a single binding event should lead to methylation of only 50% of the bound DNA duplexes. However, in both the burst and single turnover experiments, T4 Dam catalyzed complete methylation of the bound duplex following a single binding event (Fig. 5). Therefore, in order to modify all the Ade residues in the 2A/A asymmetric duplex during a burst, the T4 Dam–AdoMet complex must have either discriminated between the two strands when it initially bound (binding only to the Ade-containing productive strand), or it must have undergone rapid re-orientation after binding in the non-productive orientation. This suggests that if the initial binding is random, then there are two distinct pathways leading to a complete burst; the first pathway occurs when the enzyme binds to the productive strand, and the second pathway occurs when the enzyme binds to the non-productive strand and has to re-orient. Thus, binding to the productive strand represents one step and binding to the non-productive strand represent a second distinct intermediate step to a complete burst. To investigate this possibility, we tried fitting the experimental data to an equation that describes a two-step burst reaction (29), and continuation into a steady-state phase:
where kmeth1 is the methylation rate constant for enzyme molecules that are initially orientated to the productive strand (where methylation can occur immediately), and kmeth2 is the methylation rate constant for enzyme molecules that are initially bound to the non-productive strand and have to re-orient to the productive strand before methylation can occur. P1 is the total methylation level per bound duplex attained after the first step of the burst reaction (resulting from enzyme molecules initially bound to the productive strand), and P2 is the total methylation level per bound duplex after both steps of the burst reaction have taken place; kcat is the rate constant of the steady-state phase. An equation analogous to the one above was used to fit the single turnover data; however, the steady-state component of the equation was deleted and [3H-DNA] values were normalized to the duplex concentration. As seen in Table 1B, according to a two-step model, during the burst one-half of the duplexes were methylated at a rapid rate (the first pathway of the reaction), and the other half were methylated at a rate that was ∼17-fold slower (the second pathway of the reaction). This resulted in the complete methylation of one Ade residue per bound 2A/A duplex. Similar observations were also made in the single turnover experiment; i.e., approximately one-half of the methylation of the duplex occurred rapidly and the other half occurred at a rate ∼14-fold slower. Thus, the analysis of data by this two-step model suggests that the initial binding of the T4 Dam–AdoMet complex to the 2A/A duplex was random.
Table 1. Kinetic parameters of duplex 2A/A methylation by T4 Dam in the ‘burst’ and ‘single turnover’ experiments based on (A) a one-step model and (B) a two-step model.
| Parameter |
Burst |
Single turnover |
| A. One-step model | ||
| Pa | 0.90 ± 0.03 | 1.02 ± 0.03 |
| kmethb (s–1) | 0.67 ± 0.10 | 0.15 ±0.02 |
| kcatc (s–1) | 0.0036 ± 0.0004 | – |
| B. Two-step model | ||
| P1a | 0.55 ± 0.10 | 0.58 ± 0.06 |
| P2a | 1.01 ± 0.05 | 1.12 ± 0.03 |
| kmeth1b (s–1) | 1.83 ± 0.67 | 0.54 ± 0.12 |
| kmeth2b (s–1) | 0.11 ± 0.05 | 0.039 ± 0.008 |
| kcatc (s–1) | 0.0028 ± 0.0004 | – |
aP is the total methylation level per bound duplex; P1 is the total methylation level per bound duplex attained after the first step of the burst (or single turnover) reaction resulting from enzyme molecules initially bound to the productive strand; P2 is the total methylation level per bound duplex after both steps of the burst (or single turnover) reaction have taken place.
bkmeth is the methylation rate constant; kmeth1 is the methylation rate constant for enzyme molecules that are initially orientated to the productive strand (where methylation can occur immediately), and kmeth2 is the methylation rate constant for enzyme molecules that are initially bound to the non-productive strand and have to re-orient to the productive strand before methylation can occur.
ckcat is the steady-state rate constant.
Both experimental curves (Fig. 5) can also be fit to a classical one-step conversion (Table 1A). If the initial binding of the T4 Dam–AdoMet complex to the 2A/A duplex were random, a one-step model would predict a burst of one-half, since half of the enzyme would be bound to the non-productive 2A strand; however, we obtained a burst of one. Therefore, if the one-step model is correct, we must conclude that the T4 Dam–AdoMet complex discriminates between the two strands when it initially binds (binding only to the Ade-containing productive strand). Although both experimental curves could be fit to either a classical one- or two-step conversion scheme, the goodness-of-fit was significantly higher for the two-step burst equation. We conclude that the results favor random binding of T4 Dam–AdoMet to the 2A/A duplex, and that two distinct pathways lead to a burst of methylation. This suggests that when the T4 Dam–AdoMet complex binds to the non-productive strand, it can reorient to the productive strand without dissociating from the duplex (see Discussion). It should be noted that the results do not unequivocally rule out that T4 Dam–AdoMet is able to discriminate between the two strands during the initial binding.
Finally, it should be noted that the rate constants for single turnover were ∼3-fold lower than the kmeth determined under burst conditions. The lower rate constants for the single turnover experiments may result from an altered structural state of T4 Dam in these experiments, since the DNA duplexes contained more than one bound T4 Dam molecule (as indicated by the glutaraldehyde crosslinking experiments in Fig. 4). However, it follows that complexes containing multiply bound T4 Dam were catalytically active, although their catalytic efficiency was ∼3-fold lower than complexes containing only one bound T4 Dam.
DISCUSSION
A dual role for the substrate AdoMet in the methylation reaction with T4 Dam DNA-[N6-adenine]-methyltransferase
It is generally accepted that the strongly fluorescent Ade isomer, 2A, introduced in the target-base position of a specific oligonucleotide duplex, mimics the behavior of the Ade residue (13–17). Such a substitution has been used successfully even for a non-homologous target substitution, Cyt→2A, where a base flipping fluorescence signal was observed during interaction between the 2A-substituted duplex with the (Cyt-5)-HhaI MTase (14,18). As shown in Figure 1, in the absence of AdoMet, addition of T4 Dam MTase to 2A/A and 2A/mA duplexes elicited large increases in 2A fluorescence; the maximum signal increases were approximately the same for each duplex at equal concentrations, indicating that the interactions with T4 Dam were comparable. These results indicate extra-helical flipping of the target base, as has been reported with the (N6-Ade) MTases, EcoRI, TaqI, RsrI and EcoRV (13–17,20). With increasing T4 Dam concentration ([enzyme]/[duplex] ratios >1.0), we observed decreases in the fluorescence intensity. This is attributed to the formation of complexes containing more than one T4 Dam, but the mechanism of this fluorescence quenching is not yet understood.
Natural cofactor AdoMet and analogs AdoHcy and sinefungin increase the affinity of DNA MTases for their substrates (30–32). For example, AdoMet decreased the Kd value by 2- to 3-fold for T4 Dam binding with specific duplexes (26,33). While the effect of added sinefungin or AdoHcy was only minor (Fig. 2A), when AdoMet was added to the mixture of T4 Dam and the 2A/A (or 2A/mA) duplex, the fluorescence dropped ∼5-fold (Fig. 2A and B). This was not due to methylation of the duplex per se, but rather to some other effect of AdoMet. This conclusion follows from the fact that both the 2A/mA duplex and the 2A/A duplex behaved similarly, but the former is not a substrate for methylation. Further insight came from the T4 Dam fluorescence titration with the symmetrical duplex 2A/2A. In this experiment, added AdoMet had no effect on the final fluorescence level (Fig. 2C), indicating that there was no interference with 2A-flipping or destabilization of the flipped 2A or quenching of its fluorescence (such as by altering the environment of the flipped residue).
AdoMet binding to T4 Dam is known to induce an allosteric alteration in T4 Dam conformation, as demonstrated by tryptophan fluorescence quenching analysis (34). In addition, the methylation kinetics data (Table 1B) indicate that the enzyme–AdoMet complex was oriented ∼50% of the time to the productive strand and, thus, bound randomly to the DNA duplex. Based on this, we propose the following model. First, in the absence of AdoMet, T4 Dam binds randomly to the 2A/A or 2A/mA duplex, and 2A is flipped out into the catalytic pocket and fluoresces in half of the duplexes. Secondly, addition of AdoMet induces an allosteric conformational change in T4 Dam that causes the enzyme to favor and rapidly reorient to the ‘productive’ strand. This results in flipping of the non-fluorescing Ade (or mA) residue into the catalytic pocket, and the previously flipped 2A would no longer fluoresce when relocated in an intra-helical position. This rapid change in orientation between the non-productive and productive strand could be achieved either by dissociation–reassociation or by reorientation without dissociation of the T4 Dam–AdoMet complex from the DNA duplex. Since the recognition site within the 2A/2A duplex is symmetrical, altering the binding orientation of T4 Dam should have no influence on the fluorescence, since a 2A residue would be flipped out in every complex. As expected, addition of AdoMet to the pre-formed T4 Dam–(2A/2A) complex did not result in a decrease in fluorescence.
In addition, a burst value of one with the (2A/A) duplex (Fig. 5A) indicates that for every binding event, the Ade residue was methylated. This requires that a T4 Dam molecule oriented to the non-productive strand be able to reorient to the productive strand without dissociation. Although we have not presented direct experimental evidence that the T4 Dam–AdoMet complex does not strand-discriminate during initial binding (as opposed to binding randomly and undergoing reorientation), our data best fits the reorientation model. Thus, when a T4 Dam–AdoMet complex collides with an asymmetric 2A/A duplex under ‘burst’ conditions ([enzyme]<<[duplex]), it binds at random. Methylation proceeds rapidly only with the T4 Dam correctly oriented to the target Ade (productive strand). A slower, second phase of the burst ensues following reorienting of the T4 Dam, which was initially oriented to the non-productive 2A strand. Since burst conditions are normally found in vivo (a large excess of DNA substrate to enzyme), our model best describes how T4 Dam normally interacts with its substrate. In this regard, it is interesting to note that, in the case of the RsrI MTase, the burst value for a hemimethylated 14mer substrate duplex was also one (17). If the RsrI MTase bound randomly to the duplex and was unable to re-orient when it bound to the non-productive strand (containing mA), a burst value of 0.5 should have been observed. However, since there was complete methylation of the hemimethylated DNA substrate, as with T4 Dam (12), it appears that either the RsrI MTase is capable of re-orienting when it interacts with an asymmetrically modified recognition site or it has a preferred affinity for the productive strand. It would be interesting to determine whether other (N6-Ade) or (N4-Cyt) DNA MTases are capable, in fact, of re-orientation and whether AdoMet plays a role in strand specificity for these enzymes.
Interrelationship between binding, T4 Dam orientation, base flipping and methylation
From gel shift assays, the order of decreasing affinity of T4 Dam for duplexes with different target bases (or defects) is mismatch = deletion = abasic site > mA > A > 2A (26,33). The simplest assumption is that T4 Dam binds a duplex and orients to that strand containing a target base (or defect) for which it has a stronger affinity. Hence, we can assume that the highest orientation affinity occurs during binding in the presence of AdoMet. This assumption is strengthened by the observed ‘burst’ value close to 1.0 during pre-steady-state methylation of the 2A/A duplex (Fig. 5A). Such a value signifies that each bound T4 Dam molecule was correctly oriented with respect to the methylatable Ade residue. However, the simple assumption of the ‘highest affinity orientation’ is inconsistent with other data. For example, according to the array of affinities above, in binding the hemimethylated mA/A duplex, T4 Dam should be oriented preferentially for flipping the mA residue, so that a non-productive complex should be formed. However, the ‘burst’ value of mA/A and the native A/A duplex was the same (12). Hence, a hierarchy of preferential T4 Dam orientation, with respect to an unmodified target Ade residue, is more complicated than discussed above, and ‘higher affinity’ and preferential orientation are not related.
In the case of the more extensively studied MTase, HhaI, the interrelationship between binding and enzyme orientation about an asymmetric duplex is also ambiguous. M·HhaI binds the hemimethylated duplex, containing recognition sequence GCGC/GMGC (where M is 5-methylCyt), better than the unmethylated duplex. However, the crystal structure of M·HhaI complexed with duplex DNA, containing a hemimethylated, asymmetrical recognition sequence GCGC/GMGC, showed that the unmethylated target C residue was flipped out of the DNA helix (35). A structural basis for this preferential binding was proposed (35,36). On the other hand, in mismatched duplexes where G was not matched with its complementary target C, a mismatched Ade, uracil or abasic site was flipped out of the DNA helix, adopting the same conformation in the catalytic pocket as the normal residue (37). In contrast to the GCGC/GMGC duplex, such preferential orientation of enzyme to the strand containing the site modification conforms to the 5- to 10-fold increase in affinity to M·HhaI, if mismatches are introduced within a specific sequence context (36,38).
Two experiments show that nucleoside flipping and target base methylation by M·HhaI is most likely directly mechanistically connected. First, M·HhaI does not methylate the target C residue in the mismatched duplex GAGC/GCGC, where A is flipped-out (37,39). Secondly, it was directly shown that M·HhaI methylates the target base in the flipped state. The crystal structure of a ternary complex of M·HhaI, AdoMet and DNA containing 4′-thio-2′-deoxycytidine showed two distinguishable locations of the methyl group, which reflects partial methylation at C5 of the flipped target, 4′-thio-2′-deoxycytidine (40). A mechanistic connection between target base flipping and methylation is less clear with T4 Dam. Unlike the HhaI MTase and its asymmetric duplex, T4 Dam methylated the mismatched duplex GATC/GTAC (top strand A) and showed a burst value close to 1.0 (12). On the other hand, in the absence of AdoMet, T4 Dam titration of duplex G2ATC/GTAC produced only a low fluorescence increase at saturating enzyme concentration (data not shown). This can be ascribed to preferential enzyme orientation with respect to the mismatched strand. However, when AdoMet was added at the titration end point, a 30% increase in fluorescence level was observed. Although this effect was rather weak, it suggests that in the presence of AdoMet the ability of the enzyme to orient to the more ‘native’-like strand increased. Thus, these results suggest that there is no clear correlation between (stable) base flipping and subsequent methylation.
The role of AdoMet in the methylation reaction of T4 Dam goes beyond that of simply serving as a methyl donor. First, AdoMet-binding induces a conformational rearrangement in T4 Dam (34). This not only results in an increase in the affinity of the enzyme to bind specific DNA containing its recognition site (26,33), but it also results in an increased specificity for the strand containing the target base within the recognition site, as demonstrated in this paper. As a consequence of the AdoMet-induced conformational rearrangement and increased specificity, T4 Dam is capable of undergoing a rapid reorientation to the productive strand in an asymmetrically modified recognition site. We have demonstrated this with the asymmetric 2A/A and 2A/mA duplexes, but the situation with the physiologically significant A/mA remains to be determined. It should be mentioned, however, that we have observed a burst of approximately one using pre-formed T4 Dam–AdoMet + mA/A duplex, as well as with pre-formed T4 Dam–mA/A duplex + AdoMet (12). Biologically, it makes sense for the MTase to be able to reorient from a strand containing mA to the one containing Ade without dissociating from the DNA, so as to increase the efficiency of methylation of hemimethylated DNA produced by replication in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a US Public Health Service grant from the Fogarty International Center (No. TW00529), a grant from the Russian Foundation for Fundamental Research (No. 99-04-49868), a US Public Health Service grant GM29227 from the National Institutes of Health (to S.H.), and an NSF grant MCB-9603567 (to N.R.).
References
- 1.Cheng X. (1995) Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct., 24, 293–318. [DOI] [PubMed] [Google Scholar]
- 2.Wu J.C. and Santi,D.V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem., 262, 4778–4786. [PubMed] [Google Scholar]
- 3.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 4.Cheng X., Kumar,S., Klimasauskas,S. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase. Cold Spring Harbor Symp. Quant. Biol., 58, 331–338. [DOI] [PubMed] [Google Scholar]
- 5.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 6.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 7.Labahn J., Granzin,J., Schluckebier,G., Robinson,D.P., Jack,W.E., Schildkraut,I. and Saenger,W. (1994) Three-dimensional structure of the adenine-specific DNA methyltransferase M.TaqI in complex with the cofactor S-adenosylmethionine. Proc. Natl Acad. Sci. USA, 91, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong W., O’Gara,M., Blumenthal,R.M., and Cheng,X. (1997) Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res., 25, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran P.H., Korszun,Z.R., Cerritelli,S., Springhorn,S.S. and Lacks,S.A. (1998) Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine. Structure, 6, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 10.Scavetta R.D., Thomas,C.B., Walsh,M.A., Szegedi,S.S., Joachimiak,A., Gumport,R.I. and Churchill,M.E. (2000) Structure of RsrI methyltransferase, a member of the N6-adenine β class of DNA methyltransferases. Nucleic Acids Res., 28, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goedecke K., Pignot,M., Goody,R.S., Scheidig,A.J. and Weinhold,E. (2001) Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol., 8, 121–125. [DOI] [PubMed] [Google Scholar]
- 12.Malygin E.G., Lindstrom,W.M.,Jr, Schlagman,S.L., Hattman,S. and Reich,N.O (2000) Pre-steady-state kinetics of bacteriophage T4 Dam DNA-[N6-adenine] methyltransferase: interaction with native (GATC) or modified sites. Nucleic Acids Res., 28, 4207–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan B.W., Beechem,J.M., Lindstrom,W.M. and Reich,N.O. (1998) Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem., 273, 2368–2373. [DOI] [PubMed] [Google Scholar]
- 14.Holz B., Klimasauskas,S., Serva,S. and Weinhold,E. (1998) 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res., 26, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allan B.W. and Reich,N.O. (1996) Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry, 35, 14757–14762. [DOI] [PubMed] [Google Scholar]
- 16.Pues H., Bleimling,N., Holz,B., Woelcke,J. and Weinhold,E. (1999) Functional roles of the conserved aromatic amino acid residues at position 108 (motif IV) and position 196 (motif VIII) in base flipping and catalysis by the N6-adenine DNA methyltransferase from Thermus aquaticus. Biochemistry, 38, 1426–1434. [DOI] [PubMed] [Google Scholar]
- 17.Szegedi S.S., Reich,N.O. and Gumport,R.I. (2000) Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase. Nucleic Acids Res., 28, 3962–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilkaitis G., Dong,A., Weinhold,E., Cheng,X. and Klimasauskas,S. (2000) Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem., 275, 38722–38730. [DOI] [PubMed] [Google Scholar]
- 19.Serva S., Weinhold,E., Roberts,R.J. and Klimasauskas,S. (1998) Chemical display of thymine residues flipped out by DNA methyltransferases. Nucleic Acids Res., 26, 3473–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeltsch A., Roth,M. and Friedrich,T. (1999) Mutational analysis of target base flipping by the EcoRV adenine-N6 DNA methyltransferase. J. Mol. Biol., 285, 1121–1130. [DOI] [PubMed] [Google Scholar]
- 21.Schlagman S. and Hattman,S. (1983) Molecular cloning of a functional dam+ gene coding for phage T4 DNA adenine methylase. Gene, 22, 139–156. [DOI] [PubMed] [Google Scholar]
- 22.Kossykh V.G., Schlagman,S.L., and Hattman,S. (1995) Phage T4 DNA [N6-adenine]-methyltransferase. Overexpression, purification and characterization. J. Biol. Chem., 270, 14389–14393. [DOI] [PubMed] [Google Scholar]
- 23.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 24.Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 25.Thielking V., Du Bois,S., Eritja,R. and Guschlbauer,W. (1997) Dam methyltransferase from Escherichia coli: kinetic studies using modified DNA oligomers: nonmethylated substrates. Biol. Chem., 378, 407–415. [DOI] [PubMed] [Google Scholar]
- 26.Malygin E.G., Petrov,N.A., Gorbunov,Y.A., Kossykh,V.G. and Hattman,S. (1997) Interaction of the phage T4 DNA-[N6-adenine]-(Dam)methyltransferase with oligonucleotides containing native or modified (defective) recognition sites. Nucleic Acids Res., 25, 4393–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halvorsen Y.C., Nandabalan,K. and Dickson,R.C. (1990) LAC9 DNA-binding domain coordinates two zinc atoms per monomer and contacts DNA as a dimer. J. Biol. Chem., 265, 13283–13289. [PubMed] [Google Scholar]
- 28.Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1989) The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science, 243, 1681–1688. [DOI] [PubMed] [Google Scholar]
- 29.Fersht A. (1977) Enzyme Structure and Mechanism, 2nd Edn. W.H. Freeman and Co., New York, NY, p. 133.
- 30.Bergerat A. and Guschlbauer,W. (1990) The double role of methyl donor and allosteric effector of S-adenosyl-methionine for Dam methylase of E.coli. Nucleic Acids Res., 18, 4369–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szczelkun M.D. and Connolly,B.A. (1995) Sequence-specific binding of DNA by the EcoRV restriction and modification enzymes with nucleic acid and cofactor analogues. Biochemistry, 34, 10724–10733. [DOI] [PubMed] [Google Scholar]
- 32.Dubey A.K. and Roberts,R.J. (1992) Sequence specific DNA binding by the MspI DNA methyltransferase. Nucleic Acids Res., 20, 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malygin E.G., Zinoviev,V.V., Petrov,N.A., Evdokimov,A.A., Jen-Jacobson,L., Kossykh,V.G. and Hattman,S. (1999) Effect of base analog substitutions in the specific GATC site on binding and methylation of oligonucleotides duplexes by the bacteriophage T4 Dam DNA-[N6-adenine] methyltransferase. Nucleic Acids Res., 27, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuzikov F.V., Tuzikova,N.A., Naumochkin,A.N., Zinoviev,V.V. and Malygin,E.G. (1997) Fluorescence quenching study of equilibrium binding of phage T4 Dam DNA-(N6-adenine)-methyltransferase with substrates and ligands. Mol. Biol., 31, 73–76 (English translation). [PubMed] [Google Scholar]
- 35.O’Gara M., Roberts,R.J. and Cheng,X. (1996) A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase. J. Mol. Biol., 263, 597–606. [DOI] [PubMed] [Google Scholar]
- 36.Lindstrom W.M.,Jr, Flynn,J. and Reich,N.O. (2000) Reconciling structure and function in HhaI DNA cytosine-C-5 methyltransferase. J. Biol. Chem., 275, 4912–4919. [DOI] [PubMed] [Google Scholar]
- 37.O’Gara M., Horton,J.R., Roberts,R.J. and Cheng,X. (1998) Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat. Struct. Biol., 5, 872–877. [DOI] [PubMed] [Google Scholar]
- 38.Yang A.S., Shen,J.-C., Zingg,J.-M, Mi,S., and Jones,P.A. (1995) HhaI and HpaII DNA methyltransferases bind DNA mismatches, methylate uracil and block DNA repair. Nucleic Acids Res., 23, 1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheikhnejad G., Brank,A., Christian,J.K., Goddard,A., Alvarez,E., Ford,H., Marquez,V.E., Sufrin,J.R., O’Gara,M. and Cheng,X. (1999) Mechanism of inhibition of DNA (cytosine C5)-methyltransferase by oligodeoxyribonucleotides contaning 5,6-dihydro-5-azacytosine. J. Mol. Biol., 285, 2021–2034. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S., Horton,J.R., Jones,G.D., Walker,R.T., Roberts,R.J. and Cheng,X. (1997) DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res., 25, 2773–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]