ABSTRACT
Proton pump inhibitors (PPIs), used to treat gastro-esophageal reflux and prevent gastric ulcers, are among the most widely used drugs in the world. The use of PPIs is associated with an increased risk of enteric infections. Since the gut microbiota can, depending on composition, increase or decrease the risk of enteric infections, we investigated the effect of PPI-use on the gut microbiota. We discovered profound differences in the gut microbiota of PPI users: 20% of their bacterial taxa were statistically significantly altered compared with those of non-users. Moreover, we found that it is not only PPIs, but also antibiotics, antidepressants, statins and other commonly used medication were associated with distinct gut microbiota signatures. As a consequence, commonly used medications could affect how the gut microbiota resist enteric infections, promote or ameliorate gut inflammation, or change the host's metabolism. More studies are clearly needed to understand the role of commonly used medication in altering the gut microbiota as well as the subsequent health consequences.
keywords: gut microbiota, medication, proton pump inhibitors
Proton pump inhibitors affect the gut microbiota
Proton pump inhibitors (PPIs), used to treat gastro-esophageal reflux and to prevent gastric ulcers, are among the most commonly used drugs in the world.1,2 In the Netherlands, one PPI alone (omeprazole) was the fourth most prescribed drug in 2015. In observational studies, the use of PPIs has been associated with an increased risk of enteric infections caused by Clostridium difficile, Salmonella spp., Shigella spp and Campylobacter spp.3-5 Since the gut microbiota can, depending on composition, increase or decrease the risk of enteric infections, we investigated the effect of PPI use on the gut microbiota.6 Using the 16S rRNA sequences of stool samples from 1815 individuals spanning 3 independent cohorts, we observed profound changes in the gut microbiota of PPI users. In PPI users the relative abundance of 20% of the bacterial taxa, whereof 18 bacterial families, was statistically significantly different (either increased or decreased) compared with abundances in samples from non-users.6 Concurrently with our research, other research groups were also investigating the influence of PPIs on the gut microbiota. A small intervention study was published a few months before ours and a similar observational study was published in the same issue of Gut.7,8 In Table 1, the bacterial alterations associated with PPI use from all 3 studies are presented at the family level. There are many similarities between the results of all 3 studies and, when associations at different taxonomical levels are taken into account (e.g. the decrease of order Clostridiales and increases of class Gammaproteobacteria and order Actinomycetales), a consistent profile of alterations in gut microbiota associated with PPI use emerges.
Table 1.
PPI use associated with gut microbiota alterations in 3 studies.
| Bacterial family | Imhann et al. Gut 2016; Figure 2 Cross-sectional study 1815 individuals | Jackson et al. Gut 2016; Figure 3Cross-sectional study 1827 individuals | Freedberg et al. Gastroenterology 2015; Figure 1 Intervention, time series 12 healthy volunteers |
|---|---|---|---|
| Actinomycetaceae | Increased | NR | NR |
| Aerococcaceae | Increased | NR | NR |
| Anaeroplasmataceae | Decreased | NR | NR |
| Bifidobacteriaceae | Decreased | NR | NR |
| Burkholderiaceae | NR | Increased | NR |
| Cardiobacteriaceae | NR | Increased | NR |
| Carnobacteriaceae | Increased | Increased | NR |
| Corynebacteriaceae | NR | Increased | NR |
| Dehalobacteriaceae | Decreased | NR | NR |
| Enterobacteriaceae | Increased | NR | NR |
| Enterococcaceae | Increased | NR | Increased |
| Erysipelotrichaceae | NR | Decreased | NR |
| Gemellaceae | Increased | NR | NR |
| Lachnospiraceae | NR | Decreased | NR |
| Lactobacillaceae | Increased | Increased | NR |
| Leptotrichiaceae | Increased | NR | NR |
| Leuconostocaceae | Increased | NR | NR |
| Micrococcaceae | Increased | Increased | Increased |
| Pasteurellaceae | Increased | Increased | NR |
| Planococcaceae | Increased | NR | NR |
| Ruminococcaceae | Decreased | Decreased | NR |
| Staphylococcaceae | Increased | Increased | Increased |
| Streptococcaceae | Increased | Increased | Increased |
Note. Bacterial families associated with PPI use in 3 gut microbiota studies.6-8; Consistent changes in 2 or all studies are marked in bold; NR = Not reported.
Other commonly used drugs and the gut microbiota
To ensure that our observed alterations in gut microbiota associated with PPI use were not based the confounding effects of concomitant use of other drugs, which could also potentially influence the gut microbiota, we grouped the most commonly used drugs and analyzed possible associations with the gut microbiota. This analysis identified several changes in the gut microbiota associated with other commonly used drugs. (See the most commonly prescribed medication in the Netherlands in Table 2 and the gut microbiota associations in Table 3). However, all gut microbiota alterations associated with PPI use remained statistically significant even after statistical correction for the use of other commonly used drugs.
Table 2.
Most commonly used medication in the Netherlands (17 million inhabitants) in 2015.
| Rank | Medication name | Medication group or use | Users (millions) |
|---|---|---|---|
| 1 | diclofenac | NSAIDS | 1.29 |
| 2 | amoxicillin | Antibiotics | 1.22 |
| 3 | simvastatin | Statins | 1.17 |
| 4 | omeprazole | PPIs | 1.16 |
| 5 | metoprolol | β-blockers | 1.11 |
| 6 | macrogol | Stimulating bowel movements/anti-constipation | 1.06 |
| 7 | inert dermal creams | Skin creams for eczema | 1.02 |
| 8 | salbutamol | Dilate airways | 0.90 |
| 9 | colecalciferol | Prevent osteoporosis | 0.83 |
| 10 | acetylsalicylic acid | Platelet aggregation inhibitor | 0.81 |
Table 3.
Commonly used medication associated with gut microbiota alterations. Table from Imhann et al. Gut 2016.
| Medication category | Taxon | Direction |
|---|---|---|
| Antibiotics | g__Holdemania | Increased |
| Antidepressants (SSRI, SNRI, mirtazapine and TCA) | f__Bacteroidaceae | Increased |
| Antidepressants (SSRI, SNRI, mirtazapine and TCA) | g__Bacteroides | Increased |
| Antidiabetic medication (both oral and insulin) | o__Bacillales | Increased |
| Changes bowel movement/stool frequency* | p__Firmicutes | Decreased |
| Changes bowel movement/stool frequency* | o__Clostridiales | Decreased |
| Changes bowel movement/stool frequency* | c__Clostridia | Decreased |
| Changes bowel movement/stool frequency* | g__Coprococcus | Decreased |
| Changes bowel movement/stool frequency | o__Bacteroidales | Increased |
| Changes bowel movement/stool frequency | p__Bacteroidetes | Increased |
| Changes bowel movement/stool frequency | f__Bacteroidaceae | Increased |
| Changes bowel movement/stool frequency | g__Bacteroides | Increased |
| Cholesterol lowering medication (statins)* | o__Bacillales | Increased |
| Cholesterol lowering medication (statins) | g__Dorea|s__longicatena | Decreased |
| Cholesterol lowering medication (statins) | g__Ruminococcus | Increased |
| Triglyceride lowering medication (Fibrates)* | g__Ruminococcus|s__gnavus | Increased |
Note. All associations are statistically significant at FDR < 0.05
k__, kingdom; p__; phylum; c__, class; o__, order; f__, family; g__, genus; s__, species.
Statistically significant FDR<0.05 after correction for PPI use.
Our group published an elaborate metagenomic sequencing analysis of 1135 participants of the same general population cohort used in our first study. In this metagenomics study, we showed that several other commonly used drugs are also associated with gut microbiota alterations (See Fig. 1).9 Consistent with our earlier results, the variance in the gut microbiota that could be explained by PPI use was the largest of all the commonly used drugs. However, many other drugs also had a statistically significant effect on the gut microbiota composition.
Figure 1.
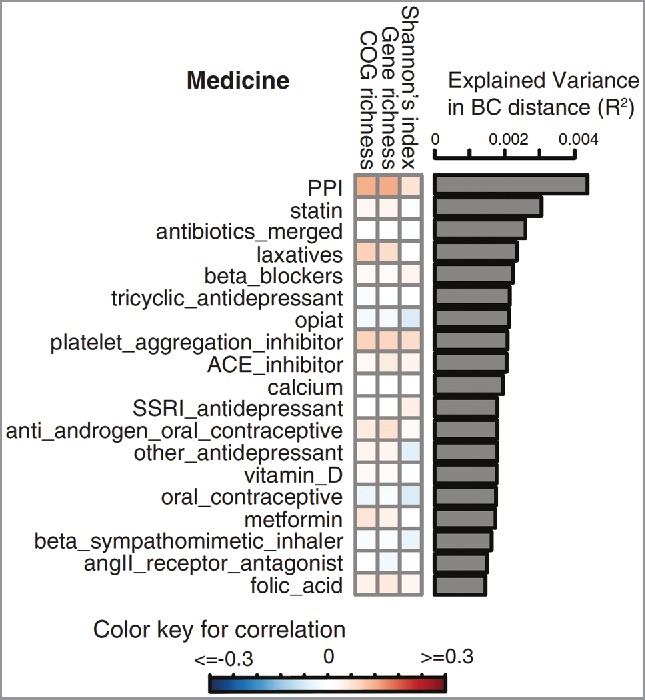
Inter-individual gut microbiota variation (Bray-Curtis distance) explained by commonly used medication at FDR < 0.1. Figure from Zhernakova et al. Science 2016. © Science. Reproduced by permission of Science. Permission to reuse must be obtained from the rightsholder.
Consequences of gut microbiota changes caused by medication
What is becoming increasingly clear is that antibiotics, PPIs and metformin affect the gut microbiota and that other commonly used medication, like statins and SSRIs and are associated with distinct gut microbiota signatures.6-11 As a consequence, these types of medication could have an effect on the risk of developing enteric infections or gut inflammation, and they may also have an effect on host metabolism.12-15
Susceptibility to enteric infections
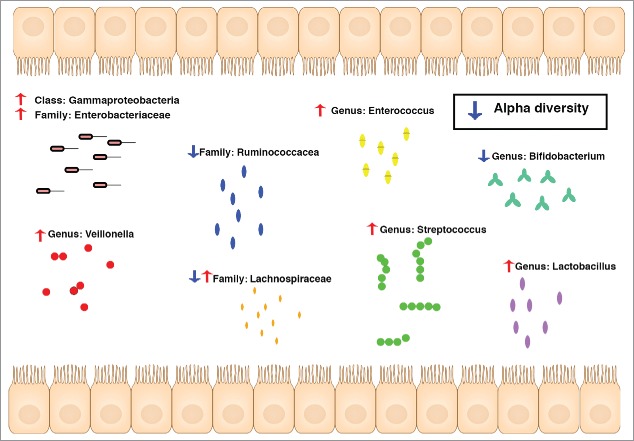
A well-known example of reduced enteric infection resistance through alteration of the gut microbiota caused by medication is the increased risk of Clostridium difficile infections after (repeated) treatment with antibiotics.16 Antibiotics kill off a large proportion of the gut microbiota, creating an empty niche that allows Clostridium difficile to colonize and overgrow.16 Recent observational studies indicated that it is not only antibiotic use, but also PPI use, that is associated with increased risk of Clostridium difficile infection.5,17 Subsequent gut microbiota studies attributed the increased risk to unfavorable gut microbiota alterations caused by PPI use.6-8 Human, animal and in vitro studies show an overlap between the specific gut microbiota alterations associated with PPI use (as found in Imhann et al. Gut 2016) and the bacterial changes that lead to increased susceptibility to Clostridium difficile (as depicted in Fig. 2).14,15,18-24 The US Food and Drug Administration (USFDA) already aims to limit the use of antibiotics to reduce the number of Clostridium difficile infections in the USA.25 Since PPIs are often prescribed, and these prescriptions renewed, without evidence-based indication, a reduction of unnecessary PPI use could also contribute to this aim.17
Figure 2.
Gut microbiota changes associated with PPI use that increase the risk of Clostridium difficile infection.14,15,18-24
PPI are also associated with an increased risk of other enteric infections caused by Salmonella, Shigella and Campylobacter species.4 In the Netherlands, recent trends toward increased incidence of campylobacter infections closely follow trends in the number of PPI prescriptions: both increased rapidly from 2004 to 2011 then showed small decrease in 2012.3 Since PPIs diminish the gastric acid barrier, pathogenic microbial species that would not otherwise survive the gastric acid could more easily be introduced into the gut microbiota.6,8 The overrepresentation of oral microbiota in the gut microbiota of PPI users supports this hypothesis.6,8 The effects of antibiotics and PPIs on the gut microbiota are currently the clearest examples of how medication can change susceptibility to enteric infections and have an effect on human health. The influence that other gut-microbiota-changing medication have on the susceptibility to enteric infections is still unclear.
Gut inflammation
Gut microbiota can promote or ameliorate gut inflammation. Favorable microbiota can ameliorate gut inflammation through the induction of regulatory T-cells (Tregs), interleukin 10 (IL-10) and the production of the short-chain fatty acid butyrate.26,27 In contrast, unfavorable microbiota can produce toxins that promote inflammation of the gut epithelium.28 A profoundly disturbed balance between favorable and unfavorable gut microbiota, resulting in gut inflammation, is seen in Inflammatory Bowel Disease (IBD).29 IBD is a common disorder of which the incidence continues to rise, and is attributed to the transition to a sedentary, high fat, high calorie ‘Western lifestyle’.30 Aside from changes in the habitual diet, prior Salmonella or Campylobacter gastroenteritis and prior use of antibiotics are also presumed IBD risk factors.30,31 Whether increased use of other commonly used medication that influence the gut microbiota could contribute to the increasing incidence of IBD is unknown. Links have been reported between the PPI use and increases in the gut inflammation marker, fecal calprotectin, but the mechanism by which PPI cause this increased fecal calprotectin, i.e. if they are due to gut microbiota changes, is not clear.32 Aside from the influence of PPIs, NSAIDs are known to sometimes cause enteropathy, ulceration and elevation of fecal calprotectin. How other commonly used medications affect inflammation in the gut still has to be elucidated.
BMI and lipid metabolism
Gut microbiota alterations have large effects on body mass index (BMI), lipid- and cholesterol-levels.12,33,34 The effects of some medications on the development of obesity have received considerable attention, for example, the effect of antibiotic use during childhood on obesity 35 Use of other drugs is also related to weight gain and obesity, including several classes of antidepressants: SSRIs, SNRIs and TCAs.36 These antidepressants are also associated with gut microbiota alterations, which could potentially be a mechanisms by which these drugs cause weight gain.6,9 PPI use is associated with an increased risk of cardiovascular disorders.37 Although other mechanisms have been proposed,38 the effects of PPIs on the microbiota could affect lipid metabolism and an thereby an individual's subsequent risk of cardiovascular events.9,12 The effects of commonly used medication and the effects on the host metabolism therefore warrant further investigation.
Intervention studies and animal studies are required to clarify the influence of commonly used medication on the gut microbiota
Intervention studies
One way to study the effects of medication on the gut microbiota is to perform an intervention study. Intervention studies in humans in which the pre-medication gut microbiota can be compared with the post-medication gut microbiota are relatively easy to implement. All commonly prescribed drugs have already been approved by the USFDA and the European Medication Agency (EMA). There are, by definition, a large number of patients who begin using these types of medication for the first time every day. Moreover, stool sampling is not an invasive procedure. One small scale intervention study investigating the role of PPI on the gut microbiota has already been performed.7
Animal studies
In addition to intervention studies in humans, mouse experiments are required in which gut microbiota changes can be studied in isolation and the effects in tissues and blood markers observed.39 The effects on enteric-infection-resistance to Clostridium difficile through gut microbiota alterations have already been tested using a mouse model in which mice were exposed to pathogens after antibiotic treatment.15 This model could be used to investigate how other commonly used drugs affect enteric infection resistance. The effects on gut epithelial inflammation and inflammatory markers can also be studied using IBD mouse models.40 Finally, mouse experiments can be used to investigate the metabolic effects of gut microbiota alterations.41
Commonly used medication should be part of the core phenotype set for human gut microbiota research
PPI use is overrepresented in many disorders and conditions, including obesity, non-alcoholic steatohepatitis (NASH), Irritable Bowel Syndrome (IBS), rheumatoid arthritis and IBD. NSAID use is overrepresented in rheumatoid arthritis. Antibiotics are much more often prescribed in Crohn's disease. Many of the current studies relating the gut microbiota and disease do not take into account the potential confounding effects of medication use. The poor reproducibility of gut microbiota studies thus far could partially be explained by lack of collection or inadequate collection of confounding phenotypes.
To begin to resolve these issues, a new core set of phenotypes needs to be used as covariates in microbiota studies. We think this new core phenotype set should at least consist of:
Conclusions
The use of PPIs, antibiotics, NSAIDs, SSRIs, metformin and other commonly used medication are associated with specific gut microbiota compositions. As a consequence, the use of these types of medication could affect how the gut microbiota can resist enteric infections, promote or ameliorate gut inflammation, or change the host metabolism. While the effects of antibiotics have been relatively well-studied, and the effects of PPI use are starting to become clear, the effects of many other commonly used medications on the gut microbiota remain unknown. More investigations of the role of commonly used medication on the gut microbiota and the subsequent health consequences is certainly needed.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank the members of the gut microbiota study group of the Departments of Gastroenterology, Genetics and Medical Microbiology in the University Medical Center Groningen for their contribution to the critical discussions of this papers' topic.
This article was edited for language and formatting by Kate McIntyre, Associate Scientific Editor in the Department of Genetics, University Medical Center Groningen.
Funding
RKW and JF are supported by VIDI grants (016.136.308 and 864.13.013) from the Netherlands Organization for Scientific Research (NWO). CW is supported by a Top Institute Food and Nutrition grant GH001, an ERC advanced grant (ERC–671274), and a Spinoza award (NWO SPI 92–266). AZ holds a Rosalind Franklin fellowship (University of Groningen) and an ERC Starting Grant (ERC–715772). JF and AZ are also supported by a CardioVasculair Onderzoek Nederland grant (CVON 2012–03).
Author contributions
FI and AGLM wrote the manuscript. AVV designed the graphics. AVV, MJB, DPYK, JF, CW, AZ and RKW critically reviewed the manuscript.
References
- [1].Dutch Foundation for Pharmaceutical Statistics (SFK) Data and facts medication use in the Netherlands in 2015. 2016. [Google Scholar]
- [2].Moayyedi P, Talley NJ. Gastro-oesophageal reflux disease. Lancet 2006; 367:2086-100; PMID:16798392; http://dx.doi.org/ 10.1016/S0140-6736(06)68932-0 [DOI] [PubMed] [Google Scholar]
- [3].Bouwknegt M, Pelt W Van, Kubbinga ME, Weda M, Havelaar AH. Potential association between the recent increase in campylobacteriosis incidence in the Netherlands and proton-pump inhibitor use – an ecological study. Eurosurveillance 2014; 19(32):20873. [DOI] [PubMed] [Google Scholar]
- [4].Leonard J, Marshall JK, Moayyedi P. Systematic review of the risk of enteric infection in patients taking acid suppression. Am J Gastroenterol 2007; 102(3):2047-56; PMID:17509031; http://dx.doi.org/ 10.1111/j.1572-0241.2007.01275.x [DOI] [PubMed] [Google Scholar]
- [5].Janarthanan S, Ditah I, Adler DG, Ehrinpreis MN. Clostridium difficile-Associated Diarrhea and Proton Pump Inhibitor Therapy: A Meta-Analysis. Am J Gastroenterol Nature Publishing Group; 2012; 107(7):1001-10; PMID:22710578; http://dx.doi.org/ 10.1038/ajg.2012.179 [DOI] [PubMed] [Google Scholar]
- [6].Imhann F, Bonder MJ, Vich Vila A, et al.. Proton pump inhibitors affect the gut microbiome. Gastroenterol 2015; 149(4):883-5; http://dx.doi.org/ 10.1053/j.gastro.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Whittier S, Wang TC, Wang HH, Abrams JA.. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: A crossover trial. Gastroenterology Elsevier, Inc; 2015; 149(4):883-5; PMID:26164495; http://dx.doi.org/ 10.1053/j.gastro.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jackson MA, Goodrich JK, Maxan ME, Freedberg DE, Abrams JA, Poole AC, Sutter JL, Welter D, Ley RE, Bell JT, et al.. Proton pump inhibitors alter the composition of the gut microbiota. Gut 2016; 65(5):749-56; PMID:26719299; http://dx.doi.org/ 10.1136/gutjnl-2015-310861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al.. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016; 352(6285):565-9; PMID:27126040; http://dx.doi.org/ 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al.. Population-level analysis of gut microbiome variation. Science 2016; 352(6285):560-4; http://dx.doi.org/ 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- [11].Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, et al.. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. Nature Publishing Group; 2015; 528:262-6; PMID:26633628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, Brandsma E, Marczynska J, Imhann F, Weersma RK, et al.. The gut microbiome contributes to a substantial proportion of the variation in blood lipidsnovelty and Significance. Circ Res 2015; 117(9):817-24; PMID:26358192; http://dx.doi.org/ 10.1161/CIRCRESAHA.115.306807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013; 14(7):685-90; PMID:23778796; http://dx.doi.org/ 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. Nature Publishing Group; 2013; 13(11):790-801; PMID:24096337; http://dx.doi.org/ 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al.. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. Nature Publishing Group; 2014; 517(7533):205-8; PMID:25337874; http://dx.doi.org/ 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. Nature Publishing Group; 2009; 7(7):526-36; PMID:19528959; http://dx.doi.org/ 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- [17].McDonald EG, Milligan J, Frenette C, Lee TC. Continuous proton pump inhibitor therapy and the associated risk of recurrent clostridium difficile infection. JAMA Intern Med. 2015; 175:784-91; PMID:25730198 [DOI] [PubMed] [Google Scholar]
- [18].Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2011; 2(March 2015):145-58; PMID:21804357 ; http://dx.doi.org/ 10.4161/gmic.2.3.16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197(3):435-8; PMID:18199029; http://dx.doi.org/ 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- [20].Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 2013; 51(9):2884-92; PMID:23804381; http://dx.doi.org/ 10.1128/JCM.00845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rea MC, O'Sullivan O, Shanahan F, O'Toole PW, Stanton C, Ross RP, Hill C. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J Clin Microbiol 2012; 50(3):867-75; PMID:22162545; http://dx.doi.org/ 10.1128/JCM.05176-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baines SD., Crowther GS, Todhunter SL, Freeman J, Chilton CH, Fawley WN, Wilcox MH.. Mixed infection by Clostridium difficile in an in vitro model of the human gut. J Antimicrob Chemother 2013; 68(1):1139-43; PMID:23354280; http://dx.doi.org/ 10.1093/jac/dks529 [DOI] [PubMed] [Google Scholar]
- [23].Schubert AM, Rogers M a M, Ring C, et al.. Microbiome data distinguish patients with clostridium difficile infection and non- C . difficile -associated diarrhea from healthy. MBio 2014; 5(3):1-9; http://dx.doi.org/ 10.1128/mBio.01021-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peterfreund GL, Vandivier LE, Sinha R, Marozsan AJ, Olson WC, Zhu J, Bushman FD. Succession in the Gut Microbiome following Antibiotic and Antibody Therapies for Clostridium difficile. PLoS One 2012; 7(10); PMID:23071679; http://dx.doi.org/ 10.1371/journal.pone.0046966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].U.S. Food and Drug Administration FDA Drug Safety Communication: Clostridium difficile-associated diarrhea can be associated with stomach acid drugs known as proton pump inhibitors (PPIs) [Internet]. 2012. Available from: http://www.fda.gov/drugs/drugsafety/ucm290510.htm.
- [26].Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al.. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500(7461):232-6; PMID:23842501; http://dx.doi.org/ 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- [27].Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al.. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. Nature Publishing Group; 2013; 504(7480):446-50; PMID:24226770; http://dx.doi.org/ 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- [28].Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol 2010; 12(1):99-113; PMID:19747213; http://dx.doi.org/ 10.1111/j.1462-5822.2009.01381.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, et al.. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2016; [Epub ahead of print] http://dx.doi.org/ 10.1136/gutjnl-2016-312135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol 2015; 12(4):205-17; http://dx.doi.org/ 10.1038/nrgastro.2015.34 [DOI] [PubMed] [Google Scholar]
- [31].Gradel KO, Nielsen HL, Schønheyder HC, Ejlertsen T, Kristensen B, Nielsen H. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. AGA Institute American Gastroenterological Association; 2009; 137(2):495-501; PMID:19361507; http://dx.doi.org/ 10.1053/j.gastro.2009.04.001 [DOI] [PubMed] [Google Scholar]
- [32].Poullis A, Foster R, Mendall MA. Proton pump inhibitors are associated with elevation of faecal calprotectin and may affect specificity. Eur J Gastroenterol Hepatol 2003; 15(5):573-4; PMID:12702920; http://dx.doi.org/ 10.1097/00042737-200305000-00021 [DOI] [PubMed] [Google Scholar]
- [33].Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al.. A core gut microbiome in obese and lean twins. Nature. Nature Publishing Group; 2009; 457(7228):480-4; PMID:19043404; http://dx.doi.org/ 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ley R, Turnbaugh P, Klein S, Gordon J. Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444:1022-3; PMID:17183309; http://dx.doi.org/ 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- [35].Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nat Rev Endocrinol. Nature Publishing Group; 2014; 11(3):182-90; PMID:25488483; http://dx.doi.org/ 10.1038/nrendo.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Blumenthal SR, Castro VM, Clements CC, Rosenfield HR, Murphy SN, Fava M, Weilburg JB, Erb JL, Churchill SE, Kohane IS, et al.. An electronic health records study of long-term weight gain following antidepressant use. JAMA Psychiatry 2014; 71(8):889; PMID:24898363; http://dx.doi.org/ 10.1001/jamapsychiatry.2014.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shah NH, LePendu P, Bauer-Mehren A, Ghebremariam YT, Iyer SV, Marcus J, Nead KT, Cooke JP, Leeper NJ. Proton pump inhibitor usage and the risk of myocardial infarction in the general population. Guo Y, editor. PLoS One 2015; 10(6):e0124653; PMID:26061035; http://dx.doi.org/ 10.1371/journal.pone.0124653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ghebremariam YT, LePendu P, Lee JC, Erlanson DA, Slaviero A, Shah NH, Leiper J, Cooke JP. Unexpected effect of proton pump inhibitors: elevation of the cardiovascular risk factor asymmetric dimethylarginine. Circulation 2013; 128(8):845-53; PMID:23825361; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.113.003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nguyen TLA, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech 2015; 8(1):1-16; PMID:25561744; http://dx.doi.org/ 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schaubeck M, Clavel T, Calasan J, et al.. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut 2016; 65(2):225-37; http://dx.doi.org/ 10.1136/gutjnl-2015-309333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al.. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology Elsevier Inc; 2012; 143(4):913-916.e7; PMID:22728514; http://dx.doi.org/ 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- [42].Tigchelaar EF, Bonder MJ, Jankipersadsing S a, Fu J, Wijmenga C, Zhernakova A. Gut microbiota composition associated with stool consistency. Gut 2016; 65(3):540-42; http://dx.doi.org/ 10.1136/gutjnl-2015-310328 [DOI] [PubMed] [Google Scholar]