ABSTRACT
The existence of an implicit living microscopic world, composed primarily of bacteria, has been known for centuries. The exact mechanisms that govern the contribution of bacteria to human health and disease have only recently become the subject of intense research efforts. Within this very evident shift in paradigms, the rational design of probiotic formulations has led to the creation of an industry that seeks to progress the engineering of probiotic bacteria that produce metabolites that may enhance human host health and prevent disease. The promotion of probiotics is often made in the absence of quality scientific and clinically plausible data. The latest incursions into the probiotic market of claims have posited the amelioration of oxidative stress via potent antioxidant attributes or limiting the administration of probiotics to those species that do not produce D-Lactic acid (i.e., claims that D-Lactic acid acidosis is linked to chronic health conditions) or are strain-specific (shaping an industry point of difference) for appraising a therapeutic effect. Evidence-based research should guide clinical practice, as there is no place in science and medicine that supports unsubstantiated claims. Extravagant industry based notions continue to fuel the imprimatur of distrust and skepticism that is leveled by scientists and clinicians at an industry that is already rife with scientific and medical distrust and questionable views on probiotics. Ignoring scientifically discordant data, when sorting through research innovations and false leads relevant to the actions of probiotics, drives researcher discomfit and keeps the bar low, impeding the progress of knowledge. Biologically plausible posits are obligatory in any research effort; companies formulating probiotics often exhibit a lack of analytical understanding that then fuels questionable investigations failing to build on research capacity.
KEYWORDS: bacteria, efficacy, D-Lactate, oxidative stress, probiotics, safety, strain
Introduction
Extensive research on lactic acid bacteria has followed on from the highly cited early work of Ellie Metchnikoff, who in 1907 insightfully reported that the health of the intestines depended on specific food intake, and that by adopting certain measures one could modify the intestinal tract (IT) cohort of bacteria helping to reduce disease and enhance health outcomes. Current probiotic formulations have attempted to target the prevention and or treatment of numerous maladies of the IT (e.g., inflammatory bowel diseases), colonic symptoms (e.g., diarrhea, constipation), as well as skin (e.g., eczema) and respiratory (e.g., asthma) conditions. Methodological variation is extensively reported in probiotic clinical research in regards to study design i.e., single or double blinded, placebo or non-placebo controlled, randomized or non-randomized; as well as the administration of single versus multi probiotic species. Furthermore, considerable discrepancy occurs in probiotic formulations, dosing, duration and mode of delivery, analysis and interpretation of results. Also of importance is the effect of inconsistent environmental conditions such as the participant's diet and whether the study is conducted in a hospital setting or with outpatients.1 Studies are often reported as having methodological limitations such as representing a high risk of bias, lacking adequate description of randomization, allocation concealment, blinding, formulation and dosages of probiotics and placebo and lack of definition of participant demographics.2 Such limitations make it difficult to draw comparisons between studies or to make firm conclusions on efficacy. As such there is a real need for the conduct of well-designed, randomized controlled trials testing for strain-specific effects that may support specific claims, especially when there is documented support for changing medical practice attitudes with recommendations that include prescriptions for the administration of probiotics.3,4
Medline provides over 1400 records in the search of ‘probiotics’ in human clinical trials. This demonstrates the extensive research that has been undertaken to assess the therapeutic activity of probiotics for certain health conditions. Systematic reviews and meta-analyses corroborate that methodological issues associated with probiotic research include trials with highly variable reports of adverse effects, outcome measures with significant data missing from multiple trials and also variable assessment of probiotic formulas, dosing, initiation of probiotic treatment and length of supplementation.5
The research upon which this review was structured has focused on the use of probiotics as formulations that may provide a pharmacotherapeutic action. It includes methodological and reporting issues. As such we have addressed issues of probiotic species and also strain specificity of probiotics investigated in clinical trials, issues surrounding dosing and duration as well as specific exemplar safety and efficacy issues that have been used to gain marketing hype (e.g., the negative health effects of D-lactate producing probiotic bacteria and the abrogation of oxidative stress with the administration of probiotics) without scientifically plausible evidence. We posit that such issues and misrepresentations need to be better deciphered by researchers and industry so as to minimize any spurious claims and implausible ideas that impede the progress of research, and that confuse academics, clinicians and the community.
The science of probiotic bacteria
The administration of probiotics
The published literature is witness to a progressive understanding of probiotics. The term probiotic was derived from the Greek meaning for life and has been associated with various connotations since its inception. The term probiotic was first used in 1965 to describe metabolites produced by one microorganism that stimulated the growth of another, essentially the opposite to that of an antibiotic. In 1971, a probiotic referred to tissue extracts that stimulated microbial growth, however it was not until 1974 when the term probiotic was introduced that was aligned to the concept of organisms and substances that contributed to intestinal microbial balance.6 The current definition of probiotics is that they are live microorganisms, which when administered in adequate amounts confer a health benefit on the host.7
Probiotic research has assessed efficacy for multiple conditions including but not limited to allergies, infections, diarrhea, functional and inflammatory bowel disorders, URTIs, atopic conditions and urogenital, metabolic, neurologic and liver disorders. Probiotics have been postulated to enhance the living conditions of commensal microbiota that reside on the body surfaces covered by mucosal epithelial cells including the gastrointestinal and respiratory tracts, the urogenital tract, nose and mouth; and the eyes, placenta as well as the skin.8 The term probiotic is sometimes erroneously used as a synonym for beneficial species of the commensal microbiota, a misconception that arises when probiotics are defined as being commonly isolated from the human commensal microbiota cohort. Until such species are isolated and then adequately characterized for content, stability, and health effects, they cannot be ascribed as being probiotics.9
The genetics of probiotic bacteria: Species and strain selection
Lactic acid bacteria (LAB) are an important group of bacteria that are inextricably linked to human health. LAB belong to the phylum Firmicutes, class Bacilli and order Lactobacillales. The families include Lactobacillaceae, Enterococcaceae, Streptococcaceae, Aerococcaceae, Carnobacteriaceae and Leuconostocaceae. In molecular biology the core genome for a bacterial species has been defined as the set of genes present in all strains of a particular species.10 Whereas the pan-genome describes the full complement of genes in a clade, this typically is applied to species of bacteria.11 Pan-genomes can have large gene content variations among closely related strains.12 Presently there are more than 1000 complete genome sequences for bacteria that have been annotated in GenBank.11 Among these are 37 strains that represent 22 species of non-pathogenic LAB and 4 Bifidobacteria species. Furthermore, complete genomic sequences from over 150 other potentially pathogenic and non-pathogenic LAB are available. Hence the available genomic information allows for a critical review of novel insights that these bacteria may provide in i) pan and core genomes, ii) the evolutionary history of LAB (with loss and gain of genetic material) and iii) their adaptation to specific ecological niches such as the IT.
The common probiotic genera used in formulations include those from Lactobacilli, Bifidobacteria and Streptococci genera with multiple species and also the beneficial yeast Saccharomyces cerevisiae. Furthermore, multiple designated strains are assigned to several species with their therapeutic activity promoted as being strain-specific. The optimal genera, species and strain selection criteria has not yet been defined according to disease indication7,13 and further there is a lack of rigorous strain-to-strain comparisons of the same species in regards to therapeutic activity and efficacy, thus confusing strain-specific activity. Different probiotic genera and species demonstrate different activity in both in vitro and in vivo analyses; therefore therapeutic activity cannot be broadly applied to all genera and species.14 Clinical evidence demonstrates that probiotic multi-species formulas may be more effective against a wide range of end points when compared with single species formulas, which may be attributed to enhanced survival due to greater bacterial diversity and functionality and also due to the synergistic effects of combining species. It must be noted however that comparisons between multi-species and single species formulas do not necessarily compare the same dose.15,16
Extensive clinical trials have been conducted assessing the efficacious use of probiotics for Irritable Bowel Syndrome (IBS). A recent systematic review and meta-analysis concluded from 43 RCTs including 3,452 patients, that probiotics are effective for IBS in improving overall symptoms as a dichotomous measure and improvement in global symptom, abdominal pain, bloating and flatulence scores.17 The authors concluded that combinations of probiotic strains with Lactobacillus plantarum DSM 9843 appeared to have the most evidence supporting their use, however which individual strains are the most beneficial still remains unclear. Considering that the mechanism of action of probiotics in different situations has not been clarified, the optimal use of one or more strains from different species and genera for therapeutic gain remains inconclusive.13
The growth, survival and colonization in the intestinal lumen of a particular probiotic strain may vary and clinical results from one study may or may not be transferable to another strain from the same species, this is yet to be proven. There is much confusion by practitioners that a therapeutic outcome can only be achieved for their patients by administering strain-specific probiotic bacteria. This is incorrect. There is sufficient evidence to demonstrate that certain probiotics have a core, widespread benefit to the host, which contradicts the notion that every strain is different and elicits a different effect in the host. Some generalizations can be made beyond strain-specific activity.7 Frequent therapeutic properties can be species-specific while strain-specific effects are rare (Fig. 1). When claiming strain-specific activity, various strains from the same species should be compared to enable a more accurate conclusion rather than claiming strain-specific activity when strains from different species are compared.18,19 It would be more accurate to state that probiotic bacteria have species-specific activity.
Figure 1.
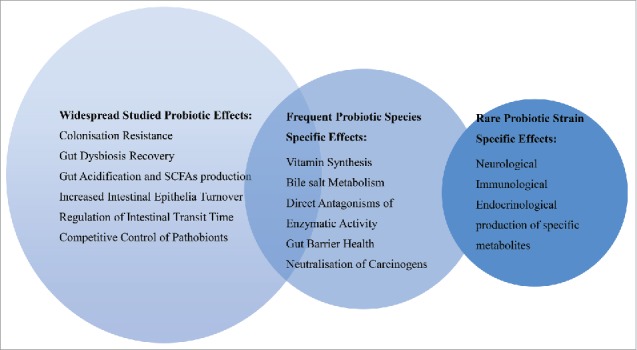
Therapeutic allocation for probiotics. Adapted from Hill, et al.7 Research demonstrates that generalised probiotic activity does exist beyond that of strain-specific effects. SCFA: short chain fatty acids.
Genomic stability of probiotics
Generic probiotics are patent-expired probiotics that can be freely used. Health and safety claims from the expired patents can be linked to the generic probiotic strains, provided that the genotype of the original strain is as identical as possible to that of the generic strain. This raises questions about the extent of genome stability in probiotic bacterial strains and its impact on probiotic functionality.20
Important factors governing the clinical effect of probiotics include their ability to adhere to the mucus membrane of the intestines. The ability to adhere is partly related to thread-like projections known as pili protruding from the cell surface.21 Poorly reproduced probiotics may produce species with genomic instability that result in the loss of genetic coding for pili. Such species, while active in vitro, may lack clinical efficacy due to lack of adhesiveness to the mucus layers of the gastrointestinal tract. Sybesma et al20 found that major genetic rearrangements occurred in some dairy product isolates, including the deletion of genes that are thought to play a role in probiotic functionality. Genomic islands deleted in 2 propagated Lactobacillus rhamnosus GG included the spaCBA gene, which encodes for pilin subunits involved in adhesion of the strains to the intestinal mucus and persistence in the human intestinal tract. These 2 species had human mucus-binding abilities of less than 1.6% compared with the LGG reference strain. It is reasonable to assume that these strains will have reduced residence in the gut and therefore reduce the clinical effects of such strains.20 SpaCBA pili has also been shown to directly and indirectly to reduce inflammatory signaling by promoting the release of anti-inflammatory proteins including Merozoite surface proteins 1 and 2 (MSP1/2).22
LGG pili are also susceptible to shearing stress. Bacteria subjected to 8000 x g centrifugal forces were found to be completely devoid of pili.23 Probiotic strains produced under strict quality control may, however, be identical to each other. A study24 that compared L. rhamnosus GG to 2 other strains of L. rhamnosus: L. rhamnosus LrV found in Freisland Campina yoghurts and L. rhamnosus LrI found in the probiotic product, Idoform (Ferrosan). The 2 strains were found to be virtually identical to L. rhamnosus GG in terms of genomes and phenotype. This is likely to be used to high manufacturing standards to maintain the genomic stability of the L. rhamnosus used.24
In a related study, L. rhamnosus was compared with over a hundred collected strains, including probiotic strains, research strains with potential probiotic properties, food starter cultures and human isolates. Genomic analysis found that the strains could be allocated into one of 7 clusters. Cluster 1 was shown to comprise L. rhamnosus GG and 7 commercial probiotic strains from 5 different companies with indistinguishable pulsed-field gel electrophoresis profiles, suggesting that all these cultures might be commercial replicates of the LGG strain.25 PFGE is considered to offer the highest resolution for strain differentiation of lactic acid bacteria and in this study was applied to a selection of probiotic strains to verify whether members of a given cluster could be identified by fluorescent amplified fragment length polymorphism that represented different strains or that they could be considered clones of the same parent strain. The results of this work emphasizes the need for quality control and assurance strategies targeting the presence and maintenance of genes involved in host-microbe interactions underlying probiotic functionality.
Although genomic instability including mutations and deletions of several genes, has been demonstrated in the studies by Sybesma et al,20 it should also be noted that the overall genome similarity of the LGG variants studied was greater than 97%, including deleted regions. Even though a single point mutation may lead to loss of functionality, it can be concluded that many of the probiotic functionalities including tolerance to bile salts, immune stimulation, production of active metabolites and other functionalities are still present. In an ideal world, significantly altered probiotic strains should be tested in clinical trials to confirm their efficacy. However, as bacteria are forever changing it would be near impossible to ever bring a one hundred percent defined probiotic to market. Sybesma et al20 therefore suggests, as a minimum, that the validation of a probiotic strain should confirm the genetic stability of the overall genome so that claims can be made based on the strain having the same genetic makeup as the strains used in clinical studies on which their health claims have been based.
The Panel on Dietetic Products, Nutrition and Allergies of EFSA has proposed species identification by DNA-DNA hybridization or 16S rRNA gene sequence analysis and/or sequence analysis of other relevant genetic markers. Strain identification is necessary to make specific health claims7 and it has been proposed that pulsed-field gel electrophoresis of genomic DNA, randomly applied polymorphic DNA analysis or other internationally accepted genetic typing molecular methods should be used.26
Dosing of probiotics
The optimal dose range, frequency and vehicle of delivery for probiotics still remains contentious. The manufacturing of probiotics is yet to be standardized in regards to product stability and verification that could potentially impact reproducibility of study results. Further, the optimal dose of probiotics likely depends on the disease indication and also the ability of the probiotic(s) to survive gastric transit to the distal small intestine and large intestine.13 An individual's phenotype, which is governed by age, genetics and exposure to environmental factors such as the endogenous microbiota, will affect the outcomes of supplementation with a probiotic formulation.21 Resistance to gastric acid and bile salts varies between probiotics, the dosing schedule may influence survivability and it is favorable to dose probiotics at least 30 minutes before eating to minimize gastric acid and bile salt exposure.18
The frequency of probiotic supplementation is typically once to twice daily. While the literature continues to indicate that probiotic species colonize the gastrointestinal tract to exert their therapeutic activity, probiotics are characteristically transient and generally do not persist past 1 to 2 weeks after supplementation has ceased,27 although others have reported that probiotics can persist for longer periods in the gastrointestinal tract.28 Transient probiotics exert therapeutic activity via the release of signaling molecules that can prevent pathobiont (bacteria capable of pathogenic activity) overgrowth through the production of bacteriocins and secondary fermentation end products and by inhibiting pathogenic adherence by stimulating mucin production. Probiotics further reinforce tight junctions between intestinal epithelial cells, stimulate host secretion of sIgA and modulate host immune responses,29 such activity representing widespread effects among probiotic bacteria.
Dosing of probiotics varies considerably in clinical research ranging from capsules, tablets, powders and liquids to yoghurt preparations. Maintenance of colony forming unit (CFU) counts is assessed before undertaking the clinical study however it is rarely reported that CFU count is analyzed at the conclusion of the study to ascertain formulation viability and stability losses.
Selective interpretation of probiotic research data
Discriminatory methodological issues associated with commensal / probiotic bacteria research is the selective interpretation of results and proposed conclusions offered while almost deliberately ignoring contentious available evidence. We cite 2 important examples that provide compelling evidence for this in spite of the overwhelming contradictory clinical data that exists. These are namely research associated with i) D-lactic acid producing probiotic bacteria and ii) probiotic species acting as antioxidants, ameliorating oxidative stress.
D(−)-lactate producing bacteria
There are several recognized LAB genera related by certain morphological, metabolic and physiologic characteristics, for example the production of lactic acid as one of the main fermentation by-products of carbohydrate metabolism. Lactobacillus and Streptococcus genera are 2 common LAB that are utilized as probiotics. The bacterial species from the LAB genera can produce L(+)-Lactic acid, D(−)-Lactic acid, the racemate DL-Lactate, or a combination of these (Table 1).30-34
Table 1.
Family and Genera of LAB and the type of lactic acid they produce.34
| Family | Genera | Type of Lactic Acid |
|---|---|---|
| Lactobacillaceae | Lactobacillus | D, L, DL |
| Pediococcus | L, DL | |
| Streptococcaceae | Lactococcus | L |
| Streptococcus | L | |
| Enterococcaceae | Enterococcus | L |
| Leuconostocaecae | Leuconostoc | D |
| Oenococcus | D | |
| Weissella | D, DL | |
| Aerococcaceae | Aerococcus | L |
| Carnobacteriaceae | Carnobacterium | L |
Note. D = D-Lactic acid; L = L-Lactic acid; DL = DL-Lactic acid
Recently there has been a deliberate misperception directed at an excess of D(−)-Lactic acid production by certain probiotic bacteria as being responsible for the symptomology associated with chronic conditions such as Chronic Fatigue Syndrome (CFS); conclusions that are made upon the selective interpretation of research results while intentionally overlooking conclusive evidence that supports quite the opposite. Sheedy et al35 proposed that an elevated fecal count of the lactic acid producing bacteria Enterococcus sp. and Streptococcus sp in CFS patients when compared with controls, is a probable link to the neurocognitive and mitochondrial dysfunction experienced by this cohort due their D(−)-Lactic acid production in the IT. However, a significant and important oversight with this study was the absence of measurements on blood levels of D(−)-Lactic acid.
In humans, elevated D(−)-Lactic acidosis is a rare metabolic occurrence that has only been reported in those with short bowel syndrome (SBS)36 and as such can present with cognitive and neurologic impairments.37 Sheedy et al35 have not addressed this important issue in light of the extensive evidence that demonstrates the association between D(−)-Lactic acidosis and SBS, which results from the excessive fermentation of carbohydrates by Lactobacilli sp and the inability of the body to effectively clear the D(−)-Lactate.36 In the Sheedy et al35 study, the aim was to determine whether there was any significant relationship between the colonization of LAB in CFS patients. The authors profiled 3 genera, including Escherichia coli, however, they failed to profile Lactobacillus genera or explain this oversight considering that Lactobacilli from the family Lactobacillaceae are the predominant commensals that produce D- and DL-Lactic acid in the IT (Table 1).35 Furthermore, Streptococcus and Enterococcus genera are considered to be L(+)-Lactic acid producers, with Lactobacillus, Pediococcus, Leuconostoc, Oenococcus and Weisella genera considered D(−)-Lactic acid producers and therefore the choice of species for analysis is perplexing.34-38 Sheedy et al35 reported that there was a marked alteration of fecal microbial bacteria in a sub-group of CFS patients that suggested a probable link between D(−)-Lactic acid and CFS symptomology without measuring blood levels of D(−)-Lactic acid. Supporting such a conclusion is scientifically flawed especially when clinical studies conducted with children demonstrate that administering probiotic bacteria that produce D(−)-Lactic acid are safe and do not cause any long-term increases in blood D(−)-Lactic acid.30 Furthermore, D(−)-Lactic acid producing bacteria have been consumed by humans for centuries from fermented foods such as yoghurt, sauerkraut and pickles and more recently from probiotic supplementation with no associated negative symptomatology.
Recently we published a study39 that analyzed stool samples from osteoarthritis (OA) patients (n = 38) in which we reported very similar bacterial counts to that from the Sheedy et al study.35 Comparative counts compared from the 2 studies showed Streptococcus 1.44 × 107 vs. 9.8 × 107, Enterococcus 7.90 × 106 vs. 3.5 × 107 and Escherichia coli 4.27 × 107 vs. 4.26 × 107, respectively.39 It would thus seem that an OA cohort demonstrates a similar altered bacterial profile to a cohort of CFS patients, albeit without the symptomology of CFS. Therefore, providing a generalized recommendation for a short course of antibiotics for CFS patients is not only scientifically unsound and clinically unsubstantiated given the current extent of antibiotic resistance. The obtuse approach in failing to make reference to previous relevant research constitutes a methodological flaw that reflects a strong bias in the Sheedy et al study35 especially since clinical publications demonstrate that D-Lactic acid bacteria pose no threat to human health except in those with SBS.33,36,40 Furthermore, numerous clinical studies have reported therapeutic efficacy with the use of D(−), L(+) and DL-Lactic acid producing bacteria without producing D-Lactic acidosis (Table 2).41-53 The misinterpretation of D-Lactate producing bacteria has created an almost deliberate atmosphere of scientific nonsense regarding D(−)-Lactate producing probiotics. The net result has some industry suppliers of probiotic formulations hastening to deliver D-Lactate free probiotic formulations together with a misinformed marketing hype that misleads and confuses healthcare practitioners and the public, for nothing more than monetary gain.
Table 2.
A selection of reports regarding the administration of specific probiotic strains for specific health indications.
| Probiotic | Therapeutic effect | Type of Lactic Acid |
|---|---|---|
| Lactobacillus plantarum | ↓cholesterol | DL–lactic acid41,64 |
| ↓glucose | ||
| ↓homocysteine | ||
| ↓metabolic syndrome ↓inflammatory markers | ||
| Lactobacillus paracasei | ↓risk of antibiotic associated diarrhoea | L(+)–lactic acid42 |
| Lactobacillus casei | ↓risk of tender swollen joints | L(+)–lactic acid43 |
| ↓inflammatory cytokines in RA* | ||
| Lactobacillus delbruekii | ↓severity and incidence of pouchitis | D(−)–lactic acid44 |
| Lactobacillus acidophillus | Assists with invasive candidiasis infections in newborn | DL–lactic acid45-49 |
| ↓the incidence of severe necrotizing enterocolitis (NEC) and mortality in preterm infants and significantly reduced hospital stay | ||
| Lactobacillus bulgaricus | ↓severity and incidence of pouchitis | D(−)-lactic acid44 |
| Lactobacillus reuteri | ↓bone loss and osteoclasts | DL–lactic acid50-53 |
| ↓Streptococcus mutans | ||
| ↓incidence of severe NEC and mortality in preterm infants and significantly reduced hospital stay |
Note.
RA = rheumatoid arthritis.
Doses for the administrations ranged from 1010 to 1011 colony-forming units per dose.
Probiotics abrogating free radicals to ameliorate oxidative stress
The second example is researchers concluding that probiotics rescue oxidative stress. The IT mucosal surface is a complex and highly metabolically active tissue in an interactional environment. The IT is continuously exposed to a range of commensal and pathobiont microorganisms. The human-microbial IT boundary is an ecosystem that shares in a variety of important roles in human health and disease. Recently it has been posited that the oral administration of select probiotic bacterial strains, in particular those from the Lactobacillus genera, exert strong antioxidant activity reducing oxidative damage, free radical scavenging rate and also modulating host antioxidant enzyme systems.54,55 However, in contrast it has been reported that redox signaling by reactive oxygen species (ROS) is in response to microbial signals via formyl peptide receptors and the gut epithelial NADPH oxidase 1 (Nox1),56,57 particularly from the Lactobacillus genera,58 and not related to the simplistic view of ROS being purely signs of oxidative stress. Reports have demonstrated that some genera of human IT bacteria can induce a rapid increase of ROS, eliciting a physiologic response through the activation of epithelial Nox1.58,59 Commensal bacterial stimulation of host ROS signaling molecules at physiologic levels has potent regulatory effects on cellular proliferation and differentiation, immune function and intracellular signaling helping to maintain gut homeostasis.57,58 Therefore it is highly improbable that probiotics can be considered as antioxidant entities that can abrogate the over production of ROS, when they in fact stimulate regulated ROS production for the maintenance of local IT homeostasis.
Probiotic bacteria from the Lactobacilli and Bifidobacteria genera can temper a range of IT physiologic functions, including control over immune responses, epithelial barrier function and cellular proliferation.59 In addition, reports site in vitro experiments with epithelial cells that, when co–cultured with specific probiotic bacteria, show an increased and rapid oxidation reaction of soluble redox sinks, namely glutathione and thioredoxin that indicate the presence of a regulated process.59,60 Therefore any laboratory or other studies that propose to advance the notion that probiotic supplementation may reduce oxidative stress is significantly implausible and chimeric. The reactions elaborated by probiotic bacteria, with the induction of ROS, define potent regulatory effects on host physiologic functions that include immune function and intracellular signaling and as such contradict any association with ameliorating oxidative stress.
Several mechanisms of action of probiotics align their activities to explain the enhancement the IT epithelial barrier, and increased bacterial adhesion to the intestinal mucus layer with an attendant inhibition of pathogen adhesion and to the competitive exclusion of pathogenic microorganisms.56,60-62 Probiotic bacteria have also been reported to generate a range of anti–microbial substances and positively affect and modulate immune system function. Lee61 reported that the enteric commensal bacteria, by rapidly generating ROS, negotiate an acceptance by the IT epithelia. Different strains of commensal bacteria can elicit markedly different levels of ROS from contacted cells. Lactobacilli are especially potent inducers of ROS generation in cultured cells and in vivo, though all bacteria tested have some ability to alter the intracellular oxido–reductase environment.62 Soluble factors that are produced by different strains of Lactobacilli that are capable of mediating beneficial effects in in vivo inflammatory models have been reported.63 It has been shown that the IT microbiota can, through redox-dependent pathways, modulate the activity of both nuclear factor-κB (NF-κB) and β-catenin within the epithelium i.e., by oxidizing crucial cysteine residues in certain enzyme systems thus altering NF-κB signaling.57 These experimental data expand our understanding of the IT microbiome's activity. ROS–stimulating bacteria that possess effective specific membrane components and/or secreted factors that activate cellular ROS production are hence important factors that maintain homeostasis.
Such examples of mechanistic reports serve to improve our understanding of the action of probiotic bacteria and should guide the development of regulatory frameworks for probiotic formulations. Moreover, scientifically proven actions should guide marketing designed for probiotics rather than adopting scientific ideas that are highly questionable, purporting to sell probiotics with unjustifiable scientific claims for a competitive financial benefit.
Discussion
The advent of the human microbiome project has strengthened the idea that there exists a strong relationship between the IT microbial biomass and gastrointestinal and overall host health. The IT mucosa is also the largest and most active immunological organ of the body where IT bacteria teach the immune system the language of molecular biology, training it to develop from an immature to a mature functioning system. Such knowledge is often cited in support of daily administration of probiotics for health maintenance. Moreover, since most probiotic bacteria have been consumed for centuries as fermented milk or vegetable products to improve health, this further accentuates the value that the community places on their daily consumption. Currently probiotics are sold as foods or dietary supplements (e.g., in liquid or powdered forms or capsules) in a global market industry that is valued in the billions of dollars. Over the last 2 to 3 decades the variety of probiotic supplements and foods has expanded exponentially.
Although there is currently no probiotic that has been approved for a specific health claim, many have undergone clinical trials and could be marketed as biologics or other drugs. There is evidence of the potential benefit of some strains and species of probiotics for a variety of indications38 most recently that proposed for species of L. plantarum as candidates to reduce cholesterol levels.64 A summary of a selection of probiotic species that have been reported efficacious in several specific health indications is presented in Table 2. The typical doses in this select group of studies varied between 1010 and 1011 CFU per dose administered.
There is a significant difference in regard to which factors trigger the impetus for an investigational new drug from probiotics as opposed to pharmaceutical compounds. A product that has been designed, tested and approved to target and address a therapeutic need in an existing health market primarily drives pharmaceutical research. It would seem reasonable to assume that similar criteria should apply to probiotic formulations purported to target specific disease outcomes for example, modulating intestinal inflammation in inflammatory bowel diseases. The development of probiotic formulations should be driven by the appreciation of a biologically plausible mechanism of action, not on unsound and misleading research and conclusions, i.e., in the case of D(−)-Lactate-free producing bacterial formulations or probiotics that ameliorate oxidative stress. It is confusing and misleading to both medical practitioners and the public.
Probiotic preparations with specific metabolic properties (e.g., species that may up-regulate mucus secretion or maintain normal bowel function) may provide clues for the future direction of clinical research. Specific probiotic species that modify the microbiome, albeit transiently, with specific beneficial actions directed at preventing or treating specific conditions (e.g., antibiotic associated diarrhea or constipation) should be examined. As an exemplar, in a recent neonatal murine study65 it was reported that probiotic bacteria indeed have target sites in the intestine that significantly influence early development. The 2 Lactobacillus reuteri strains investigated (i.e., DSM 17938 and ATCC PTA 6475) showed differences in the amount of enterocyte migration, proliferation and crypt height effects. However, importantly both strains increased the phylogenetic diversity and evenness between the taxa of the fecal microbiome. In effect shifts in the intestinal microbiome promoted by probiotic bacteria may constitute the most crucially important factor that enhances intestinal health and maintains gut homeostasis. A further study66 provided significant insight into the molecular mechanisms as to how a specific probiotic namely Lactobacillus rhamnosus GG may stimulate intestinal cell proliferation and protect against intestinal injury. Ardita et al66 report that the probiotic effect was dependent on the bacterial-epithelial interaction mediated by the SpaC pilin subunit. The study66 reported that the pillin SpaC was an essential element for the probiotic strain to adhere to the intestinal mucosa. As such the probiotic triggered the pillin-induced epithelial generation of ROS that contributes to the capacity to stimulate extracellular signal-regulated kinase/mitogen-activated protein kinase signaling in enterocytes that maintains homeostasis or assists with the recovery of intestinal homeostasis following an external insult such as radiotherapy induced enterocyte injury. Such research will lead to the wider acceptance of live bacterial cultures as efficacious therapeutics. As is the case, currently the strongest clinical evidence for the administration of probiotics is as an adjunctive supportive therapy for IT-related and other end-organ disorders, especially in preventing or treating antibiotic associated diarrhea.
Conclusion
The intense public interest and acceptance of probiotics for general health benefits and for specific disorders continues to drive current research but unfortunately also leads to misleading industry marketing hype. This has created significant confusion in regards to the effects of different probiotic bacteria, the method by which they should be administered, their therapeutic dose and for which indications. For suppliers of probiotic supplements, formulations should be evidenced-based and not based on a current fad or slick marketing.
Careful selection of appropriate probiotic bacteria is essential for any probiotic product formulated for a specific condition, such as gastrointestinal mucosal immune response or gut barrier function. A consensus by the relevant research communities to define and validate research biomarkers is also warranted. There is an additional requisite to provide meaningful measurements of physiologic changes that probiotic formulations may induce and to report these findings without ambiguity.
The effect of the widespread use of safe and effective probiotic formulations on society-wide economic and quality-of-life indicators should be assessed with clearly defined end-points. The implementation and administration of probiotics to reduce the risk of disease is a promising intervention that is biologically plausible; it is however mechanistically complex and thus requires continued comprehensive targeted research.
Disclosure of potential conflicts of interest
Luis Vitetta has received National Institute of Complementary Medicine and National Health and Medical Research Council of Australia competitive funding and Industry support for research into probiotics and is Director of Medical Research at Medlab Clinical. Sean Hall is CEO with Medlab Clinical a facility that is conducting research with probiotic bacteria.
References
- [1].Rogers NJ, Mousa SA. The shortcomings of clinical trials assessing the efficacy of probiotics in irritable bowel syndrome. J Alt Comp Med (New York, NY) 2012; 18(2):112-9; http://dx.doi.org/ 10.1089/acm.2011.0015 [DOI] [PubMed] [Google Scholar]
- [2].Dhingra K. Methodological issues in randomized trials assessing probiotics for periodontal treatment. J Periodontal Res 2012; 47(1):15-26; PMID:21777405; http://dx.doi.org/ 10.1111/j.1600-0765.2011.01399.x [DOI] [PubMed] [Google Scholar]
- [3].AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evid Based Child Health 2014; 9(3):584-671; http://dx.doi.org/ 10.1002/ebch.1976 [DOI] [PubMed] [Google Scholar]
- [4].Hao Q, Dong BR, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev 2015; 2:Cd006895. [DOI] [PubMed] [Google Scholar]
- [5].Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, Guyatt GH, Johnston BC. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev 2013; 5:Cd006095. [DOI] [PubMed] [Google Scholar]
- [6].Fuller, R (Ed). Probiotics: The scientific basis. Berlin/Heidelberg: Springer Science+Business Media; 1992, pp. 377–85. [Google Scholar]
- [7].Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al.. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014; 11(8):506-14; PMID:24912386; http://dx.doi.org/ 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- [8].Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Trans Med 2014; 6(237):237ra65; http://dx.doi.org/ 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sanders ME. Probiotics: definition, sources, selection, and uses. Clin Infect Dis 2008; 46(Suppl 2):S58-61; discussion S144–51; PMID:18181724 [DOI] [PubMed] [Google Scholar]
- [10].Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol 2007; 189(4):1199-208; PMID:17085562; http://dx.doi.org/ 10.1128/JB.01351-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2017; 45(D1):D12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fu J, Qin QW. Pan-genomics analysis of 30 Escherichia coli genomes]. Yi Chuan 2012; 34(6):765-72; PMID:22698749; http://dx.doi.org/ 10.3724/SP.J.1005.2012.00765 [DOI] [PubMed] [Google Scholar]
- [13].Shanahan F. Probiotics: a perspective on problems and pitfalls. Scand J Gastroenterol Suppl 2003; 237:34-6 [DOI] [PubMed] [Google Scholar]
- [14].Boyle RJ, Robins-Browne RM, Tang ML. Probiotic use in clinical practice: what are the risks? Am J Clin Nutr 2006; 83(6):1256-64; PMID:16762934 [DOI] [PubMed] [Google Scholar]
- [15].Chapman CM, Gibson GR, Rowland I. Health benefits of probiotics: are mixtures more effective than single strains? Eur J Nutr 2011; 50(1):1-17; PMID:21229254; http://dx.doi.org/ 10.1007/s00394-010-0166-z [DOI] [PubMed] [Google Scholar]
- [16].Marteau P, Shanahan F. Basic aspects and pharmacology of probiotics: an overview of pharmacokinetics, mechanisms of action and side-effects. Best Pract Res Clin Gastroenterol 2003; 17(5):725-40; PMID:14507584; http://dx.doi.org/ 10.1016/S1521-6918(03)00055-6 [DOI] [PubMed] [Google Scholar]
- [17].Hill C, Sanders ME. Rethinking “probiotics”. Gut Microbes 2013; 4(4):269-70; PMID:23778363; http://dx.doi.org/ 10.4161/gmic.25143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Timmerman HM, Koning CJ, Mulder L, Rombouts FM, Beynen AC. Monostrain, multistrain and multispecies probiotics–A comparison of functionality and efficacy. Int J Food Microbiol 2004; 96(3):219-33; PMID:15454313; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2004.05.012 [DOI] [PubMed] [Google Scholar]
- [19].Kekkonen RA, Lummela N, Karjalainen H, Latvala S, Tynkkynen S, Jarvenpaa S, Kautiainen H, Julkunen I, Vapaatalo H, Korpela R. Probiotic intervention has strain-specific anti-inflammatory effects in healthy adults. World J Gastroenterol 2008; 14(13):2029-36; PMID:18395902; http://dx.doi.org/ 10.3748/wjg.14.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sybesma W, Molenaar D, van IJcken W, Venema K, Kort R. Genome instability in Lactobacillus rhamnosus GG. App Environ Microbiol 2013; 79(7):2233-9; http://dx.doi.org/ 10.1128/AEM.03566-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Segers ME, Lebeer S. Towards a better understanding of Lactobacillus rhamnosus GG–host interactions. Microb Cell Fact 2014; 13(Suppl 1):S7; PMID:25186587; http://dx.doi.org/ 10.1186/1475-2859-13-S1-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, et al.. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 2012; 78(1):185-93; PMID:22020518; http://dx.doi.org/ 10.1128/AEM.06192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tripathi P, Beaussart A, Alsteens D, Dupres V, Claes I, von Ossowski I, de Vos WM, Palva A, Lebeer S, Vanderleyden J, et al.. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 2013; 7(4):3685-97; PMID:23531039; http://dx.doi.org/ 10.1021/nn400705u [DOI] [PubMed] [Google Scholar]
- [24].Vancanneyt M, Huys G, Lefebvre K, Vankerckhoven V, Goossens H, Swings J. Intraspecific genotypic characterization of Lactobacillus rhamnosus strains intended for probiotic use and isolates of human origin. Appl Environ Microbiol 2006; 72(8):5376-83; PMID:16885289; http://dx.doi.org/ 10.1128/AEM.00091-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Douillard FP, Ribbera A, Jarvinen HM, Kant R, Pietilä TE, Randazzo C, Paulin L, Laine PK, Caggia C, von Ossowski I, et al.. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiology 2013; 79(6):1923-33; http://dx.doi.org/ 10.1128/AEM.03467-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). General guidance for stakeholders on the evaluation of Article 13.1, 13.5 and 14 health claims. EFSA 2011; 9:2135; http://dx.doi.org/ 10.2903/j.efsa.2011.2135 [DOI] [Google Scholar]
- [27].Ouwehand AC SS. In vitro adhesion assays for probiotics and their in vivo relevance: a review. Microb Ecol Health Dis 2003; 15:175-84; http://dx.doi.org/ 10.1080/08910600310019886 [DOI] [Google Scholar]
- [28].Alander M, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett Appl Microbiol 1997; 24(5):361-4; PMID:9172443; http://dx.doi.org/ 10.1046/j.1472-765X.1997.00140.x [DOI] [PubMed] [Google Scholar]
- [29].Sanders ME. Impact of probiotics on colonizing microbiota of the gut. J Clin Gastroenterol 2011; 45(Suppl):S115-9; PMID:21992949; http://dx.doi.org/ 10.1097/MCG.0b013e318227414a [DOI] [PubMed] [Google Scholar]
- [30].Papagaroufalis K, Fotiou A, Egli D, Tran LA, Steenhout P. A randomized double blind controlled safety trial evaluating D-lactic acid production in healthy infants fed a lactobacillus reuteri-containing formula. Nutr Metab Insights 2014; 7:19-27; PMID:24812520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Haschke-Becher E, Brunser O, Cruchet S, Gotteland M, Haschke F, Bachmann C. Urinary D-lactate excretion in infants receiving Lactobacillus johnsonii with formula. Ann Nutr Metab 2008; 53(3–4):240-4; PMID:19088469; http://dx.doi.org/ 10.1159/000185642 [DOI] [PubMed] [Google Scholar]
- [32].Cekola PL, Czerkies LA, Storm HM, Wang MH, Roberts J, Saavedra JM. Growth and Tolerance of Term Infants Fed Formula With Probiotic Lactobacillus reuteri. Clin Pediatr (Phila) 2015; 54(12):1175-84; PMID:25758426; http://dx.doi.org/ 10.1177/0009922815574076 [DOI] [PubMed] [Google Scholar]
- [33].Connolly E, Abrahamsson T, Bjorksten B. Safety of D(−)-lactic acid producing bacteria in the human infant. J Pediatr Gastroenterol Nutr 2005; 41(4):489-92; PMID:16205524; http://dx.doi.org/ 10.1097/01.mpg.0000176179.81638.45 [DOI] [PubMed] [Google Scholar]
- [34].von Wright A AL. Lactic acid bacteria: microbiological and functional aspects. 4th ed. Lahtinen S OA, Salminen S, von Wright A, editor. Florida, U.S.A.: Taylor and Francis; 2012. 2012 pp.761 p [Google Scholar]
- [35].Sheedy JR, Wettenhall RE, Scanlon D, Gooley PR, Lewis DP, McGregor N, Stapleton DI, Butt HL, DE Meirleir KL. Increased D-lactic acid intestinal bacteria in patients with chronic fatigue syndrome. In vivo 2009; 23(4):621-8; PMID:19567398 [PubMed] [Google Scholar]
- [36].Ewaschuk JB, Naylor JM, Zello GA. D-lactate in human and ruminant metabolism. J Nutr 2005; 135(7):1619-25; PMID:15987839 [DOI] [PubMed] [Google Scholar]
- [37].Uribarri J, Oh MS, Carroll HJ. D-lactic acidosis. A review of clinical presentation, biochemical features, and pathophysiologic mechanisms. Medicine (Baltimore) 1998; 77(2):73-82; PMID:9556700; http://dx.doi.org/ 10.1097/00005792-199803000-00001 [DOI] [PubMed] [Google Scholar]
- [38].Carr FJ, Chill D, Maida N. The lactic acid bacteria: a literature survey. Crit Rev Microbiol 2002; 28(4):281-370; PMID:12546196; http://dx.doi.org/ 10.1080/1040-840291046759 [DOI] [PubMed] [Google Scholar]
- [39].Coulson S, Butt H, Vecchio P, Gramotnev H, Vitetta L. Green-lipped mussel extract (Perna canaliculus) and glucosamine sulphate in patients with knee osteoarthritis: therapeutic efficacy and effeects on gastrointestinal microbiota profiles. Inflammopharmacology 2013; 21:79-90; PMID:22821424; http://dx.doi.org/ 10.1007/s10787-012-0146-4 [DOI] [PubMed] [Google Scholar]
- [40].de Vrese M, Barth CA. Postprandial plasma D-lactate concentrations after yogurt ingestion. Z Ernahrungswiss 1991; 30(2):131-7; PMID:1897274; http://dx.doi.org/ 10.1007/BF01610068 [DOI] [PubMed] [Google Scholar]
- [41].Barreto FM, Colado Simao AN, Morimoto HK, Batisti Lozovoy MA, Dichi I, Helena da Silva Miglioranza L. Beneficial effects of Lactobacillus plantarum on glycemia and homocysteine levels in postmenopausal women with metabolic syndrome. Nutrition 2013; 30:939-942; PMID:24613434; http://dx.doi.org/ 10.1016/j.nut.2013.12.004 [DOI] [PubMed] [Google Scholar]
- [42].Lenoir-Wijnkoop I, Nuijten MJ, Craig J, Craig J, Butler CC. Nutrition economic evaluation of a probiotic in the prevention of antibiotic associated diarrhea. Front Pharmacol 2014; 5(13):1-8; PMID:24596556; http://dx.doi.org/ 10.3389/fphar.2014.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Alipour B, Homayouni-Rad A, Vaghef-Mehrabany E, Sharif SK, Vaghef-Mehrabany L, Asghari-Jafarabadi M, Nakhjavani MR, Mohtadi-Nia J. Effects of Lactobacillus casei supplementation on disease activity and inflammatory cytokines in rheumatoid arthritis patients: A randomized double-blind clinical trial. Int J Rheum Dis 2014; 17:519-527; PMID:24673738 [DOI] [PubMed] [Google Scholar]
- [44].Tomasz B, Zoran S, Jroslaw W, Ryszard M, Marcin G, Robert B, Piotr K, Lukasz K, Jacek P, Piotr G, et al.. Long-term use of probiotics Lactobacillus and Bifidobacterium has a prophylactic effect on the occurrence and severity of pouchitis: A randomized prospective study. Biomed Res Int 2014; 2014:208064; PMID:24579075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Betta P. Not all probiotics are the same: Gut microbiota modulation with a multistrain probiotics. N Am J Med Sci 2014; 6:58-59; PMID:24678480; http://dx.doi.org/ 10.4103/1947-2714.125871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sepp E, Lõivukene K, Julge K, Voor T, Mikelsaar M. The association of gut microbiota with body weight and body mass index in preschool children of Estonia. Microb Ecol Health Dis 2013; 24:1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lin HC, Su BH, Chen AC, Lin TW, Tsai CH, Yeh TF, Oh W. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics 2005; 115(1):1-4; PMID:15629973 [DOI] [PubMed] [Google Scholar]
- [48].Lin HC, Hsu CH, Chen HL, Chung MY, Hsu JF, Lien RI, Tsao LY, Chen CH, Su BH. Oral probiotics prevent necrotizing enterocolitits in very low birth weight preterm infants: a multicenter, randomized controllled trial. Pediatrics 2008; 122(4):693-700; PMID:18829790; http://dx.doi.org/ 10.1542/peds.2007-3007 [DOI] [PubMed] [Google Scholar]
- [49].Samanta M, Sarkar M, Ghosh P, Jk Ghosh, Mk Sinha, Chatterjee S. Prophylatic probioitcs for prevention of necrotizinfg enterocolitis in very low birth weight newborns. J Trop Pediatr 2009; 55(2):128-31; PMID:18842610; http://dx.doi.org/ 10.1093/tropej/fmn091 [DOI] [PubMed] [Google Scholar]
- [50].Britton RA, Irwin R, Quach D, Schaefer L, Zhang J, Lee T, Parameswaran N, McCabe LR, Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J Cell Physiol 2014; 229:1822-30; PMID:24677054; http://dx.doi.org/ 10.1002/jcp.24636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Laleman I, Detailleur V, Slot DE, Slomka V, Quirynen M, Teughels W. Probiotics reduce mutans streptococci counts in humans: A systematic review and meta-analysis. Clin Oral Investig 2014; 18:1539-52; PMID:24663813; http://dx.doi.org/ 10.1007/s00784-014-1228-z [DOI] [PubMed] [Google Scholar]
- [52].Rojas MA, Lozano JM, Rojas MX, Rodriguez VA, Rondon MA, Bastidas JA, Perez LA, Rojas C, Ovalle O, Garcia-Harker JE, Tamayo ME, et al.. Prophylactic probiotics to prevent death and nosocomial infection in preterm infants. Pediatrics 2012; 130(5):e1113-20; PMID:23071204; http://dx.doi.org/ 10.1542/peds.2011-3584 [DOI] [PubMed] [Google Scholar]
- [53].Romeo MG, Romeo DM, Trovato L, Oliveri S, Palermo F, Cota F, Betta P. Role of probiotics in the prevention of the enteric colonization by Candida in preterm newborns: incidence of late-onset sepsis and neurological outcome. J Perinatol 2011; 31(1):63-9; PMID:20410904; http://dx.doi.org/ 10.1038/jp.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Martarelli D, Verdenelli MC, Scuri S, Cocchioni M, Silvi S, Cecchini C, Pompei P. Effect of a probioitc intake on oxidant and antioxidant paramters in plasma of athletes during intense exercise training. Curr Microbiol 2011; 62(6):1689-96; PMID:21400082; http://dx.doi.org/ 10.1007/s00284-011-9915-3 [DOI] [PubMed] [Google Scholar]
- [55].Mishra V, Shah C, Mokashe N, Chavan R, Yadav H, Prajapati J. Probiotics as potential antioxidants: a systematic review. J Agric Food Chem 2015; 63(14):3615-26; PMID:25808285; http://dx.doi.org/ 10.1021/jf506326t [DOI] [PubMed] [Google Scholar]
- [56].Vitetta L, Briskey D, Hayes E, Shing C, Peake J. A review of the pharmacobiotic regulation of gastrointestinal inflammation by probiotics, commensal bacteria and prebiotics. Inflammopharmacology 2012; 20(5):251-66; PMID:22427210; http://dx.doi.org/ 10.1007/s10787-012-0126-8 [DOI] [PubMed] [Google Scholar]
- [57].Holstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol 2014; 15(6):411-21; PMID:24854789; http://dx.doi.org/ 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- [58].Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al.. Symbiotic lactobacillu stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J 2013; 32(23):3017-28; PMID:24141879; http://dx.doi.org/ 10.1038/emboj.2013.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Neish AS. Redox signaling mediated by the gut microbiota. Free Radic Res 2013; 47(11):950-7; PMID:23937589; http://dx.doi.org/ 10.3109/10715762.2013.833331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bermudez-Brito M, Plaza-Diaz J, Munoz-Quezada S, Gómez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab 2012; 61(2):160-74; PMID:23037511; http://dx.doi.org/ 10.1159/000342079 [DOI] [PubMed] [Google Scholar]
- [61].Lee WJ. Bacterial-modulated signaling pathways in gut homeostasis. Sci Signal 2008; 1(21):pe24; PMID:18506033; http://dx.doi.org/ 10.1126/stke.121pe24 [DOI] [PubMed] [Google Scholar]
- [62].Lin PW, Myers LE, Ray L, Song SC, Nasr TR, Berardinelli AJ, Kundu K, Murthy N, Hansen JM, Neish AS. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic Biol Med 2009; 47(8):1205-11; PMID:19660542; http://dx.doi.org/ 10.1016/j.freeradbiomed.2009.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007; 132(2):562-75; PMID:17258729; http://dx.doi.org/ 10.1053/j.gastro.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bosch M, Fuentes MC, Audivert S, Bonachera MA, Peiró S, Cuñé J. Lactobacillus plantarum CECT 7527, 7528 and 7529: probiotic candidates to reduce cholesterol levels. J Sci Food Agric 2014; 94(4):803-9; PMID:24186773; http://dx.doi.org/ 10.1002/jsfa.6467 [DOI] [PubMed] [Google Scholar]
- [65].Preidis GA, Saulnier DM, Blutt SE, Mistretta TA, Riehle KP, Major AM, Venable SF, Finegold MJ, Petrosino JF, Conner ME, et al.. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine’. FASEB J 2012; 26(5):1960-9; PMID:22267340; http://dx.doi.org/ 10.1096/fj.10-177980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ardita CS, Mercante JW, Kwon YM, Luo L, Crawford ME, Powell DN, Jones RM, Neish AS. Epithelial adhesion mediated by pilin SpaC is required for Lactobacillus rhamnosus GG-induced cellular responses. Appl Environ Microbiol 2014; 80(16):5068-77; PMID:24928883; http://dx.doi.org/ 10.1128/AEM.01039-14 [DOI] [PMC free article] [PubMed] [Google Scholar]


