ABSTRACT
The intestinal tract provides ideal niches for several different microbial species, which are collectively called the gut microbiota. A key host immune effector that controls the microbiota and prevents mucosal infection is IgA. Gut microbiota-derived factors are largely classified into molecular pattern recognition receptor ligands and nutrient-derived metabolites including short-chain fatty acids and adenosine triphosphate. Along with host-derived factors such as retinoic acid, various cytokines and cytokine-like molecules, gut microbial products profoundly shape B cell responses. Gut microbial products can directly regulate B cell activation and differentiation. They can also indirectly affect B cells through epithelial cells, T cells, and myeloid cell subsets. We highlight the various direct and indirect mechanisms by which microbial products regulate humoral immunity.
KEYWORDS: Gut microbiota, humoral immunity, short-chain fatty acids, IgA, B cells
Introduction
The gastrointestinal tract, rich in dietary materials, provides ideal niches for a myriad of microbes, which are called commensal microbiota. Roughly, a thousand of microbial operational taxonomic units (OTUs) dominated by the bacteria kingdom are found in various niches of the gut, defined by complex gradients of chemical, nutritional, and immunological factors. The gut microbiota provide important health benefits to the host such as enhanced energy harvest from diet and homeostatic activation of the immune system. Importantly, the members of the gut microbiota and their relative frequencies are finely controlled by diet and the host immune system to ensure mutually beneficial host-microbial symbiosis. This equilibrium can be altered in pathological conditions caused by adverse metabolic, dietary and immunological changes in hosts, leading to dysbiosis conditions. A key component of the host immune system important in regulating the microbiota is the mucosal antibody IgA. The intestine has many tissue and immune cell types in various compartments to regulate the production and/or secretion of antibodies into the intestinal lumen. Secreted host enzymes process dietary materials in the upper elementary tract, and the bulk of nutrients are absorbed by epithelial cells in the small intestine. Some of these metabolites have immune-regulatory functions. For example, the roles of versatile mucosal immune regulators, vitamin A metabolites (i.e. retinoic acids or RA), are well established.1,2 In the colon, the gut microbiota produce a myriad of cellular constituents and metabolites. Some of these microbial products promote the population and maturation of immune cells in gut-associated lymphoid tissues and effector sites such as intestinal lamina propria and intraepithelial compartments.
Gut microbial products include the ligands that activate pathogen-associated molecular pattern receptors (PAMPRs) and nutrient-derived microbial metabolites. Each day, dietary fiber (20–50 g on average), protein (5–10 g), fat, and other dietary constituents (vitamins, catechins, tannins, lignin, polyphenols and other micronutrients) pass into the colon for microbial fermentation. As a consequence, various metabolites ranging from short-chain fatty acids (SCFAs) to secondary bile acids and vitamin B12 are produced. These metabolites are used by the host and gut microbes to maintain nutritional, physiologic and immunological homeostasis. One of the immunological targets of the microbial factors is the humoral immunity. We highlight here the recent progress in our understanding of roles of SCFAs and other microbial products in regulating humoral immunity.
IgA and commensal bacteria regulate each other
IgA is secreted into the human gut lumen at a rate of 40–60 mg/kg/day of weight,3 which is translated into several grams of IgA protein per day. Secreted IgA binds commensal and pathogenic microbes and their products including toxins. Generally, 20–50% of gut bacteria are coated with IgA. IgA-coated commensal bacteria appear to be more inflammatory than IgA-noncoated bacteria in inducing colitis.4 IgA coating of bacteria can be mediated by either antigen binding domains or glycan moieties of IgA. In general, pathogens and pathogenic commensal bacteria induce the production of IgA with high affinity antigen-binding domains. Defective IgA production or secretion due to loss of function mutations in the genes encoding activation induced cytidine deaminase (AID), IgA or polymeric immunoglobulin receptor (pIgR) leads to dysbiosis, bacterial invasion, and inflammatory diseases.5,6 For example, Aicda mutant mice produce IgA with decreased affinity for gut bacteria due to defective somatic hypermutation and have an altered microbial community such as abnormal overgrowth of segmented filamentous bacteria (SFB) in the upper small intestine.5 IgA coating induces immune exclusion of gut microbes via agglutination, microbial entrapment in mucus, and peristalsis-mediated clearance. IgA coating interferes with the attachment of pathogenic microbes and their products to the intestinal epithelial surface, thus decreasing their pathogenicity, as evidenced by IgA binding-mediated inhibition of shigella type 3-secretion (T3S) system.7 IgA also facilitates the engulfment of pathogens by Peyer's patch M cells and phagocytes, such as neutrophils, dendritic cells and macrophages, to mount effective local immune responses to pathogens.8-10
Gut microbiota induce the development of gut-associated lymphoid tissues (GALT), such as isolated lymphoid follicle (ILF) and Peyer's patches (PPs), which are major inductive sites for IgA-producing plasma B cells.11 Germ-free (GF) animals have reduced numbers of IgA-producing plasma cells, and the gut microbiota are required for normal levels of class switch recombination (CSR) from IgM to IgA. B cells, particularly marginal zone (MZ) B cells, are negatively affected in certain restricted flora (RF) mice which have altered commensal microbiota.12 The impaired plasma B cell responses in GF or RF animals were restored by conventionalization of the mice. Also, somatic hypermutation and IgA repertoire diversification were greatly suppressed in GF mice.13 It has been shown that certain microbiota, especially Escherichia coli and Bifidobacteria, promote B cell maturation.14 In gnotobiotic mice, SFB colonization increased the numbers of IgA-producing cells.15 Our current understanding of the mechanisms by which the gut microbiota regulate IgA production is discussed below.
General effects of gut microbiota on B cells
The gut microbiota influence the activation and differentiation of B cells via direct and indirect mechanisms. B cell activation and differentiation are triggered by activation of B cell receptor (BCR), CD40, Toll-like receptors (TLRs), cytokine receptors (e.g. IL-21R), and/or receptors for B-cell activating factor (BAFF) and A Proliferation Inducing Ligand (APRIL). T-dependent B cell antibody production is triggered by activation of BCR, CD40 and IL-21R, which can be further potentiated by BAFF and APRIL signaling. T-independent B cell antibody production is largely triggered by activation of TLRs and BAFF/APRIL receptors. The gut microbiota produce protein and carbohydrate antigens that activate BCR. Also, many microbial products activate TLRs and NOD-like receptors (NLRs). B cells express TLR1, 2, 4, 6, 7, and 9, and activation of these TLRs increase B cell survival, antigen presentation, and antibody production.16 In steady-states, the gut microbiota produce TLR ligands that activate B cells. For example, low levels of lipopolysaccharides from gram-negative bacteria activate B cells to induce CSR of the immunoglobulin (Ig) heavy chain gene to produce IgA.17 Interestingly, the gut microbiota also increase the number of regulatory B cells (Bregs), which have anti-inflammatory functions. Gut microbiota colonization induces the production of IL-1β and IL-6 by dendritic cells (DCs) and tissue cells to promote naïve B cell differentiation into Bregs in mesenteric lymph node (MLN).18 Bregs produce IL-10 to suppress inflammatory T cells and tissue inflammation. Thus, the microbiota can exert various effects on B cells beyond antibody production.
Impact of gut microbiota on B cell metabolism
Mammals use a significant portion of available energy and metabolic building blocks to maintain immune cells. In this regard, production of antibodies by B cells is a metabolically demanding process. Lymphocytes metabolize nutrients for energy (e.g., Adenosine Triphosphate, ATP) production, biogenesis of cellular building blocks, and production of effector molecules such as antibodies and cytokines. Glucose, along with fatty acids and amino acids, provides the bulk of required energy and building blocks for B cells. Upon activation of BCR or TLR4, B cells undergo both glycolysis and mitochondrial oxidative phosphorylation via a c-Myc dependent pathway.19 In contrast, activated T cells mainly utilize glycolysis to become effector T cells.20 Both glycolysis and oxidative phosphorylation are required for B cell activation and plasma cell differentiation. Increased cell metabolism generates higher levels of cellular ATP, some of which is secreted out of cells. Necrotic cells also leak ATP into tissue environments. Moreover, the gut microbiota themselves produce ATP. Extracellular ATP, produced by host and microbes, activates purinergic P2X and P2Y of colonic DCs to produce IL-6 and TGF-β,21 which can promote CSR for IgA production. Secreted extracellular ATP is hydrolyzed eventually to adenosine by host and microbial ectonucleotidases. The resulting product, adenosine, can activate adenosine receptors on B cells to promote Ig gene CSR to produce IgG and IgA (Fig. 1).22
Figure 1.
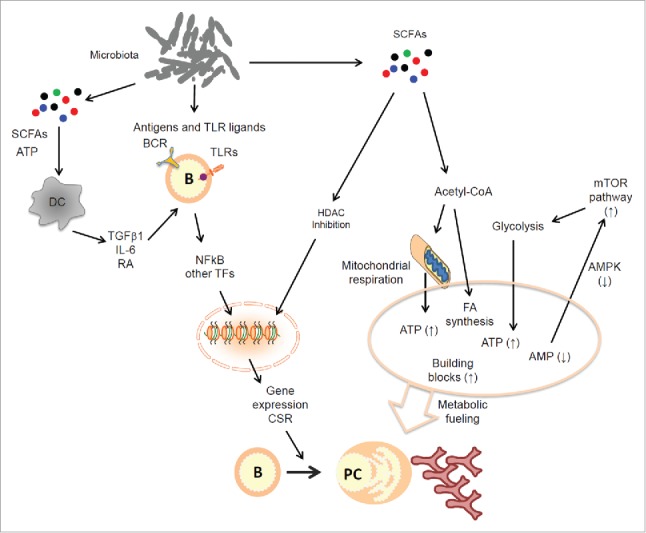
Regulation of B cells by microbial products. The gut microbiota generate a wide variety of products that can activate BCR and TLRs. This activation triggers B cell differentiation into plasma B cells. Moreover, gut microbial metabolites such as SCFAs play important roles by serving as nutrients for B cells undergoing activation. SCFAs inhibit HDACs to facilitate gene expression, and this supports plasma B cell differentiation. At the same time, SCFAs are converted into acetyl-CoA which fuels mitochondrial oxidative phosphorylation and fatty acid synthesis. Moreover, The SCFA effect on B cells increases ATP levels but decreases that of AMP (the AMPK ligand), leading to sustained mTOR activation. This boosts glycolysis in B cells for further providing essential energy and building blocks. These events promote plasma B cell generation and increased production of antibodies particularly IgA, which plays a central role in maintaining the host-microbial symbiosis. Abbreviations: ATP, adenosine triphosphate; AMP, adenosine monophosphate; AMPK, AMP-activated protein kinase; BCR, B cell receptor; CSR, class switch recombination; HDAC, histone deacetylase; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; PC, plasma cells; SCFA, short-chain fatty acid; TF, transcription factor; TLR, toll-like receptor.
Regulation of antibody production by SCFAs
The gut microbiota process dietary materials and intestine-derived host products such as mucins to produce diverse metabolites. SCFAs, produced from dietary fiber (DF), are the most abundant microbial metabolites in the gut. Recently, we reported that SCFAs support plasma B cell differentiation.23 SCFA deficiency occurs when DF levels in diet are low or when the microbiota are suppressed by antibiotics or altered due to dysbiosis. In GF mice, the SCFA levels in the colon are decreased to about one percent of normal levels.24 In low SCFA conditions due to low DF consumption, serum antibody and mucosal IgA levels are significantly decreased.23,25 Germinal center responses in PPs are suppressed in low SCFA conditions but SCFA administration restored antibody production in mice fed a DF-free diet (Fig. 1). SCFAs increase cellular levels of acetyl-CoA and energy (e.g., ATP) and lipid biogenesis to drive plasma B cell differentiation.23 SCFAs are efficient histone deacetylase (HDAC) inhibitors, and this function boosts the expression of genes such as Xbp-1, Aicda and Prdm1, involved in plasma B cell differentiation. Activation of the mTOR pathway increases glycolysis in B cells. SCFAs increase the level of ATP but decrease that of adenosine monophosphate (AMP), which is the agonist of 5′ AMP-activated protein kinase (AMPK). Thus, SCFAs inhibit AMPK, and this leads to activation of the mTOR pathway. Lipid biogenesis is important for plasma B cell differentiation. In this regard, dietary palmitic acid enhances intestinal IgA responses.26 Additionally, the roles of Retinaldehyde Dehydrogenase 2 (RALDH2)-expressing mucosal DCs in inducing IgA responses have been suggested.25,27 Interestingly, it is not just intestinal IgA but also systemic IgG responses that are supported by SCFAs.23 We will discuss the roles of non-B cells in mediating the regulatory effects of the gut microbiota on B cells responses in the next section.
Indirect mechanisms of B cell regulation by microbiota: Role of epithelial cells
Intestinal epithelial cells provide not only a physical barrier between the gut microbiota and host tissues but actively detect invading microbes and initiate immune responses. The intestinal epithelial cells use various PAMPRs to monitor the luminal environment for the presence of pathogenic versus non-pathogenic microbial signals. When triggered by microbial PAMPR ligands, epithelial cells produce acute inflammatory cytokines and chemokines to recruit and activate immune cells. To regulate B cells, epithelial cells produce B cell-activating or attracting molecules such as IL-6, BAFF, APRIL, CCL20, CCL28, and thymic stromal lymphopoietin (TSLP) (Fig. 2). We found that SCFAs activate gut epithelial cells through SCFA receptors, GPR41 and GPR43, and this activation makes epithelial cells highly efficient in producing inflammatory mediators upon TLR4 activation.28 TSLP, produced by gut epithelial cells upon TLR activation, activates a specialized DC subset that produces TNF-α and iNOS. These DC products induce the expression of APRIL and IL-10, which in turn induce CSR in B cells to produce IgA. Moreover, the development of lymphoid tissue inducer (LTi) cells, a subset of group 3 innate lymphoid cells (ILC3), requires the gut microbiota. LTi cells express LTα1β2 to activate stromal cells to produce TGFβ, which acts on B cells to promote IgA production.29 Thus, gut microbial factors activate epithelial cells to produce B cell-activating factors in steady-states.
Figure 2.
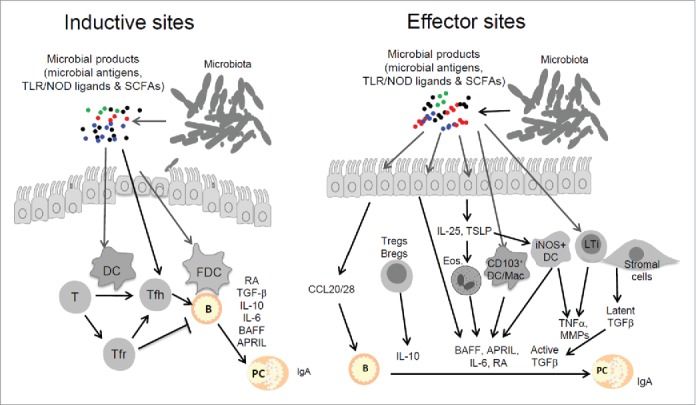
Indirect regulation of B cells by gut microbial products through various non-B cell types. In inductive sites such as PPs, epithelial cells actively sample antigens and microbial products for homeostatic activation of immune cells such as T cells, macrophages and DCs. Microbial products stimulate DCs and FDCs to produce immune regulatory molecules that activate T cells and B cells. Key molecules that are produced include TGFβ1, IL-10, IL-6, and IL-21, which collectively induce Tfh cell generation and facilitate B cell activation and differentiation into IgA-producing B cells. Retinoic acid (RA), produced by mucosal epithelial cells and DCs, skews B cell differentiation into IgA-producing cells. In effector sites such as intestinal lamina propria (LP), microbial factors activate lymphocytes, eosinophils, macrophages, DCs, ILCs, and tissue stromal cells. Host cells that are activated by microbial factors produce chemokines, cytokines and other molecules such as CCL20, CCL28, IL-10, BAFF, APRIL, IL-6, TGFβ1, and TNFα to promote IgA production by B cells. The B cell-activating molecules and cell types that are activated by microbial factors affect most B cell types including B2, MZ and B1 cells. Abbreviations: APRIL, A Proliferation Inducing Ligand; BAFF, B-cell Activating Factor; Breg, Regulatory B cells; DC, dendritic cell; Eos, eosinophils; FDC, follicular DC; GALT, gut-associated lymphoid tissue; ILCs, innate lymphoid cells; iNOS, Inducible Nitric Oxide Synthase; LP, lamina propria; LTi, lymphoid tissue inducer; Mac, macrophage; MMPs, matrix metalloproteinases; RA, retinoic acid; TLR/NOD, toll-like receptor/NOD-like receptor; Tfh, T follicular helper cells; Tfr, T follicular regulatory cells; Treg, T regulatory cells; TSLP, Thymic Stromal Lymphopoietin.
Indirect mechanisms of B cell regulation by microbiota: Role of T cells
Depending on functional specialization, T cells either positively or negatively regulate B cell differentiation into plasma B cells. While at lower than the TLR expression levels of phagocytes and DCs, T cell subsets, such as Tregs, express TLRs and sense microbial products.30 For example, TLRs (TLR2, 4, 5, 7, and 8) are expressed by CD4+CD25+ T regulatory cells (Tregs), and TLR2 ligands can expand the number of CD4+CD25+ Tregs.31 The effects of TLR activation on Tregs may either positively or negatively affect B cell responses. T follicular helper (Tfh) cells positively regulate plasma B cell differentiation in lymphoid follicles. Tfh cells activate B cells with CD40L and inducible co-stimulator (ICOS) along with various cytokines such as IL-21, IL-4, IL-10, and IFNγ. The gut microbiota promote Tfh cell development in the intestine. Tfh cell development is deficient in GF mice and can be restored by administration of TLR2 ligands in a MyD88 signaling-dependent manner.32 T-cell specific deletion of MyD88 decreased high-affinity IgA binding of the microbiota, resulting in microbial dysbiosis.32 Some FoxP3+ T cells can differentiate into Tfh cells in PPs to promote, rather than suppress, antibody production,33 which can shift the local immunological balance from immune tolerance toward activation during infection. SCFAs increase the Treg pool in the MLN by HDAC inhibition-mediated FoxP3 expression.34 In line with this, SCFAs increase Tfh cell differentiation in vitro via metabolic regulation.23 Thus, microbial products support B cell-regulating T cells.
Indirect mechanisms of B cell regulation by microbiota: Roles of myeloid cell populations
DCs produce cytokines and present antigens to T cells, and this function is also required to generate Tfh and T follicular regulatory (Tfr) cells. DCs express several TLRs and are activated by TLR ligands. DCs sense microbial products not only within tissues but also in the gut lumen using their membrane extensions across the epithelial barrier. DCs also directly activate B cells with cytokines and cell-surface ligands (e.g., BAFF and APRIL). Therefore, the effect of microbial products on DCs can indirectly regulate B cell activation and differentiation. For example, MyD88 is required for DCs to enhance antibody responses by enhancing the production of cytokines (IL-6, IL-10 and TGFβ1) and other B cell-activating molecules.35,36 TLR activation also enhances follicular dendritic cells (FDCs) to activate B cells by secreting BAFF, APRIL, and TGF-β137 Moreover, SCFAs up-regulate RALDH2 in DCs to increase RA production. Thus, it is possible that RA produced by SCFA-activated DCs can promote IgA-producing plasma B cells. RA is produced mainly in the small intestine, whereas SCFAs are mainly produced in the colon.25,27 Therefore, this pathway is likely to be effective mainly in MLN which drains metabolites from both small and large intestines rather than in effector sites of the intestines where SCFAs and RA are produced in different locations.
SCFAs activate GPCRs such as GPR41, GPR43 and GP109A. Myeloid cells, such as neutrophils, macrophages and DCs, variably express GPR43 and GP109A. Therefore, SCFAs have the potential to indirectly regulate B cells through myeloid cells. For example, SCFAs increase IL-10 production by macrophages and DCs.38,39 via either SCFA-receptor signaling or HDAC inhibition (Fig. 1). RA and IL-10, produced from SCFA-regulated myeloid cells, can promote antibody production, particularly IgA. In contrast, T and B cells do not significantly express SCFA receptors.
Eosinophils, abundant in the intestinal lamina propria, sense microbial signals and regulate B cells. Eosinophils, when activated by commensal bacterial products, produce various B cell-activating molecules, such as BAFF, APRIL, IL-6 and matrix metalloproteinase 9 (MMP9), which can promote the differentiation and survival of IgA+ plasma cells. Eosinophil-deficient mice (ΔdblGATA-1 and PHIL mice) have reduced numbers of IgA+ plasma cells but increased numbers of IgG1+ cells in PPs. Eosinophil-deficient mice also have decreased numbers of CD103+ DC and Tregs but increased production of Th2 cytokines (IL-4 and IL-5) by Tfh cells in PPs40 The gut microbiota activate epithelial cells for production of IL-25, which in turn activates eosinophils. For example, Tritrichomonas muris, a symbiotic protozoon in mice, induces IL-25 production by specialized epithelial cells called Tuft cells. Tuft cell-derived IL-25 mounts a type 2 innate lymphoid cell (ILC2) response and produce IL-5 and IL-13,41 which can increase eosinophil responses and affect B cell responses in the gut.
Antibodies are produced by several B cell types including B2, B1 and marginal zone (MZ) B cells. TLR ligands activate TLRs of B1 and MZ B cells for antibody production. IFN-γ and type I IFNs are produced following TLR activation, and these cytokines promote the production of antigen-specific antibodies (IgG2c, IgG2a and IgM) by MZ B cells.42 Microbial products activate neutrophils to regulate B cells. For example, the neutrophils that reside in the peri-MZ of the spleen support MZ B cells.43 The microbiota are required for colonization of neutrophils in the peri-MZ and their maturation into B cell-helping neutrophils. These neutrophils produce BAFF, APRIL and IL-21 to induce T-independent antibody responses to type 2 thymus-independent (TI-2) antigens such as bacterial capsular antigens. Thus, many cell types mediate the regulatory effects of the gut microbiota on B cells.
Conclusions and questions
IgA is a key regulator of the gut microbiota. We have witnessed a significant progress in our understanding of the diverse functions of gut host and microbial factors in regulating IgA production. There are many direct and indirect mechanisms by which the gut microbiota regulate antibody production. The gut microbiota produce many factors that activate B and non-B cell types. A major group of microbial factors important for B cell regulation includes TLR and related microbial PAMPR ligands. Another emerging group of microbial factors important for mucosal antibody production includes microbial metabolites such as SCFAs and ATP. SCFAs exert their functions through GPCR receptors primarily expressed by epithelial cells and some myeloid cells. Also, SCFAs are integrated into cellular metabolism following their conversion to acetyl-CoA, and this greatly affects the cellular metabolism in T and B cells. Because SCFAs work through several different pathways in SCFA receptor-dependent and independent pathways in many cell types, it is not easy to determine the functional significance of individual pathways in vivo. More studies are required in the future to dissect the functions of SCFAs and other metabolites in regulating B cell responses. It appears that these microbial metabolites not only affect the immune cells in the intestine but also the cells in systemic sites and organs. This is because microbial metabolites can be transported through the blood circulation. Whether antibody responses to specific pathogens are effectively supported by any of the metabolites is an important question to answer. More studies are needed to establish the roles of these microbial metabolites in regulating antibody-mediated inflammatory diseases such as lupus and rheumatoid arthritis. Moreover, it is important to devise strategies to prevent infection and enhance immunity using the B cell-helping microbial factors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported, in part, from grants from NIH (R01AI121302 and R01AI080769) to CHK.
References
- [1].Kim CH. Retinoic acid, immunity, and inflammation. Vitam Horm 2011; 86:83-101; PMID:21419268 [DOI] [PubMed] [Google Scholar]
- [2].Larange A, Cheroutre H. Retinoic acid and retinoic acid receptors as pleiotropic modulators of the immune system. Ann Rev Immunol 2016; 34:369-94; PMID:27168242; http://dx.doi.org/ 10.1146/annurev-immunol-041015-055427 [DOI] [PubMed] [Google Scholar]
- [3].Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol 2008; 1:11-22; PMID:19079156; http://dx.doi.org/ 10.1038/mi.2007.6 [DOI] [PubMed] [Google Scholar]
- [4].Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al.. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000-10; PMID:25171403; http://dx.doi.org/ 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA 2004; 101(7):1981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Suzuki K, Nakajima A. New aspects of IgA synthesis in the gut. Int Immunol 2014; 26:489-94; PMID:24872116; http://dx.doi.org/ 10.1093/intimm/dxu059 [DOI] [PubMed] [Google Scholar]
- [7].Forbes SJ, Bumpus T, McCarthy EA, Corthesy B, Mantis NJ. Transient suppression of Shigella flexneri type 3 secretion by a protective O-antigen-specific monoclonal IgA. Mbio 2011; 2:e00042-11; PMID:21610121; http://dx.doi.org/ 10.1128/mBio.00042-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kadaoui KA, Corthesy B. Secretory IgA mediates bacterial translocation to dendritic cells in mouse Peyer's patches with restriction to mucosal compartment. J Immunol 2007; 179:7751-7; PMID:18025221; http://dx.doi.org/ 10.4049/jimmunol.179.11.7751 [DOI] [PubMed] [Google Scholar]
- [9].Mantis NJ, Cheung MC, Chintalacharuvu KR, Rey J, Corthesy B, Neutra MR. Selective adherence of IgA to murine Peyer's patch M cells: evidence for a novel IgA receptor. J Immunol 2002; 169:1844-51; PMID:12165508; http://dx.doi.org/ 10.4049/jimmunol.169.4.1844 [DOI] [PubMed] [Google Scholar]
- [10].Morton HC, van den Herik-Oudijk IE, Vossebeld P, Snijders A, Verhoeven AJ, Capel PJ, van de Winkel JG. Functional association between the human myeloid immunoglobulin A Fc receptor (CD89) and FcR gamma chain. Molecular basis for CD89/FcR gamma chain association. J Biol Chem 1995; 270:29781-7 [DOI] [PubMed] [Google Scholar]
- [11].Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008; 456:507-10; PMID:18987631; http://dx.doi.org/ 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- [12].Wei B, Su TT, Dalwadi H, Stephan RP, Fujiwara D, Huang TT, Brewer S, Chen L, Arditi M, Borneman J, et al.. Resident enteric microbiota and CD8+ T cells shape the abundance of marginal zone B cells. Euro J Immunol 2008; 38:3411-25; PMID:19009526; http://dx.doi.org/ 10.1002/eji.200838432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, Prinz I, Pabst O. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med 2012; 209:365-77; PMID:22249449; http://dx.doi.org/ 10.1084/jem.20111980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lundell AC, Bjornsson V, Ljung A, Ceder M, Johansen S, Lindhagen G, Törnhage CJ, Adlerberth I, Wold AE, Rudin A. Infant B cell memory differentiation and early gut bacterial colonization. J Immunol 2012; 188:4315-22; PMID:22490441; http://dx.doi.org/ 10.4049/jimmunol.1103223 [DOI] [PubMed] [Google Scholar]
- [15].Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun 1999; 67:1992-2000; PMID:10085047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Buchta CM, Bishop GA. Toll-like receptors and B cells: functions and mechanisms. Immunol Res 2014; 59:12-22; PMID:24847763; http://dx.doi.org/ 10.1007/s12026-014-8523-2 [DOI] [PubMed] [Google Scholar]
- [17].Coffman RL, Lebman DA, Shrader B. Transforming growth factor beta specifically enhances IgA production by lipopolysaccharide-stimulated murine B lymphocytes. J Exp Med 1989; 170:1039-44; PMID:2788703; http://dx.doi.org/ 10.1084/jem.170.3.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rosser EC, Oleinika K, Tonon S, Doyle R, Bosma A, Carter NA, Harris KA, Jones SA, Klein N, Mauri C. Regulatory B cells are induced by gut microbiota-driven interleukin-1beta and interleukin-6 production. Nat Med 2014; 20:1334-9; PMID:25326801; http://dx.doi.org/ 10.1038/nm.3680 [DOI] [PubMed] [Google Scholar]
- [19].Caro-Maldonado A, Wang R, Nichols AG, Kuraoka M, Milasta S, Sun LD, Gavin AL, Abel ED, Kelsoe G, Green DR, et al.. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol 2014; 192:3626-36; PMID:24616478; http://dx.doi.org/ 10.4049/jimmunol.1302062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wahl DR, Byersdorfer CA, Ferrara JL, Opipari AW Jr, Glick GD. Distinct metabolic programs in activated T cells: opportunities for selective immunomodulation. Immunol Rev 2012; 249:104-15; PMID:22889218; http://dx.doi.org/ 10.1111/j.1600-065X.2012.01148.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al.. ATP drives lamina propria T(H)17 cell differentiation. Nature 2008; 455:808-12; PMID:18716618; http://dx.doi.org/ 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- [22].Schena F, Volpi S, Faliti CE, Penco F, Santi S, Proietti M, Schenk U, Damonte G, Salis A, Bellotti M, et al.. Dependence of immunoglobulin class switch recombination in B cells on vesicular release of ATP and CD73 ectonucleotidase activity. Cell Rep 2013; 3:1824-31; PMID:23770243; http://dx.doi.org/ 10.1016/j.celrep.2013.05.022 [DOI] [PubMed] [Google Scholar]
- [23].Kim M, Qie Y, Park J, Kim CH. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016; 20:202-14; PMID:27476413; http://dx.doi.org/ 10.1016/j.chom.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr 1986; 116:1772-6; PMID:3761032 [DOI] [PubMed] [Google Scholar]
- [25].Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, Macia L, Mackay CR. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 2016; 15:2809-24; PMID:27332875; http://dx.doi.org/ 10.1016/j.celrep.2016.05.047 [DOI] [PubMed] [Google Scholar]
- [26].Kunisawa J, Hashimoto E, Inoue A, Nagasawa R, Suzuki Y, Ishikawa I, Shikata S, Arita M, Aoki J, Kiyono H. Regulation of intestinal IgA responses by dietary palmitic acid and its metabolism. J Immunol 2014; 193:1666-71; PMID:25031459; http://dx.doi.org/ 10.4049/jimmunol.1302944 [DOI] [PubMed] [Google Scholar]
- [27].Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, Huang X, Xiao Y, Yao S, Zhao Q, et al.. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol 2016; [Epub ahead of print] http://dx.doi.org/ 10.1038/mi.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013; 145:396-406 e1-10; PMID:23665276 [DOI] [PubMed] [Google Scholar]
- [29].Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, Seleznik GM, Eberl G, Littman DR, Heikenwalder M, Tumanov AV, et al.. Nonredundant function of soluble LTalpha3 produced by innate lymphoid cells in intestinal homeostasis. Science 2013; 342:1243-6; PMID:24311691; http://dx.doi.org/ 10.1126/science.1243364 [DOI] [PubMed] [Google Scholar]
- [30].Liu G, Zhang L, Zhao Y. Modulation of immune responses through direct activation of Toll-like receptors to T cells. Clin Exp Immunol 2010; 160:168-75; PMID:20128825; http://dx.doi.org/ 10.1111/j.1365-2249.2010.04091.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sutmuller RP, den Brok MH, Kramer M, Bennink EJ, Toonen LW, Kullberg BJ, Joosten LA, Akira S, Netea MG, Adema GJ. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest 2006; 116:485-94; PMID:16424940; http://dx.doi.org/ 10.1172/JCI25439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O'Connell RM, Round JL. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 2015; 17:153-63; PMID:25620548; http://dx.doi.org/ 10.1016/j.chom.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, Hori S, Fagarasan S. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science 2009; 323:1488-92; PMID:19286559; http://dx.doi.org/ 10.1126/science.1169152 [DOI] [PubMed] [Google Scholar]
- [34].Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569-73; PMID:23828891; http://dx.doi.org/ 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hou B, Saudan P, Ott G, Wheeler ML, Ji M, Kuzmich L, Lee LM, Coffman RL, Bachmann MF, DeFranco AL. Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity 2011; 34:375-84; PMID:21353603; http://dx.doi.org/ 10.1016/j.immuni.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tezuka H, Abe Y, Iwata M, Takeuchi H, Ishikawa H, Matsushita M, Shiohara T, Akira S, Ohteki T. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature 2007; 448:929-33; PMID:17713535; http://dx.doi.org/ 10.1038/nature06033 [DOI] [PubMed] [Google Scholar]
- [37].Suzuki K, Maruya M, Kawamoto S, Sitnik K, Kitamura H, Agace WW, Fagarasan S. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity 2010; 33:71-83; PMID:20643338; http://dx.doi.org/ 10.1016/j.immuni.2010.07.003 [DOI] [PubMed] [Google Scholar]
- [38].Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al.. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014; 40:128-39; PMID:24412617; http://dx.doi.org/ 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 2014; 111:2247-52; PMID:24390544; http://dx.doi.org/ 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chu VT, Beller A, Rausch S, Strandmark J, Zanker M, Arbach O, Kruglov A, Berek C. Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity 2014; 40:582-93; PMID:24745334; http://dx.doi.org/ 10.1016/j.immuni.2014.02.014 [DOI] [PubMed] [Google Scholar]
- [41].Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, et al.. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016; 351:1329-33; PMID:26847546; http://dx.doi.org/ 10.1126/science.aaf1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Oganesyan G, Saha SK, Pietras EM, Guo B, Miyahira AK, Zarnegar B, Cheng G. IRF3-dependent type I interferon response in B cells regulates CpG-mediated antibody production. J Biol Chem 2008; 283:802-8; PMID:17925397; http://dx.doi.org/ 10.1074/jbc.M704755200 [DOI] [PubMed] [Google Scholar]
- [43].Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, Comerma L, Chorny A, Shan M, Xu W, et al.. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol 2011; 13:170-80; PMID:22197976; http://dx.doi.org/ 10.1038/ni.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]


