ABSTRACT
Bile salts act as a stressor to bacteria that transit the intestinal tract. Enteric pathogens have hijacked bile as an intestinal signal to regulate virulence factors. We recently demonstrated that Vibrio parahemolyticus senses bile salts via a heterodimeric receptor formed by the periplasmic domains of inner-membrane proteins VtrA and VtrC. Crystal structures of the periplasmic complex reveal that VtrA and VtrC form a β-barrel that binds bile salts in its hydrophobic interior to activate the VtrA cytoplasmic DNA-binding domain. Proteins with the same domain arrangement as VtrA and VtrC are widespread in Vibrio and related bacteria, where they are involved in regulating virulence and other unknown functions. Here we discuss our findings and review current knowledge on VtrA and VtrC homologs. We propose that signaling by these membrane-bound transcription factors can be advantageous for the regulation of membrane and secretory proteins.
KEYWORDS: bile, bile salts, intestinal pathogens, signaling, T3SS, T3SS2, type-three secretion, Vibrio, Vibrio cholerae, Vibrio parahaemolyticus
Bile as a stressor and a signal
Bile is a secretory mixture that plays a key role in digestion. It is synthesized in the liver, stored in the gallbladder, and secreted into the small intestine after a meal. Its principal components are bile salts, bilirubin, cholesterol, phospholipids, and inorganic salts.1 Of these, bile salts play a major role in solubilizing lipids and fat-soluble vitamins to facilitate their absorption. Bile salts are surfactant molecules that are synthesized from cholesterol and conjugated to glycine or taurine to increase their solubility.1 The detergent properties of bile salts render them antimicrobial, as they can disrupt cell membranes via their interaction with lipids and proteins, damage nucleic acids, and cause redox stress.2 Commensal and pathogenic intestinal bacteria will inevitably come into contact with bile salts and must evolve strategies to cope with their damaging effects. Many intestinal bacteria have adapted to bile by decreasing membrane permeability, inducing efflux pumps, inducing biofilm formation, and upregulating redox and DNA damage repair genes.2 Others are able to use bile salts as a cue for intestinal location to regulate virulence factors. These responses can be complex, vary among pathogens, and often depend on specific bile salts. For example, while deoxycholate has been shown to induce virulence gene expression in Campylobater jejuni,3 this same bile acid represses invasion genes in Salmonella.4 Although bile salt induced phenotypes in intestinal pathogens have been thoroughly documented,5 the mechanisms used for sensing of bile salts, whether directly by binding to signaling proteins or indirectly by sensing cell damage, remain poorly characterized.
Virulence gene regulation by bile salts in Vibrio parahaemolyticus
V. parahaemolyticus is a halophilic bacterium that inhabits marine environments and enters the human body mainly through the consumption of contaminated water or undercooked seafood.6 Pathogenic strains of V. parahaemolyticus are able to colonize and invade the digestive track, resulting in acute gastroenteritis.7,8 Disease is primarily caused by a set of virulence determinants: pore-forming hemolysins and a Type-III Secretion System (T3SS2) that are encoded by a pathogenicity island (Vp-PAI) in chromosome II of V. parahaemolyticus.9,10 The T3SS2 is a needle-like apparatus that spans the inner and outer bacterial membranes and translocates toxic effector proteins into eukaryotic cells. Several T3SS2 effectors have been shown to manipulate actin and hijack host signaling pathways; their functions are thoroughly reviewed by de Souza Santos et al.11
A regulatory network that is specifically responsive to bile salts restricts expression of Vp-PAI genes to when V. parahaemolyticus encounters the small intestine.12,13 This network comprises 3 inner-membrane proteins: VtrA, VtrB, and VtrC.13 VtrA and VtrB contain an N-terminal winged helix-turn-helix (wHTH) DNA-binding domain of the OmpR family that is attached to the inner membrane by a single transmembrane helix; VtrA also has a C-terminal periplasmic domain.14 (Fig. 1A). We recently uncovered the third and essential protein component of this pathway, VtrC, which is encoded by a gene located downstream of and in an operon with vtrA.13 VtrC is anchored to the inner membrane by a single transmembrane helix and contains a C-terminal periplasmic domain like VtrA, but lacks a cytoplasmic domain (Fig. 1A). We found that VtrA and VtrC form a 1:1 complex through their periplasmic domains to form a membrane-bound receptor that allows V. parahaemolyticus to sense bile salts.13 X-ray structures of this complex reveal an obligate heterodimer where 8 β-strands from VtrC and a single β-strand from VtrA form a β-barrel that harbors a hydrophobic inner chamber with a bile salt binding pocket (Fig. 1B). Upon bile salt binding by this complex, VtrA is able to induce transcription from the vtrB promoter. Newly synthesized VtrB then binds to Vp-PAI promoters to induce the expression of T3SS2-related genes. 12,14
Figure 1.
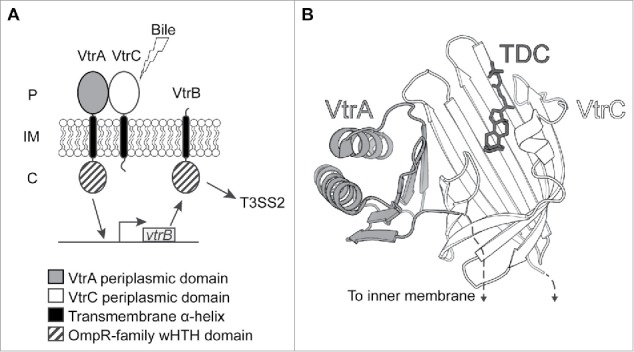
Bile salt sensing by VtrA/VtrC. (A) Schematic of bile salt signaling network formed by VtrA, VtrB and VtrC. The VtrA/VtrC complex senses bile salts in the periplasm, which activates the VtrA cytoplasmic DNA-binding domain to promote vtrB transcription, resulting in T3SS expression. P: periplasm; IM: inner membrane, C: cytoplasm. (B) Structure of the VtrA/VtrC periplasmic domain complex with the ligand taurodeoxycholate (TDC).
Various Vibrio species possess VtrA and VtrC homologs and/or more diverse protein pairs with the same domain arrangement as VtrA and VtrC. These include VtrA/VtrC homologs of unknown function, as well as previously characterized regulators of virulence gene expression TcpP/TcpH and ToxR/ToxS.15-17 Below, we summarize what is known about the function and mechanism of these proteins.
VtrA/VtrC homologs
Pathogenic non-O1/O139 V. cholerae that lack cholera toxin (CT) and toxin coregulated pilus (TCP), such as strain AM-19226, cause enterotoxicity via a T3SS pathogenicity island similar to Vp-PAI.18 This pathogenicity island is controlled by VtrA and VtrB homologs VttRA and VttRB, respectively.18 A VtrC homolog present in V. cholerae AM-19226 is likely to be part of this regulatory pathway as well.13,19 Variations in the genes in this pathogenicity island can be reduced to 2 lineages, T3SS2α and T3SS2β, both of which are distributed among V. parahaemolyticus and V. cholerae strains and were acquired through independent horizontal gene transfer events.20 While strain AM-19226 and the V. parahaemolyticus strain used in our study (RIMD2210633) contain T3SS2α, we found genes encoding VtrA, VtrB, and VtrC homologs in strains with T3SS2β (V. parahaemolyticus TH3996 and V. cholerae strains 1587 and 623–39, unpublished). This suggests that the regulatory pathway controlling T3SS2s from both lineages has been conserved through evolution. There is evidence that genes outside of the V. cholerae strain AM-19226 T3SS pathogenicity island that are involved in flagellar biosynthesis, chemotaxis, metabolism, and type 6 secretion are influenced by VttRA and VttRB,21 supporting the idea that the this regulatory cascade goes beyond controlling T3SS.
VtrA, VtrB, and VtrC are also conserved in species with an incomplete set of T3SS genes, or no T3SS genes at all.13 Remarkably, we found close VtrA/VtrC homologs in a group of species that lack a membrane-bound VtrB homolog.13 These species have a gene encoding a predicted sphingomyelinase, an enzyme that hydrolyses sphingomyelin,22 downstream of the vtrA/vtrC operon. Sphingomyelin is abundant in eukaryotic cell membranes and several bacterial pathogens produce sphingomyelinases that contribute to their virulence.23-25 Some of the species that contain a vtrA/vtrC operon followed by a sphingomyelinase gene, like Moritella marina (formerly V. marinus), V. campbellii, V. harveyi, and V. splendidus, are pathogens of marine animals,26-29 so it is possible that this enzyme could play a role in virulence toward aquatic organisms. More work is needed to determine if this sphingomyelinase is a virulence factor and whether VtrA/VtrC control its expression in response to an environmental or host-derived signal. The amino acid sequences of these VtrA/VtrC homologs diverge significantly from the V. parahaemolyticus VtrA/VtrC sequences and given that bacteria are exposed to diverse environments, we think it likely that these homologs will respond to signals other than bile salts. Further work will determine whether these homologs use sphingomyelin or a similar compound as a signal.
TcpP/TcpH
V. cholerae TcpP and TcpH adopt the same topology as VtrA and VtrC (Fig. 2), respectively. TcpP and TcpH are also encoded in a bicistronic operon as overlapping genes in the Vibrio pathogenicity island (VPI),30 but lack homology to the periplasmic domains of VtrA and VtrC. They regulate the expression of cytoplasmic transcription factor ToxT, which activates the transcription of VPI genes encoding factors involved in colonization, such as TCP and CT genes.31 The TcpP DNA-binding domain binds a site immediately upstream of the predicted RNA polymerase binding site in the toxT promoter, suggesting that it might interact with RNA polymerase to activate toxT transcription.15,32 TcpP's interaction with TcpH, as well as the formation of a disulfide bond between the 2 cysteines in the TcpP periplasmic domain, protect it from degradation by regulated intramembrane proteolysis (RIP).33-35
Figure 2.
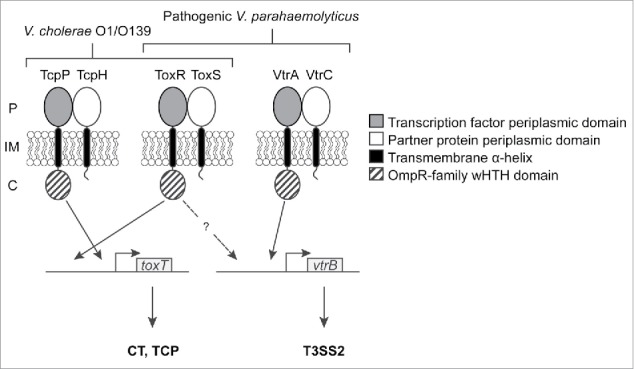
Proposed model for coordinate regulation of V. cholerae and V. parahaemolyticus virulence by inner-membrane proteins. In V. cholerae O1/O139 strains, TcpP, TcpH, ToxR and ToxS activate the transcription of toxT, whose gene product controls the expression of TCP and CT genes. In pathogenic V. parahaemolyticus strains, VtrA, VtrC, ToxR and ToxS control vtrB transcription. VtrB then activates the expression of T3SS genes. Known interactions between DNA-binding domains and promoters are indicated by solid arrows. A dashed arrow indicates that an interaction between ToxR and the vtrB promoter is pressumed but has not been experimentally confirmed. P: periplasm; IM: inner membrane, C: cytoplasm.
While a signal that binds the TcpP or TcpH periplasmic domains has not been identified, the bile salt taurocholate has been reported to induce TcpP homodimerization and TcpP-dependent induction of toxT expression.36 Recent data suggest that this effect is indirectly mediated by the thiol:disulfide interchange protein DsbA, rather than by bile salt binding by TcpP or TcpH.37 V. cholerae DsbA was shown to bind taurocholate, which correlates with a shift in the redox equilibrium of this enzyme toward the reduced state in the presence of taurocholate.37 This is proposed to interfere with its ability to catalyze intramolecular disulfide bond formation in TcpP, which favors the formation of active TcpP homodimers linked, instead, by intermolecular disulfide bonds.
ToxR/ToxS
ToxR and ToxS adopt the same domain topology as VtrA and VtrC (Fig. 2) respectively, and are expressed from a bicistronic operon that is part of the V. cholerae ancestral genome, but is also widespread among Vibrio species. In V. cholerae, ToxR controls the expression of the outer membrane proteins important for surviving in the small intestine 38,39 and is also involved in the regulation of toxT by binding to a site upstream of the TcpP binding site on the toxT promoter.32 Overexpressed TcpP can activate toxT transcription in the absence of ToxR,15 suggesting that TcpP is the main player in activation whereas ToxR's role is to enhance TcpP activity by recruiting it to the toxT promoter.32 The involvement of ToxS in ToxR-dependent regulation is not completely clear, however, studies have shown that ToxS enhances ToxR function as a transcriptional activator16 and that it is able to dimerize with ToxR.40 ToxS also decreases ToxR degradation through RIP under starvation conditions and after alkaline pH shock.41 ToxR has been reported to respond to a variety of stimuli, such as pH, osmolarity, presence of amino acids, bile, and cyclo(Phe-Pro),16,17,42-44 but it remains to be determined whether these signals act on ToxR (and/or ToxS) directly or indirectly via additional factors. The sequence similarity between the periplasmic domains of ToxR/ToxS and VtrA/VtrC is limited (< 25%), hindering inferences about ligand binding based on the VtrA/VtrC structure. Additional biochemical and biophysical studies are needed to determine what, if any, compounds bind ToxR/ToxS.
ToxR and ToxS have adopted alternative roles in Vibrio species that lack TCP and CT, like non-O1/O139 V. cholerae strains and V. parahaemolyticus. Studies with lacZ reporter fusions indicate that ToxR moderately affects T3SS expression in V. cholerae strain AM-19226.19 The V. parahaemolyticus ToxR homolog, Vp-ToxR, has recently been shown to be necessary for vtrB expression, after it was identified in a genetic screen for factors contributing to colonization of the mammalian intestine.45 This evidence suggests that ToxR/ToxS works with VtrA/VtrC (or its homologs) to control vtrB promoter expression in V. parahaemolyticus and V. cholerae non-O1/O139 strains, analogous to what has been proposed for ToxR/ToxS and TcpP/TcpH to control toxT expression.32 Further evidence that VttRA or VtrA overexpression in the absence of ToxR restores T3SS and T3SS2 expression, respectively,45,46 supports a scenario where ToxR plays a secondary role by enhancing VtrA's transcription factor activity. A possible mechanism that could explain this role is that ToxR binding to the vtrB promoter recruits VtrA to the promoter or affects VtrA's ability to activate transcription by other mechanism. Future studies will determine if ToxR binds the vtrB promoter and whether protein-protein interactions between ToxR and VtrA are involved in this process.
Homology beyond Vibrio spp.
Although the VtrA and VtrC periplasmic domains lack sequence homology with proteins of known structure, their tertiary structure bears striking structural similarity the calycin superfamily β-barrel fold.47 Calycins have diverse biologic functions and are found in both prokaryotes and eukaryotes. Many members of this superfamily bind small hydrophobic molecules such as fatty acids, retinol, and biotin inside their characteristic β-barrel.47 Thus, it is not surprising that the structure of VtrA/VtrC in complex with taurodeoxycholate reveals that bile salts bind inside this β-barrel, displacing a disordered loop that covers the binding site in the apo structure.13 Interestingly, the VtrA/VtrC complex is the first example of a calycin that forms an obligate heterodimer and can transmit a signal.
Our finding raises the question of how bile salt binding to the VtrA/VtrC complex in the periplasm activates VtrA's function as a transcription factor in the cytoplasm. While VtrA's domain topology with a DNA-binding domain anchored to the inner membrane is atypical – only 3% of prokaryotic transcritptional regulators incorporating a sensing domain and a HTH domain have transmembrane segments 48 – it is also found in other proteins with diverse periplasmic sensing domains. Examples of these are the CadC pH-responsive regulator found in Salmonella enterica serovar Typhimurium,49 Escherichia coli,50 and V. cholerae,51 Yersinia pestis PsaE, a regulator for the production of the pH6 antigen,52 V. cholerae TfoS, a regulator for compentence in response to chitin,53,54 and the Pseudoalteromonas tunicata WmpR, a regulator for antifouling compound production.55 Further work to determine how signals are transduced across the membrane by VtrA/VtrC might reveal mechanistic similarities between these distinct systems.
Conclusion
The prominent roles of inner-membrane bound transcription factors and their partner proteins in Vibrio spp suggest that this arrangement has a functional advantage over other systems that couple sensing of the external environment to gene regulation, such as 2-component systems. Both membrane-bound transcription factors and 2-component systems, the latter of which are composed of a histidine kinase and a response regulator, allow for transduction of signals received from the environment. We speculate that membrane-bound transcription factors have the additional feature in that they localize DNA transcription to the cytoplasmic membrane by a mechanism reminiscent of, but distinct from, transertion (simultaneous transcription, translation and membrane insertion, resulting in DNA attachment to the membrane; for a review, see ref. 56). Membrane-anchored transcription factors promote the enrichment of mRNAs encoding membrane-bound and secreted proteins at the membrane, potentially facilitating co-translational insertion and assembly of T3SS apparatus components, as well as effector secretion. This is analogous to the idea that certain mRNAs encoding membrane, polar and septal proteins are targeted to their site of function in the prokaryotic cell via information contained within the untranslated region of mRNA transcripts.57 Studies suggest that the 3'-untranslated region of some mRNA transcripts of flagellar and pathogenic T3SS proteins in E. coli and Salmonella, respectively, are important for membrane protein localization.58,59 Evidence of mRNA localization goes beyond transcripts encoding flagellar or T3SS proteins and does not always involve the unstranslated region of mRNAs. An example of this is the bglF mRNA transcript from E. coli, encoding a membrane-bound β-glucoside permease, that was shown to localize to the membrane by way of a signal within its coding sequence, independently of translation.60 It is important to note that not all mRNAs encoding membrane-bound or secreted proteins localize to the membrane. Nonetheless, membrane localization of some mRNAs may have evolved to minimize non-specific interactions between newly translated proteins thereby facilitating the assembly of multiprotein complexes.57 Thus, it is conceivable that a similar approach of restricting T3SS gene transcription to the cytoplasmic membrane is used in Vibrio spp to expedite the assembly of the needle apparatus and effector secretion.
Despite significant advancements in our understanding of how environmental signals control virulence in Vibrios, key questions remain to be answered. Our recent results demonstrate that V. parahemolyticus senses bile salts via the periplasmic domains of VtrA and VtrC. This pair of inner-membrane proteins form a signaling unit that regulates virulence in the mammalian gut. To fully understand this system, we need to determine how the binding of bile salt to their periplasmic domains affects the VtrA DNA-binding domain. Since DNA recognition sites for OmpR DNA-binding domains tend to be direct repeats,61 one possibility is that ligand binding promotes VtrA dimerization. Testing this hypothesis will require further work to identify the VtrA recognition site and to determine if dimerization is part of the mechanism by which VtrA activates transcription from the vtrB promoter. Nevertheless, mechanisms that do not involve a monomer to dimer transition are also possible.
Another aspect that needs further study is the crosstalk between ToxR/S and TcpP/H in V. cholerae and VtrA/C and ToxR/S in V. parahaemolyticus. While it has been established that these receptor pairs influence the same virulence pathways, the actual mechanism of how this happens remains elusive. Last but not least, it would be interesting to know if the periplasmic domains of ToxR/S, TcpP/H and VtrA/C homologs from species that lack T3SS bind small molecules. The identity such signals, if any, would shed light on how these receptors are stimulated and could open an avenue for therapeutic development.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Orth Lab for helpful discussions and Dr. Nicole De Nisco for critical reading of the manuscript.
Funding
This work was supported by NIH under Grant R01-AI087808; Welch Research Foundation under Grant I-1561; and Once Upon A Time…
References
- [1].Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005; 29:625-51; PMID:16102595; http://dx.doi.org/ 10.1016/j.femsre.2004.09.003 [DOI] [PubMed] [Google Scholar]
- [2].Hay AJ, Zhu J. In Sickness and in Health: The relationships between bacteria and bile in the human gut. Adv Appl Microbiol 2016; 96:43-64; PMID:27565580 [DOI] [PubMed] [Google Scholar]
- [3].Malik-Kale P, Parker CT, Konkel ME. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol 2008; 190:2286-97; PMID:18223090; http://dx.doi.org/ 10.1128/JB.01736-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Eade CR, Hung CC, Bullard B, Gonzalez-Escobedo G, Gunn JS, Altier C. Bile Acids Function Synergistically To Repress Invasion Gene Expression in Salmonella by Destabilizing the Invasion Regulator HilD. Infect Immun 2016; 84:2198-208; PMID:27185788; http://dx.doi.org/ 10.1128/IAI.00177-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sistrunk JR, Nickerson KP, Chanin RB, Rasko DA, Faherty CS. Survival of the fittest: How bacterial pathogens utilize bile to enhance infection. Clin Microbiol Rev 2016; 29:819-36; PMID:27464994; http://dx.doi.org/ 10.1128/CMR.00031-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, et al.. Vibrio parahaemolyticus infections in the United States, 1973-1998. J Infect Dis 2000; 181:1661-6; PMID:10823766; http://dx.doi.org/ 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- [7].O'Boyle N, Boyd A. Manipulation of intestinal epithelial cell function by the cell contact-dependent type III secretion systems of Vibrio parahaemolyticus. Front Cell Infect Microbiol 2014; 3:114; PMID:24455490; http://dx.doi.org/ 10.3389/fcimb.2013.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, Gilpin CJ, Orth K. Type III effector VopC mediates invasion for Vibrio species. Cell Rep 2012; 1:453-60; PMID:22787576; http://dx.doi.org/ 10.1016/j.celrep.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Izutsu K, Kurokawa K, Tashiro K, Kuhara S, Hayashi T, Honda T, Iida T. Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kgbase pathogenicity island and pathogenicity in kanagawa phenomenon-positive Vibrio parahemolyticus strains. Infect Immun 2008; 76:1016-23; PMID:18195030; http://dx.doi.org/ 10.1128/IAI.01535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sugiyama T, Iida T, Izutsu K, Park KS, Honda T. Precise region and the character of the pathogenicity island in clinical Vibrio parahaemolyticus strains. J Bacteriol 2008; 190:1835-7; PMID:18156272; http://dx.doi.org/ 10.1128/JB.01293-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].de Souza Santos M, Salomon D, Li P, Krachler A-M, Orth K. Vibrio parahaemolyticus virulence determinants. In: Alouf J, Ladant D, Popoff MR, eds. The Comprehensive Sourcebook of Bacterial Protein Toxins. Waltham: Elsevier, 2015:230-60. [Google Scholar]
- [12].Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park KS, Dryselius R, Akeda Y, Honda T, Iida T. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One 2010; 5:e13365; PMID:20967223; http://dx.doi.org/ 10.1371/journal.pone.0013365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, Grishin NV, Orth K. Bile salt receptor complex activates a pathogenic type III secretion system. Elife 2016; 5:e15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, Akeda Y, Park KS, Cantarelli VV, Dryselius R, Iida T, et al.. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One 2010; 5:e8678; PMID:20084267; http://dx.doi.org/ 10.1371/journal.pone.0008678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Häse CC, Mekalanos JJ. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 1998; 95:730-4; PMID:9435261; http://dx.doi.org/ 10.1073/pnas.95.2.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Miller VL, DiRita VJ, Mekalanos JJ. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol 1989; 171:1288-93; PMID:2646275; http://dx.doi.org/ 10.1128/jb.171.3.1288-1293.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 1987; 48:271-9; PMID:3802195; http://dx.doi.org/ 10.1016/0092-8674(87)90430-2 [DOI] [PubMed] [Google Scholar]
- [18].Shin OS, Tam VC, Suzuki M, Ritchie JM, Bronson RT, Waldor MK, Mekalanos JJ. Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. MBio 2011; 2:e00106-11; PMID:21673189; http://dx.doi.org/ 10.1128/mBio.00106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Alam A, Tam V, Hamilton E, Dziejman M. vttR(A) and vttR(B) encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect Immun 2010; 78:2554-70; PMID:20385759; http://dx.doi.org/ 10.1128/IAI.01073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Okada N, Iida T, Park KS, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun 2009; 77:904-13; PMID:19075025; http://dx.doi.org/ 10.1128/IAI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chaand M, Dziejman M. Vibrio cholerae VttR(A) and VttR(B) regulatory influences extend beyond the type 3 secretion system genomic island. J Bacteriol 2013; 195:2424-36; PMID:23524608; http://dx.doi.org/ 10.1128/JB.02151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Goñi FM, Alonso A. Sphingomyelinases: enzymology and membrane activity. FEBS Lett 2002; 531:38-46; PMID:12401200; http://dx.doi.org/ 10.1016/S0014-5793(02)03482-8 [DOI] [PubMed] [Google Scholar]
- [23].Gonzalez-Zorn B, Dominguez-Bernal G, Suarez M, Ripio MT, Vega Y, Novella S, Vázquez-Boland JA. The smcL gene of Listeria ivanovii encodes a sphingomyelinase C that mediates bacterial escape from the phagocytic vacuole. Mol Microbiol 1999; 33:510-23; PMID:10417642; http://dx.doi.org/ 10.1046/j.1365-2958.1999.01486.x [DOI] [PubMed] [Google Scholar]
- [24].Oda M, Hashimoto M, Takahashi M, Ohmae Y, Seike S, Kato R, Fujita A, Tsuge H, Nagahama M, Ochi S, et al.. Role of sphingomyelinase in infectious diseases caused by Bacillus cereus. PLoS One 2012; 7:e38054; PMID:22701599; http://dx.doi.org/ 10.1371/journal.pone.0038054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rivas AJ, Balado M, Lemos ML, Osorio CR. Synergistic and additive effects of chromosomal and plasmid-encoded hemolysins contribute to hemolysis and virulence in Photobacterium damselae subsp. damselae. Infect Immun 2013; 81:3287-99; PMID:23798530; http://dx.doi.org/ 10.1128/IAI.00155-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Austin B, Zhang XH. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol 2006; 43:119-24; PMID:16869892; http://dx.doi.org/ 10.1111/j.1472-765X.2006.01989.x [DOI] [PubMed] [Google Scholar]
- [27].Benediktsdóttir, Helgason, Sigurjónsdóttir. Vibrio spp. isolated from salmonids with shallow skin lesions and reared at low temperature. J Fish Dis 1998; 21:19-28; http://dx.doi.org/ 10.1046/j.1365-2761.1998.00065.x [DOI] [PubMed] [Google Scholar]
- [28].Haldar S, Chatterjee S, Sugimoto N, Das S, Chowdhury N, Hinenoya A, Asakura M, Yamasaki S. Identification of Vibrio campbellii isolated from diseased farm-shrimps from south India and establishment of its pathogenic potential in an Artemia model. Microbiology 2011; 157:179-88; PMID:20847009; http://dx.doi.org/ 10.1099/mic.0.041475-0 [DOI] [PubMed] [Google Scholar]
- [29].Sugumar G, Nakai T, Hirata Y, Matsubara D, Muroga K. Vibrio splendidus biovar II as the causative agent of bacillary necrosis of Japanese oyster Crassostrea gigas larvae. Dis Aquat Organ 1998; 33:111-8; PMID:9684317; http://dx.doi.org/ 10.3354/dao033111 [DOI] [PubMed] [Google Scholar]
- [30].Yu RR, Dirita VJ. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol 1999; 181:2584-92; PMID:10198025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].DiRita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 1991; 88:5403-7; PMID:2052618; http://dx.doi.org/ 10.1073/pnas.88.12.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Krukonis ES, Yu RR, DiRita VJ. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: Distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol 2000; 38:67-84; PMID:11029691; http://dx.doi.org/ 10.1046/j.1365-2958.2000.02111.x [DOI] [PubMed] [Google Scholar]
- [33].Beck NA, Krukonis ES, DiRita VJ. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J Bacteriol 2004; 186:8309-16; PMID:15576780; http://dx.doi.org/ 10.1128/JB.186.24.8309-8316.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Matson JS, DiRita VJ. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc Natl Acad Sci U S A 2005; 102:16403-8; PMID:16254052; http://dx.doi.org/ 10.1073/pnas.0505818102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Morgan SJ, French EL, Thomson JJ, Seaborn CP, Shively CA, Krukonis ES. Formation of an Intramolecular Periplasmic Disulfide Bond in TcpP Protects TcpP and TcpH from Degradation in Vibrio cholerae. J Bacteriol 2016; 198:498-509; http://dx.doi.org/ 10.1128/JB.00338-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 2013; 110:2348-53; PMID:23341592; http://dx.doi.org/ 10.1073/pnas.1218039110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xue Y, Tu F, Shi M, CQ Wu, Ren G, Wang X, Fang W, Song H, Yang M. Redox pathway sensing bile salts activates virulence gene expression in Vibrio cholerae. Mol Microbiol 2016; 102:909-924 [DOI] [PubMed] [Google Scholar]
- [38].Crawford JA, Kaper JB, DiRita VJ. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol 1998; 29:235-46; PMID:9701817; http://dx.doi.org/ 10.1046/j.1365-2958.1998.00925.x [DOI] [PubMed] [Google Scholar]
- [39].Li CC, Merrell DS, Camilli A, Kaper JB. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol Microbiol 2002; 43:1577-89; PMID:11952906; http://dx.doi.org/ 10.1046/j.1365-2958.2002.02845.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ottemann KM, Mekalanos JJ. The ToxR protein of Vibrio cholerae forms homodimers and heterodimers. J Bacteriol 1996; 178:156-62; PMID:8550410; http://dx.doi.org/ 10.1128/jb.178.1.156-162.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Almagro-Moreno S, Root MZ, Taylor RK. Role of ToxS in the proteolytic cascade of virulence regulator ToxR in Vibrio cholerae. Mol Microbiol 2015; 98:963-76; PMID:26316386; http://dx.doi.org/ 10.1111/mmi.13170 [DOI] [PubMed] [Google Scholar]
- [42].Bina XR, Taylor DL, Vikram A, Ante VM, Bina JE. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). mBio 2013; 4:e00366-13; PMID:23982069; http://dx.doi.org/ 10.1128/mBio.00366-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hung DT, Mekalanos JJ. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci U S A 2005; 102:3028-33; PMID:15699331; http://dx.doi.org/ 10.1073/pnas.0409559102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Provenzano D, Schuhmacher DA, Barker JL, Klose KE. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun 2000; 68:1491-7; PMID:10678965; http://dx.doi.org/ 10.1128/IAI.68.3.1491-1497.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, Zhou X, Davis BM, Waldor MK. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci U S A 2016; 113:6283-8; PMID:27185914; http://dx.doi.org/ 10.1073/pnas.1601718113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Miller KA, Sofia MK, Weaver JWA, Seward C, Dziejman M. Regulation by ToxR-like proteins converges on vttR(B) expression to control T3SS-dependent Caco2-BBE cytotoxicity in V. cholerae. J Bacteriol 2016; 198:JB.00130-16; http://dx.doi.org/ 10.1128/JB.00130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta 2000; 1482:9-24; PMID:11058743; http://dx.doi.org/ 10.1016/S0167-4838(00)00148-5 [DOI] [PubMed] [Google Scholar]
- [48].Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 2005; 13:52-6; PMID:15680762; http://dx.doi.org/ 10.1016/j.tim.2004.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee YH, Kim JH, Bang IS, Park YK. The membrane-bound transcriptional regulator CadC is activated by proteolytic cleavage in response to acid stress. J Bacteriol 2008; 190:5120-6; PMID:18487329; http://dx.doi.org/ 10.1128/JB.00012-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Watson N, Dunyak DS, Rosey EL, Slonczewski JL, Olson ER. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol 1992; 174:530-40; PMID:1370290; http://dx.doi.org/ 10.1128/jb.174.2.530-540.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Merrell DS, Camilli A. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the "ToxR-like" family of transcriptional regulators. J Bacteriol 2000; 182:5342-50; PMID:10986235; http://dx.doi.org/ 10.1128/JB.182.19.5342-5350.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yang Y, Isberg RR. Transcriptional regulation of the Yersinia pseudotuberculosis pH6 antigen adhesin by two envelope-associated components. Mol Microbiol 1997; 24:499-510; PMID:9179844; http://dx.doi.org/ 10.1046/j.1365-2958.1997.3511719.x [DOI] [PubMed] [Google Scholar]
- [53].Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. Mbio 2014; 5:e01028-13; PMID:24473132; http://dx.doi.org/ 10.1128/mBio.01028-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yamamoto S, Mitobe J, Ishikawa T, Wai SN, Ohnishi M, Watanabe H, Izumiya H. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Mol Microbiol 2014; 91:326-47; PMID:24236404; http://dx.doi.org/ 10.1111/mmi.12462 [DOI] [PubMed] [Google Scholar]
- [55].Egan S, James S, Holmstrom C, Kjelleberg S. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ Microbiol 2002; 4:433-42; PMID:12153584; http://dx.doi.org/ 10.1046/j.1462-2920.2002.00322.x [DOI] [PubMed] [Google Scholar]
- [56].Roggiani M, Goulian M. Chromosome-Membrane Interactions in Bacteria. Annu Rev Genet 2015; 49:115-29; PMID:26436460; http://dx.doi.org/ 10.1146/annurev-genet-112414-054958 [DOI] [PubMed] [Google Scholar]
- [57].Kannaiah S, Amster-Choder O. Protein targeting via mRNA in bacteria. Biochim Biophys Acta 2014; 1843:1457-65; PMID:24263243; http://dx.doi.org/ 10.1016/j.bbamcr.2013.11.004 [DOI] [PubMed] [Google Scholar]
- [58].Majander K, Anton L, Antikainen J, Lang H, Brummer M, Korhonen TK, Westerlund-Wikström B. Extracellular secretion of polypeptides using a modified Escherichia coli flagellar secretion apparatus. Nat Biotechnol 2005; 23:475-81; PMID:15806100; http://dx.doi.org/ 10.1038/nbt1077 [DOI] [PubMed] [Google Scholar]
- [59].Niemann GS, Brown RN, Mushamiri IT, Nguyen NT, Taiwo R, Stufkens A, Smith RD, Adkins JN, McDermott JE, Heffron F. RNA type III secretion signals that require Hfq. J Bacteriol 2013; 195:2119-25; PMID:23396917; http://dx.doi.org/ 10.1128/JB.00024-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science 2011; 331:1081-4 [DOI] [PubMed] [Google Scholar]
- [61].Martinez-Hackert E, Stock AM. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol 1997; 269:301-12; PMID:9199401; http://dx.doi.org/ 10.1006/jmbi.1997.1065 [DOI] [PubMed] [Google Scholar]


