Abstract
Objective
Monogenic diabetes, a young-onset form of diabetes, is often misdiagnosed as Type 1 diabetes, resulting in unnecessary treatment with insulin. A screening approach for monogenic diabetes is needed to accurately select suitable patients for expensive diagnostic genetic testing. We used C-peptide and islet autoantibodies, highly sensitive and specific biomarkers for discriminating Type 1 from non-Type 1 diabetes, in a biomarker screening pathway for monogenic diabetes.
Research Design and Methods
We studied patients diagnosed ≤30y, currently <50y, in two UK regions with existing high detection of monogenic diabetes. The biomarker screening pathway comprised 3 stages: 1) Assessment of endogenous insulin secretion using urinary C-peptide/creatinine ratio (UCPCR); 2) If UCPCR≥0.2nmol/mmol, measurement of GAD and IA2 islet autoantibodies; 3) If negative for both autoantibodies, molecular genetic diagnostic testing for 35 monogenic diabetes subtypes.
Results
1407 patients participated (1365 no known genetic cause, 34 monogenic diabetes, 8 cystic-fibrosis-related diabetes). 386/1365(28%) had UCPCR≥0.2nmol/mmol. 216/386(56%) of these patients were negative for GAD and IA2 and underwent molecular genetic testing. 17 new cases of monogenic diabetes were diagnosed (8 common MODY (Sanger sequencing), 9 rarer causes (next generation sequencing)) in addition to the 34 known cases (estimated prevalence of 3.6% (51/1407) (95%CI: 2.7-4.7%)). The positive predictive value was 20%, suggesting a 1-in-5 detection rate for the pathway. The negative predictive value was 99.9%.
Conclusions
The biomarker screening pathway for monogenic diabetes is an effective, cheap, and easily implemented approach to systematically screening all young-onset patients. The minimum prevalence of monogenic diabetes is 3.6% of patients diagnosed ≤30y.
Registered on Clinicaltrials.gov ref NCT01238380
Introduction
Correct classification of a patient’s diabetes is important to ensure they receive the most appropriate treatment and ongoing management. The most common form of diabetes in children and young adults is Type 1 diabetes, accounting for over 90% of cases(1; 2). Other forms of diabetes in this age group, such as monogenic diabetes (including Maturity Onset Diabetes of the Young (MODY)), or young-onset Type 2, are not often considered. It is estimated that at least 80% of patients with MODY are misdiagnosed(3), and other rarer forms of monogenic diabetes often go unrecognized due to lack of awareness(4). Patients with MODY or Type 2 diabetes misclassified as Type 1 diabetes will be treated with insulin, whereas non-insulin therapy would be more appropriate. Diet and metformin are the treatment of choice in young Type 2 diabetes(5). Patients with MODY due to mutations in the HNF1A or HNF4A genes respond well to low dose sulphonylureas(6; 7) and those with MODY due to mutations in the GCK gene require no pharmacological treatment(8). Getting a correct diagnosis for all forms of monogenic diabetes has important implications for management of an individual’s diabetes, their prognosis, and recognition of associated clinical features; it also allows appropriate counselling of other family members regarding likely inheritance (4).
Identifying patients with monogenic diabetes, particularly MODY, can be challenging. Monogenic diabetes is confirmed by molecular genetic testing, but this is expensive so testing all patients is not feasible. An approach that could be used to enrich for monogenic diabetes, increasing the proportion identified in those who undergo genetic testing, would be helpful. Clinical features can aid identification of those who may have an alternative diagnosis, and a probability calculator has been developed to help determine which patients are likely to have the most common forms of MODY(9). However, this will not pick up other forms of monogenic diabetes and its performance is weaker for detecting MODY in insulin treated patients compared to non-insulin treated patients.
An alternative approach to enrich for monogenic diabetes is to use biomarkers which have been shown to discriminate well between Type 1 and other forms of young onset diabetes. Type 1 diabetes is characterized by autoimmune destruction of the beta-cells in the pancreas leading to absolute insulin deficiency so two tests that could be used to diagnose Type 1 diabetes are islet autoantibodies (markers of the autoimmune process) and C-peptide (a marker of insulin deficiency). C-peptide has been shown to be a highly sensitive and specific biomarker for discriminating between Type 1 and Type 2 diabetes and MODY 3-5 years after diagnosis(10; 11). Urine C-peptide-Creatinine ratio (UCPCR) can be used to remove the need for blood samples, which may be of particular concern in the pediatric population, and means that the sample can easily be taken at home and posted to the laboratory(12). GAD and IA2 islet autoantibodies also discriminate well between Type 1 and MODY, with cross sectional studies showing they are present in 80% of patients with Type 1 diabetes and in less than 1% of patients with MODY(13). These biomarkers have been used to screen for MODY in other studies(14; 15), but have been limited to pediatric cases only. Given the median age at diagnosis for MODY is 20 years (from UK referrals data(3)), and there is on average a delay of 13 years from diabetes diagnosis to a confirmed genetic diagnosis(16), it is crucial to study adults as well. Furthermore the combined diagnostic performance of the two biomarkers as a screening pathway has not been formally assessed.
By excluding those with Type 1 diabetes using these two biomarkers we can obtain a smaller percentage of patients in whom diagnostic molecular testing for monogenic diabetes could be performed. We tested a screening pathway using both C-peptide and islet autoantibodies to exclude Type 1 diabetes in two populations with previously high pick-up rates of MODY(3), and performed genetic testing on all patients with significant endogenous insulin and absence of islet autoantibodies. This allowed us to determine the prevalence of all monogenic diabetes subtypes in those diagnosed ≤30 years, and to calculate the positive and negative predictive values for the pathway.
Research Design and Methods
Subjects
Patients diagnosed aged 30 years or under, and currently aged under 50, in the catchment areas of the Royal Devon and Exeter NHS Foundation Trust (Exeter, UK) and Ninewells Hospital (Dundee, UK) were invited to take part in the study via the doctors looking after their medical care. All patients with diabetes in this age group were eligible regardless of cause. Both regions had existing high pick-up rates for MODY prior to the study due to research interests(3). Patients that consented provided samples as part of the biomarkers screening pathway (the UNITED study (Using pharmacogeNetics to Improve Treatment in Early-onset Diabetes (UNITED)), clinicaltrials.gov ref NCT01238380).
Biomarker Screening Pathway
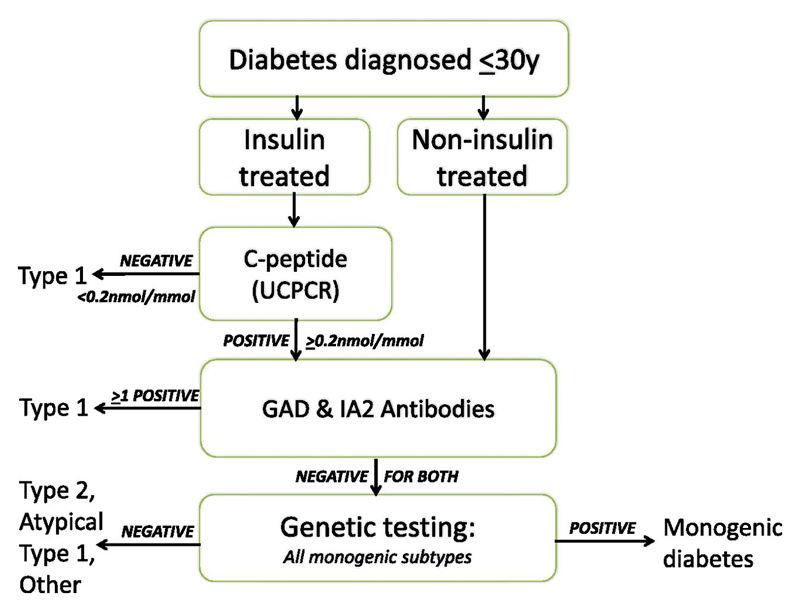
All recruited patients followed the biomarker screening pathway (Figure 1):
-
Assessment of endogenous insulin, in insulin treated patients, using urinary C-peptide creatinine ratio (UCPCR)
A key determinant of requirement for insulin treatment is lack of endogenous insulin secretion, and UCPCR is an easy screening test that can be done at home. UCPCR was used to rule out the majority of Type 1 patients in the first stage of screening, with minimal patient burden.
Insulin treated patients were asked to collect a urine sample two hours after the largest, carbohydrate containing meal of the day and to post this direct to the laboratory in a pack provided to allow analysis within 72 hours of sample collection, in line with assay stability(12).
Urinary C-peptide was measured by an electrochemiluminescence immunoassay (intra-assay coefficient of variation, 3.3%; interassay coefficient of variation, 4.5%) on a Roche Diagnostics E170 analyzer (Mannheim, Germany)(12). The lower limit of the C-peptide assay was 0.03nmol/l. Urinary creatinine was analyzed on the Roche P800 platform using creatinine Jaffé reagent (standardized against isotope dilution mass spectrometry) and used to calculate UCPCR (nmol/mmol). Patients with UCPCR≥0.2nmol/mmol were considered to have significant endogenous insulin secretion(10).
-
Islet autoantibody measurement in patients with significant endogenous insulin
Islet autoantibodies (GAD and IA2) were measured in patients who tested positive for UCPCR (UCPCR≥0.2nmol/mmol) or who were non-insulin treated. In order to minimize taking blood samples, particularly in children, the local pathology databases were checked for previous GAD and IA2 results and these were used if available. Patients with no previous islet autoantibody results were invited to attend an appointment with the study’s research nurse to provide blood samples for islet autoantibody testing and DNA.
GAD and IA2 antibody analysis was performed using commercial ELISA assays (RSR Ltd., Cardiff, UK) and a Dynex DSX automated ELISA system (Launch Diagnostics, Longfield, UK)(13). Both methods are highly specific and sensitive, (GAD antibodies 98% and 84% and IA-2 antibodies 99% and 74%, respectively). The laboratory participates in the Diabetes Autoantibody Standardization Programme. Patients were considered positive for antibodies if their results were >99th centile (64 WHOunits/ml for GAD and 15 WHOunits/ml for IA2)(13).
-
Diagnostic molecular genetic testing for monogenic diabetes in patients with significant endogenous insulin and negative antibody results
-
a)
Sequencing of three MODY genes, the most common forms of monogenic diabetes.
For all patients who were negative for both GAD and IA2 antibodies with significant endogenous insulin, DNA sequencing of HNF1A, HNF4A and GCK was performed by PCR amplification of purified genomic DNA, followed by Sanger DNA sequencing of each gene’s exons and flanking intronic regions. Dosage analysis of HNF1A, HNF4A and GCK for partial and whole gene deletions was also performed by multiplex ligation-dependent probe amplification (MLPA) using the MRC-Holland MODY MLPA kit-P241-B1.
-
b)
Targeted next generation sequencing for 35 genes in which mutations are known to cause monogenic diabetes. If no pathogenic mutation was identified in HNF1A, HNF4A or GCK, further targeted next generation sequencing was performed for mutations in 35 monogenic diabetes genes (all genes where mutations are known to cause MODY, neonatal diabetes, and other genetic diabetes syndromes), using a custom Agilent SureSelect exon-capture assay (Agilent Technologies, Santa Clara, CA, USA)(17) (see supplemental materials and Supplemental Table S1 for methodology, sensitivity and details of genes tested).
-
a)
Figure 1.
The UNITED biomarker screening pathway to investigate etiology of diabetes in patients diagnosed ≤30y: Genetic testing is carried out on all patients who have endogenous insulin (UCPCR≥0.2nmol/mmol) and who do not have either GAD or IA2 islet autoantibodies. Patients without endogenous insulin or have GAD and/or IA2 islet autoantibodies are classed as having Type 1 diabetes.
Statistical analysis
For comparing new cases diagnosed through the screening pathway to known cases of monogenic diabetes, and for comparing the biomarker screening pathway with an approach using clinical features (including the MODY probability calculator(9)) to detect monogenic diabetes, variables were categorical and so chi-squared and Fisher’s exact tests were used.
Prevalence of MODY
The prevalence of MODY in this population was determined as the proportion of positive cases, (including both known MODY that were recruited and those identified through the study), out of the total recruited.
To determine whether there was any potential bias in recruitment of MODY patients that may affect our prevalence estimate, we also obtained summary data on the number of patients with previously confirmed monogenic diabetes in each study area who had not been recruited into this study.
Positive and Negative Predictive Values of Pathway
Calculating the prevalence in this population allows us to determine the positive and negative predictive values for the pathway, the most important statistics for the clinician. Positive and negative predictive values were calculated as:
where pre-test odds is prevalence/(1-prevalence) and positive likelihood ratio is sensitivity/(1-specificity). Positive predictive value (PPV; equivalent to post-test probability) is post-test odds/(1+post-test odds). Negative predictive value (NPV) was calculated similarly, but using a negative likelihood ratio (1-sensitivity/specificity), with negative post-test probability equal to 1 - NPV. Number needed to test was calculated as 1/PPV.
Performance of the pathway – sensitivity and specificity
The key question is how well, if applied to a whole population, do the biomarkers perform in a pathway for identifying new cases. Screening literature emphasises the difference between programme sensitivity/specificity and test sensitivity/specificity, where assessing the sensitivity/specificity of a screening programme such as this, necessarily requires approximation using multiple data sources(18). As this was a population based study, rather than a case-control study, formal assessments of sensitivity and specificity (as normally conducted using a 2x2 table) of the pathway were limited due to the rarity of monogenic diabetes (meaning a small sample size of true positive cases of monogenic diabetes), and the expense of genetic testing (restricting confirmation of all the true negative non-monogenic cases).
Assessments of sensitivity of the components of the pathway for detecting monogenic diabetes have been carried out in larger case control cohorts (n=508 monogenic diabetes cases for islet autoantibodies (99% sensitivity)(13), n=160 for UCPCR (99% sensitivity, both studies combined(10; 11))), so it is more appropriate to use these estimates. We assumed a 98% sensitivity for both combined, based on these larger studies (assuming 1% missed due to false negative UCPCR and 1% due to false positive islet autoantibodies). However, the detection rate in all true monogenic cases in this pathway will be calculated for comparison.
Calculation of the specificity is limited as we have not performed genetic testing on all C-peptide negative patients. Previous larger studies have shown <1% of patients are missed(10; 11; 13). However, specificity of the biomarkers in these studies was assessed using gold standard Type 1 diabetes as the comparison group, rather than all non-MODY patients in this age range, and so likely overestimates the performance due to spectrum bias(19). We therefore, calculated specificity based on one minus the false positive rate of the pathway (i.e. proportion UCPCR positive/antibody negative, but not having a confirmed diagnosis of monogenic diabetes on subsequent genetic testing). This assumes all patients negative according to the pathway are true negatives. As an additional test of this assumption, a subset of patients negative for islet autoantibodies received genetic testing for the 3 main MODY genes and the proportion of MODY was calculated.
Health economic evaluation of the pathway is addressed in a separate paper (Peters et al. manuscript under review, protocol and conference abstract available(20; 21)).
Results
Subjects
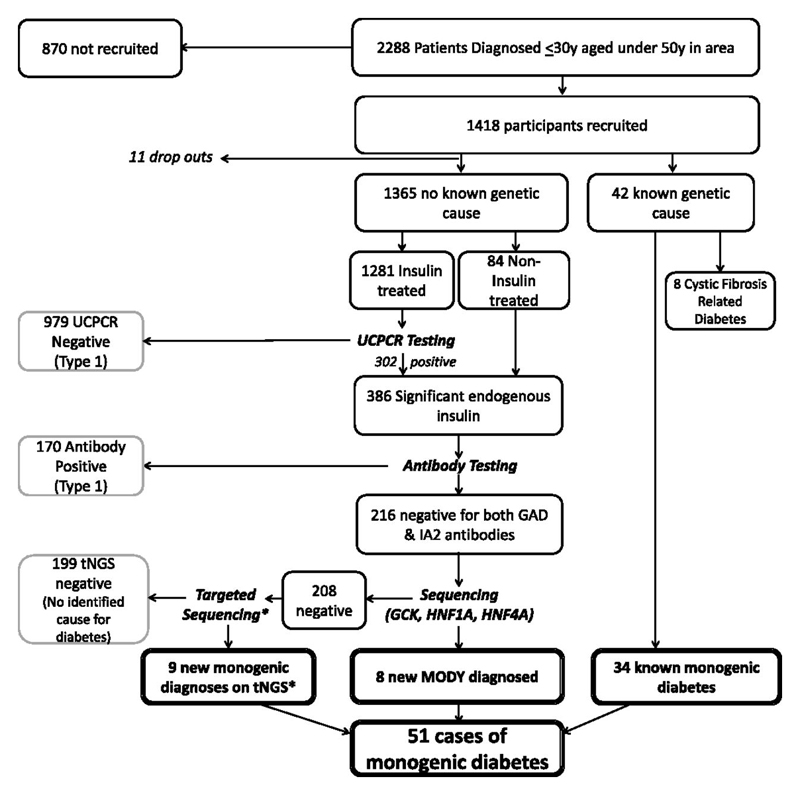
The flow of subjects through the study is shown in Figure 2. 2288 patients were eligible in area and 1418 subjects (62%) in total consented to the study and were recruited: 716 from the Exeter area, 702 from Dundee. 11 patients dropped out (9 did not provide blood samples for antibody testing, and 2 did not provide samples for DNA testing). Of the 1407 remaining patients, 1365 had no known genetic cause for their diabetes. Characteristics of these patients are shown in Supplemental Table S2 and subsequent results on the screening pathway are based on these patients. 42 patients had a known genetic cause for their diabetes prior to participating in this pathway: 34 patients had confirmed monogenic diabetes (see Table 2 for details) and 8 patients had Cystic-fibrosis related diabetes.
Figure 2.
Flow chart of patients recruited as part of UNITED. Biomarker screening pathway in 1376 patients with no known genetic cause for their diabetes in Exeter and Tayside. 11 dropped out. 17 new cases of monogenic diabetes detected (*one case identified through exome sequencing)
Table 2.
Positive and negative predictive values for the biomarker pathway, traditional MODY criteria (age at diagnosis <25y, non-insulin treated, parent affected with diabetes) and the MODY probability calculator (using a probability >25%, the pick-up rate for the diagnostic laboratory). Prevalence is the proportion of diagnosed monogenic diabetes, PPV is the positive predictive value, NPV is the negative predictive value, percentage of monogenic cases missed is the proportion of monogenic cases not picked up by the approach, and number needed to test is 1/PPV.
| N | Prevalence of monogenic diabetes | PPV (%) | NPV (%) | % of monogenic cases missed | Number needed to test | |
| Biomarker pathway | 1407 | 3.6% (51/1407) | 20.0% | 99.91% | 0% | 5 |
| Traditional MODY criteria | 1362 | 3.6% (49/1362) | 57.6% | 97.7% | 63% | 2 |
| MODY Probability calculator | 1347 | 3.3% (45/1347) | 40.4% | 98.3% | 55% | 3 |
Biomarker screening pathway identifies 17 new cases of monogenic diabetes (Figure 2)
Excluding drop-outs, 1281 (94%) of 1365 patients with no known genetic cause for their diabetes were insulin treated and provided a sample for UCPCR testing. 2 patients were anuric due to renal failure and so went straight on to antibody testing. 979 of these patients (76%) had minimal endogenous insulin secretion (UCPCR <0.2nmol/mmol) indicating a diagnosis of Type 1 diabetes, so received no further testing
Islet autoantibodies were tested in the 84 non-insulin treated patients, 300 UCPCR positive patients, and the 2 anuric patients. 170/386 (44%) tested positive for GAD and/or IA2 antibodies, confirming islet autoimmunity and hence a diagnosis of Type 1 diabetes. So these patients received no further testing.
Sanger sequencing for the 3 commonest MODY genes was undertaken in 216 patients (16% of the whole cohort). 8 patients tested positive confirming a diagnosis of MODY: 5 HNF1A, 2 HNF4A, and 1 GCK (Table 1, Figure 2).
Table 1.
Characteristics of patients diagnosed with monogenic diabetes and details of mutations found for a) those recruited but diagnosed with monogenic diabetes prior to the study, and b) those diagnosed as a result of the biomarker pathway. References for the genes and further details of the mutations are in Supplemental Table S3.
| a) Recruited but diagnosed prior to the UNITED study | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic characteristics | Clinical characteristics | ||||||||||
| ID | Gene | Method | Zygosity | DNA level desc | Age dx | Treatment | Parent DM | BMI | HbA1c | Age rec | Additional clinical features |
| 211 | GCK | Sanger | Het | c.97_117dup | 3 | Diet | Yes | 34.0 | 48 | 16 | |
| 523 | GCK | Sanger | Het | c.97_117dup | 27 | Diet | No | 51.7 | 55 | 43 | |
| 537 | GCK | Sanger | Het | c.683C>T | 11 | Diet | Yes | - | - | 13 | |
| 538 | GCK | Sanger | Het | c.683C>T | 9 | Diet | Yes | - | - | 11 | |
| 542 | GCK | Sanger | Het | c.184G>A | 29 | Diet | Yes | 38.9 | 48 | 39 | |
| 543 | GCK | Sanger | Het | c.184G>A | 4 | Diet | Yes | - | - | 4 | |
| 544 | GCK | Sanger | Het | c.184G>A | 3 | Diet | Yes | - | - | 5 | |
| 1155 | GCK | Sanger | Het | c.1343G>T | 25 | Diet | Yes | 19.3 | 50 | 25 | |
| 82095 | GCK | Sanger | Het | c.1019G>T | 9 | Diet | Yes | 21.9 | 45 | 14 | |
| 535 | HNF1A | Sanger | Het | c.379_381del | 24 | OHA | Yes | 25.9 | 51 | 47 | |
| 547 | HNF1A | Sanger | Het | c.1748G>A | 22 | Diet | Yes | 24.4 | 40 | 30 | Low renal threshold |
| 554 | HNF1A | Sanger | Het | c.872dup | 18 | OHA | Yes | 30 | 86 | 39 | |
| 566 | HNF1A | Sanger | Het | c.872dup | 17 | OHA | Yes | 29.2 | 51 | 42 | Sulphonylurea sensitivity, low renal threshold |
| 603 | HNF1A | Sanger | Het | c.1420C>T | 20 | OHA | Yes | 26.5 | 56 | 42 | Low renal threshold |
| 617 | HNF1A | Sanger | Het | c.779C>T | 25 | Diet | Yes | 25.4 | 44 | 26 | |
| 892 | HNF1A | Sanger | Het | c.476G>A | 14 | Insulin | Yes | 30.0 | 63 | 40 | |
| 1370 | HNF1A | Sanger | Het | c.872dup | 21 | Diet | Yes | 36.1 | 83 | 21 | |
| 1409 | HNF1A | Sanger | Het | c.872dup | 21 | OHA+Ins | Yes | 32.8 | 95 | 42 | Sulphonylurea sensitivity |
| 80480 | HNF1A | Sanger | Het | c.1093_1107+6del | 19 | OHA | - | 22.9 | 73 | 40 | |
| 82261 | HNF1A | Sanger | Het | c.185del | 12 | OHA+Ins | Yes | 23.7 | 73 | 25 | Low renal threshold |
| 82276 | HNF1A | Sanger | Het | c.434C>T | 13 | Insulin | Yes | 23.8 | 60 | 27 | |
| 82301 | HNF1A | Sanger | Het | c.1340C>T | 20 | OHA | Yes | 27.4 | 91 | 37 | |
| 82310 | HNF1A | Sanger | Het | c.185del | 18 | OHA | Yes | 24.4 | 48 | 45 | Low renal threshold |
| 82374 | HNF1A | Sanger | Het | c.1093_1107+6del | 19 | OHA | Yes | 23.8 | 83 | 20 | Sulphonylurea sensitivity |
| 82258 | HNF4A | Sanger | Het | c.322G>A | 28 | Insulin | Yes | 20.9 | 60 | 31 | |
| 600 | HNF1B | Sanger | Het | c.982_986del | 20 | Insulin | No | 23.4 | 122 | 35 | Renal cysts |
| 82033 | HNF1B | Sanger | Het | c.466A>G | 17 | Insulin | No | 25.3 | 54 | 35 | Genital tract malformations, renal hypoplasia |
| 82006 | KCNJ11 | Sanger | Het | c.601C>T | 0 | OHA | No | 26.6 | 33 | 35 | Diagnosed at 12 weeks of age |
| 539 | LMNA | Sanger | Het | c.1930C>T | 17 | OHA+Ins | Yes | 24.2 | 114 | 49 | Lipodystrophy |
| 595 | LMNA | Sanger | Het | c.1444C>T | 21 | OHA+Ins | Yes | 25.1 | 62 | 34 | |
| 604 | 3243 | Hp | m.3243A>G | 27 | Insulin | Yes | 26.9 | 54 | 36 | ||
| 80541 | 3243 | Hp | m.3243A>G | 28 | Insulin | Yes | 26.4 | 83 | 48 | ||
| 82399 | 3243 | Hp | m.3243A>G | 29 | Insulin | Yes | 26.4 | 56 | 41 | Deafness | |
| 540 | NEUROD1 | Sanger | Het | c.616dup | 21 | OHA+Ins | Yes | 49.8 | 83 | 36 | Lipodystrophy and necrobiosis |
| b) Identified as part of the biomarker pathway | |||||||||||
| 82372 | GCK | Sanger | Het | c.1340G>A | 18 | Diet | No | 25.5 | 46 | 19 | |
| 82316 | HNF4A | Sanger | Het | c.1064-5_1070del | 14 | Diet | Yes | 32.3 | 38 | 33 | |
| 377 | HNF4A | Sanger | Het | c.-12G>A | 11 | Insulin | Yes | 28.4 | 104 | 14 | |
| 80089 | HNF1A | Sanger | Het | c.1349dup | 30 | Insulin | Yes | 31.0 | 72 | 48 | |
| 80170 | HNF1A | Sanger | Het | c.391C>T | 21 | Insulin | No | 23.5 | 52 | 35 | Low renal threshold |
| 80173 | HNF1A | Sanger | Het | c.495G>C | 17 | Insulin | Yes | 24.5 | 56 | 46 | |
| 82003 | HNF1A | Sanger | Het | c.28A>C | 26 | Diet | Yes | 29.8 | 73 | 26 | |
| 82352 | HNF1A | Sanger | Het | c.814C>T | 13 | Insulin | Yes | 32.3 | 91 | 45 | |
| 82013 | HNF1A | tNGS | Het | c.-258A>G | 24 | OHA | Yes | 39.6 | 75 | 43 | |
| 307 | HNF1B | tNGS | Het | c.1-?_1674+?del | 29 | Insulin | No | 22.7 | 62 | 31 | Aspergers, renal cysts, low fecal elastase low magnesium |
| 82014 | NEUROD1 | tNGS | Het | c.616dup | 21 | OHA | No | 35.3 | 88 | 31 | |
| 183 | NEUROD1 | tNGS | Het | c.616dup | 29 | Insulin | No | 27.1 | 55 | 46 | |
| 82010 | 3243 | tNGS | Hp | m.3243A>G | 27 | OHA+Ins | Yes | 28.6 | 91 | 46 | |
| 82038 | PPARG | tNGS | Het | c.1154G>A | 22 | OHA | No | 26.6 | 53 | 36 | Lipodystrophy, acanthosis |
| 80925 | TRMT10A | tNGS | Hom | c.79G>T | 23 | OHA+Ins | No | 33.0 | 69 | 28 | Microcephaly, learning difficulties, epilepsy |
| 17 | WFS1 | tNGS | C/Het | c.874C>T & c.877del | 20 | Insulin | n/k | 21.8 | 42 | 24 | Bilateral Optic atrophy, neurogenic bladder, diet treatment, muscle pain on exercise |
| 175 | POLD1 | Exome | Het | c.1812-1814del | 14 | OHA | no | 18.6 | 30 | 21 | total lipodystrophy, sensori-neural deafness, mandibular hypoplasia, hypogonadism, undescended testes, severe insulin resistance |
Of the 208 who tested negative for the common MODY genes, additional testing by targeted next generation sequencing identified mutations in genes associated with monogenic diabetes in a further 8 patients and 1 patient had a mutation in POLD1 identified through exome sequencing (see Table 1, Figure 2).
New cases of monogenic diabetes identified were more likely to be rarer causes and atypical (Supplemental Figure S1)
More of the new cases of monogenic diabetes identified had mutations in genes other than the 3 most common forms of MODY. (25/34 (74%) of those diagnosed prior to the study had mutations in HNF1A, HNF4A, or GCK compared with 8/17 (47%) identified from Sanger sequencing as part of the biomarker screening pathway, p=0.06). Those diagnosed with monogenic diabetes as part of the study were less likely to have a parent known to be affected than those with a previous known monogenic diagnosis (8/17 (47%) v 29/34 (85%), p=0.007).
Minimum prevalence of monogenic diabetes of 3.6% in those diagnosed <30y, currently under 50y
We found 51 cases of monogenic diabetes (which represents a further 50% (n=17) in addition to the 34 previously diagnosed) out of 1407 recruited patients providing a prevalence of 3.6% (95% CI: 2.7 to 4.7%) in patients diagnosed under 30 and currently under 50 years.
From the database of UK referrals, we identified 26 patients with a diagnosis of monogenic diabetes in the Exeter and Tayside regions, who met study inclusion criteria, but were not recruited to the UNITED study. Therefore, the proportion of known monogenic diabetes prior to the study in the recruited population (34/1407 (2.4%)) was similar to the proportion in the non-recruited population (26/870 (3.0%)) (p=0.4), suggesting no overall bias in recruitment. More of the non-recruited cases had MODY caused by mutations in the GCK gene but this was not significant given the small numbers (46% v 26%, p=0.1). There was no difference in terms of age at diagnosis (mean 18 v 19y, p=0.5), age at time of recruitment (using 2011 for non-recruited patients) (32 v 32y, p=0.98) or gender (35% v 45% male, p=0.4).
Performance of the pathway (Table 2)
In line with what was expected given larger studies of the diagnostic accuracy of UCPCR and islet autoantibodies and the known pathophysiology of monogenic diabetes, all cases with previously diagnosed monogenic diabetes who provided all samples for the pathway (n=21) were UCPCR positive and antibody negative. Similarly, all antibody positive patients with DNA available (n=47) tested negative for the three main MODY genes so no additional MODY cases were picked up in this group.
199/1348 (15%) of patients were put forward for genetic testing who were not found to have monogenic diabetes (i.e. 15% false positive rate, so 85% specificity). Assuming a 98% sensitivity and 85% specificity, the positive predictive value for the pathway is 20%, suggesting a 1 in 5 pick-up rate for monogenic diabetes, a 5.6-fold increase in probability over the background prevalence alone. The negative predictive value was 99.9%, indicating the probability of having monogenic diabetes if you are UCPCR negative or islet autoantibody positive is 0.1% (1 in 1000).
Comparison of biomarker screening pathway with clinical features (Table 2)
If genetic testing had been limited to the standard clinical criteria for MODY (age at diagnosis <25y, non insulin requiring and a parent known to be affected with diabetes), fewer patients would have required testing (n=33) leading to a higher pick-up rate and positive predictive value (PPV=57.6%) than the biomarker pathway, but the majority of monogenic cases would have been missed (63% compared with 0% for the biomarker pathway). The MODY probability calculator also had a higher positive predictive value (PPV=40.4%), but missed more cases (55%) compared with the biomarker pathway.
Conclusions
The biomarker screening pathway for monogenic diabetes is a systematic, cheap (UK UCPCR cost=£10.80, antibodies cost=£20), and easily implemented approach to screening all patients with young-onset diabetes in a clinic or population that helps identify suitable patients for molecular diagnostic genetic testing. The pathway picked up new cases of monogenic diabetes, even in areas of existing high detection due to research interests in the regions. We found 3.6% of patients diagnosed less than 30 years of age have monogenic diabetes. In areas where no cases have been identified, we estimate that 1 in 5 patients referred for genetic testing as a result of the pathway will have monogenic diabetes which is a 5.6-fold higher detection rate than if all patients in this age range received genetic testing. The high negative predictive value of 99.9% indicates it is an extremely effective approach for ruling out monogenic diabetes.
There have been relatively few studies that have systematically screened whole populations for monogenic diabetes. The majority of studies have been in pediatric populations only(14; 15; 22–26), with only two studies that have screened adults(27; 28). No other study has systematically screened a whole population of both adults and children together. Only 8/51 (16%) of patients with a genetic diagnosis of monogenic diabetes in our cohort were in the pediatric age range (<20y) at the time of recruitment, highlighting the importance of looking for monogenic diabetes in adult diabetes clinics. This may explain why the prevalence we find is higher than any of the previous pediatric studies.
The strength of our pathway is the integration of two biomarkers (C-peptide and islet autoantibodies (both GAD and IA2)), rather than relying on clinical features. This offers a simple approach that does not require specific clinician interpretation or complex algorithms of different combinations of features. We showed that by using clinical features alone over half the cases of monogenic diabetes would be missed. By combining the two biomarkers we increase the discriminatory ability and allow the clinician to pick up even atypical cases and rarer forms of monogenic diabetes, which traditional criteria may miss. The use of clinical features, however, results in fewer cases being sent for genetic testing that are negative, which clearly has cost implications. The most cost effective approach is likely to involve a combination of biomarkers and clinical features. Further studies are needed to determine whether the pick-up rate could be further improved by integrating the pathway with clinical features, such as the MODY calculator, or whether this would result in more missed patients as a consequence of reduced testing.
In this study we also systematically tested all known genes for monogenic diabetes, rather than just the most common MODY genes (GCK, HNF1A and HNF4A). 9/17 (53%) of the cases identified as part of our cohort had mutations identified through additional testing on the targeted capture and 17/51 (33%) of all the monogenic diabetes cases found in total had mutations in other genes, highlighting the advantage of further testing using targeted next generation sequencing.
Health economic evaluation of the pathway for detecting the common forms of MODY (GCK, HNF1A and HNF4A) has been carried out as a separate project, which has shown the pathway to be cost-saving (Peters et al, manuscript under review, abstract/protocol available(20; 21)). The cost-effectiveness of additional testing for other forms of monogenic diabetes has not been assessed. Due to the rarity of other monogenic diabetes there are little data available to inform such analyses. Treatment change from insulin to sulphonylureas is still possible in cases diagnosed with ABCC8 and KCNJ11(29; 30), and for other genes where treatment change is not an option, a confirmed diagnosis can still help with management, prognosis and advice on risk to other family members(4). The decision whether to pay for the more expensive, but more comprehensive next generation sequencing, rather than Sanger sequencing for MODY genes only, would depend on assessing the trade-offs of additional costs with long term benefits to the patient. The presence of additional clinical features (e.g. renal cysts associated with HNF1B) may also point to specific monogenic diagnoses and increase the likelihood of a positive genetic test result.
A limitation of our study was that we had small numbers of patients with monogenic diabetes on which to evaluate the sensitivity of the pathway. Considerably larger studies have shown the biomarkers individually to be highly sensitive for monogenic diabetes (99% for UCPCR(10; 11) and >99% for islet autoantibodies(13)), and by using both of these markers in a pathway the number of missed cases should be minimal at a population level (2% of 3.6% = 0.07%, reflected in the NPV of 99.9%). Although there have been reports of MODY patients who are positive for islet autoantibodies (reviewed in (13)), these are rare and are likely to be cases with coincidental Type 1 diabetes. Previous studies reporting high prevalence of positive autoantibodies in their cohort have included clinically defined, rather than genetically confirmed MODY(31) or use low cutoffs for antibody positivity, which can be inappropriate(32), so are likely to represent an overestimate. There is also the potential for missed cases based on UCPCR, but again, the number of these patients will be small, and as they have insulin levels suggestive of Type 1 diabetes(33) they are unlikely to be able to transfer off insulin even if a genetic diagnosis is made.
A further limitation is that despite screening using C-peptide and antibody testing the positive predictive value is still fairly low at 20%, indicating 4 out of 5 screened will not have a monogenic cause identified on diagnostic molecular genetic testing. However, the aim of our screening pathway is that it is used purely as a tool to narrow down those individuals who would be more appropriate for genetic testing. This approach is a vast improvement over no screening (which would represent a PPV at the background prevalence rate of 3.6%), misses fewer cases than using clinical features alone, and is at a level that has been shown to be cost-effective (manuscript under review, protocol and abstract available(20; 21)). Furthermore, the screening pathway still provides useful test results for this age group that offer additional information to support patient care. Patients with severe insulin deficiency as determined by very low C-peptide values, will not respond to non-insulin therapy(33). Positive C-peptide and negative antibody results are important clinically to highlight atypical cases of Type 1 diabetes or where other forms of diabetes, such as young-onset Type 2 diabetes should be considered. Patients with very high endogenous insulin without islet autoantibodies and no mutations in monogenic diabetes genes, are likely to have Type 2 diabetes, and may be able to manage on non-insulin treatment.
Finally, this study comprised a 98% White Caucasian population and assesses patients at a median of 14 years after diagnosis. Assessment of the pathway in other racial groups and in patients close to diagnosis is needed.
In conclusion, we have demonstrated a simple, cheap, effective screening pathway that could be implemented at a population level to help correctly diagnose patients with monogenic diabetes.
Supplementary Material
Acknowledgements
This study was funded by the Department of Health and Wellcome Trust Health Innovation Challenge Award (HICF-1009-041; WT-091985). ATH and SE are Wellcome Trust Senior Investigators. ATH is an NIHR Senior Investigator. BS, ATH, MH, SE, and BK are core members of the NIHR Exeter Clinical Research Facility. EP is a Wellcome Trust New Investigator. TM is supported by NIHR CSO Fellowship. JP is partly funded by the NIHR Collaboration for Leadership in Applied Health Research and Care for the South West (PenCLAHRC).
Footnotes
Author contributions:
BS carried out all analysis and drafted the manuscript. MS collected data and reviewed/edited the manuscript. MH coordinated the UNITED study and reviewed/edited the manuscript. TM coordinated the C-peptide and islet autoantibody testing and reviewed/edited the manuscript. KC coordinated the genetic testing and reviewed/edited the manuscript. JP provided input on analysis and reviewed/edited the manuscript. BK wrote the protocol and ethics application and reviewed/edited the manuscript. CH provided input on study design and analysis and reviewed/edited the manuscript. SE led the genetic testing and reviewed/edited the manuscript. EP designed the study and led the Tayside arm of the project and reviewed/edited the manuscript. AH designed the study and led the Exeter arm of the project and reviewed/edited the manuscript.
Declaration of interests: No conflicts of interest.
BS had full access to all the data in the study and had final responsibility for the decision to submit for publication
References
- 1.Diabetes UK. Diabetes in the UK 2012: Key statistics on diabetes. 2012 [Google Scholar]
- 2.Scottish Diabetes Survey Monitoring Group NS. Scottish Diabetes Survey 2014. 2014 [Google Scholar]
- 3.Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53:2504–2508. doi: 10.1007/s00125-010-1799-4. [DOI] [PubMed] [Google Scholar]
- 4.Rubio-Cabezas O, Hattersley AT, Njolstad PR, Mlynarski W, Ellard S, White N, Chi DV, Craig ME, International Society for P, Adolescent D ISPAD Clinical Practice Consensus Guidelines 2014. The diagnosis and management of monogenic diabetes in children and adolescents. Pediatric diabetes. 2014;15(Suppl 20):47–64. doi: 10.1111/pedi.12192. [DOI] [PubMed] [Google Scholar]
- 5.Zeitler P, Fu J, Tandon N, Nadeau K, Urakami T, Barrett T, Maahs D, International Society for P, Adolescent D ISPAD Clinical Practice Consensus Guidelines 2014. Type 2 diabetes in the child and adolescent. Pediatric diabetes. 2014;15(Suppl 20):26–46. doi: 10.1111/pedi.12179. [DOI] [PubMed] [Google Scholar]
- 6.Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabetic medicine : a journal of the British Diabetic Association. 2009;26:437–441. doi: 10.1111/j.1464-5491.2009.02690.x. [DOI] [PubMed] [Google Scholar]
- 7.Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362:1275–1281. doi: 10.1016/S0140-6736(03)14571-0. [DOI] [PubMed] [Google Scholar]
- 8.Stride A, Shields B, Gill-Carey O, Chakera AJ, Colclough K, Ellard S, Hattersley AT. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia. 2014;57:54–56. doi: 10.1007/s00125-013-3075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields BM, McDonald TJ, Ellard S, Campbell MJ, Hyde C, Hattersley AT. The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia. 2012;55:1265–1272. doi: 10.1007/s00125-011-2418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besser RE, Shepherd MH, McDonald TJ, Shields BM, Knight BA, Ellard S, Hattersley AT. Urinary C-peptide creatinine ratio is a practical outpatient tool for identifying hepatocyte nuclear factor 1-{alpha}/hepatocyte nuclear factor 4-{alpha} maturity-onset diabetes of the young from long-duration type 1 diabetes. Diabetes care. 2011;34:286–291. doi: 10.2337/dc10-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besser RE, Shields BM, Hammersley SE, Colclough K, McDonald TJ, Gray Z, Heywood JJ, Barrett TG, Hattersley AT. Home urine C-peptide creatinine ratio (UCPCR) testing can identify type 2 and MODY in pediatric diabetes. Pediatric diabetes. 2013;14:181–188. doi: 10.1111/pedi.12008. [DOI] [PubMed] [Google Scholar]
- 12.McDonald TJ, Knight BA, Shields BM, Bowman P, Salzmann MB, Hattersley AT. Stability and reproducibility of a single-sample urinary C-peptide/creatinine ratio and its correlation with 24-h urinary C-peptide. Clinical chemistry. 2009;55:2035–2039. doi: 10.1373/clinchem.2009.129312. [DOI] [PubMed] [Google Scholar]
- 13.McDonald TJ, Colclough K, Brown R, Shields B, Shepherd M, Bingley P, Williams A, Hattersley AT, Ellard S. Islet autoantibodies can discriminate maturity-onset diabetes of the young (MODY) from Type 1 diabetes. Diabetic medicine : a journal of the British Diabetic Association. 2011;28:1028–1033. doi: 10.1111/j.1464-5491.2011.03287.x. [DOI] [PubMed] [Google Scholar]
- 14.Pihoker C, Gilliam LK, Ellard S, Dabelea D, Davis C, Dolan LM, Greenbaum CJ, Imperatore G, Lawrence JM, Marcovina SM, Mayer-Davis E, et al. Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. The Journal of clinical endocrinology and metabolism. 2013;98:4055–4062. doi: 10.1210/jc.2013-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd M, Shields B, Hammersley S, Hudson M, McDonald TJ, Colclough K, Oram RA, Knight B, Hyde C, Cox J, Mallam K, et al. Systematic Population Screening, Using Biomarkers and Genetic Testing, Identifies 2.5% of the U.K. Pediatric Diabetes Population With Monogenic Diabetes. Diabetes care. 2016;39:1879–1888. doi: 10.2337/dc16-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thanabalasingham G, Owen KR. Diagnosis and management of maturity onset diabetes of the young (MODY) BMJ. 2011;343:d6044. doi: 10.1136/bmj.d6044. [DOI] [PubMed] [Google Scholar]
- 17.Ellard S, Lango Allen H, De Franco E, Flanagan SE, Hysenaj G, Colclough K, Houghton JA, Shepherd M, Hattersley AT, Weedon MN, Caswell R. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013;56:1958–1963. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole P, Morrison AS. Basic issues in population screening for cancer. J Natl Cancer Inst. 1980;64:1263–1272. [PubMed] [Google Scholar]
- 19.Ransohoff DF, Feinstein AR. Problems of spectrum and bias in evaluating the efficacy of diagnostic tests. N Engl J Med. 1978;299:926–930. doi: 10.1056/NEJM197810262991705. [DOI] [PubMed] [Google Scholar]
- 20.Peters JL, Anderson R, Hyde C. Development of an economic evaluation of diagnostic strategies: the case of monogenic diabetes. BMJ open. 2013;3 doi: 10.1136/bmjopen-2013-002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters JL, Anderson R, Shields BM, King SM, Hudson M, Shepherd M, McDonald TJ, Pearson E, Hattersley AT, Hyde C. Strategies to identify individuals with MODY: results of a health economic model. Diabetic Medicine 2016. 2016;33:158. [Google Scholar]
- 22.Chambers C, Fouts A, Dong F, Colclough K, Wang Z, Batish SD, Jaremko M, Ellard S, Hattersley AT, Klingensmith G, Steck AK. Characteristics of maturity onset diabetes of the young in a large diabetes center. Pediatric diabetes. 2015 doi: 10.1111/pedi.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandica RG, Chung WK, Deng L, Goland R, Gallagher MP. Identifying monogenic diabetes in a pediatric cohort with presumed type 1 diabetes. Pediatric diabetes. 2015;16:227–233. doi: 10.1111/pedi.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irgens HU, Molnes J, Johansson BB, Ringdal M, Skrivarhaug T, Undlien DE, Sovik O, Joner G, Molven A, Njolstad PR. Prevalence of monogenic diabetes in the population-based Norwegian Childhood Diabetes Registry. Diabetologia. 2013;56:1512–1519. doi: 10.1007/s00125-013-2916-y. [DOI] [PubMed] [Google Scholar]
- 25.Rubio-Cabezas O, Edghill EL, Argente J, Hattersley AT. Testing for monogenic diabetes among children and adolescents with antibody-negative clinically defined Type 1 diabetes. Diabetic medicine : a journal of the British Diabetic Association. 2009;26:1070–1074. doi: 10.1111/j.1464-5491.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- 26.Johansson BB, Irgens HU, Molnes J, Sztromwasser P, Aukrust I, Juliusson PB, Sovik O, Levy S, Skrivarhaug T, Joner G, Molven A, et al. Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia. 2016 doi: 10.1007/s00125-016-4167-1. [DOI] [PubMed] [Google Scholar]
- 27.Kropff J, Selwood MP, McCarthy MI, Farmer AJ, Owen KR. Prevalence of monogenic diabetes in young adults: a community-based, cross-sectional study in Oxfordshire, UK. Diabetologia. 2011;54:1261–1263. doi: 10.1007/s00125-011-2090-z. [DOI] [PubMed] [Google Scholar]
- 28.Thanabalasingham G, Pal A, Selwood MP, Dudley C, Fisher K, Bingley PJ, Ellard S, Farmer AJ, McCarthy MI, Owen KR. Systematic assessment of etiology in adults with a clinical diagnosis of young-onset type 2 diabetes is a successful strategy for identifying maturity-onset diabetes of the young. Diabetes care. 2012;35:1206–1212. doi: 10.2337/dc11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson ER, Flechtner I, Njolstad PR, Malecki MT, Flanagan SE, Larkin B, Ashcroft FM, Klimes I, Codner E, Iotova V, Slingerland AS, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med. 2006;355:467–477. doi: 10.1056/NEJMoa061759. [DOI] [PubMed] [Google Scholar]
- 30.Rafiq M, Flanagan SE, Patch AM, Shields BM, Ellard S, Hattersley AT, Neonatal Diabetes International Collaborative Group Effective treatment with oral sulfonylureas in patients with diabetes due to sulfonylurea receptor 1 (SUR1) mutations. Diabetes care. 2008;31:204–209. doi: 10.2337/dc07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schober E, Rami B, Grabert M, Thon A, Kapellen T, Reinehr T, Holl RW, DP-WIotGWGfPD Phenotypical aspects of maturity-onset diabetes of the young (MODY diabetes) in comparison with Type 2 diabetes mellitus (T2DM) in children and adolescents: experience from a large multicentre database. Diabetic medicine : a journal of the British Diabetic Association. 2009;26:466–473. doi: 10.1111/j.1464-5491.2009.02720.x. [DOI] [PubMed] [Google Scholar]
- 32.Bingley PJ. Clinical applications of diabetes antibody testing. The Journal of clinical endocrinology and metabolism. 2010;95:25–33. doi: 10.1210/jc.2009-1365. [DOI] [PubMed] [Google Scholar]
- 33.Jones AG, Hattersley AT. The clinical utility of C-peptide measurement in the care of patients with diabetes. Diabetic medicine : a journal of the British Diabetic Association. 2013;30:803–817. doi: 10.1111/dme.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.