Summary
The liver plays a pivotal role in metabolism and xenobiotic detoxification, processes that must be particularly efficient when animals are active and feed. A major question is how the liver adapts to these diurnal changes in physiology. Here, we show that, in mice, liver mass, hepatocyte size, and protein levels follow a daily rhythm, whose amplitude depends on both feeding-fasting and light-dark cycles. Correlative evidence suggests that the daily oscillation in global protein accumulation depends on a similar fluctuation in ribosome number. Whereas rRNA genes are transcribed at similar rates throughout the day, some newly synthesized rRNAs are polyadenylated and degraded in the nucleus in a robustly diurnal fashion with a phase opposite to that of ribosomal protein synthesis. Based on studies with cultured fibroblasts, we propose that rRNAs not packaged into complete ribosomal subunits are polyadenylated by the poly(A) polymerase PAPD5 and degraded by the nuclear exosome.
Introduction
Most mammals sequester their activity to certain time windows during the day. Accordingly, physiology follows daily rhythms, which are driven by the photoperiod and an endogenous circadian timing system. The latter is organized in a hierarchical network, composed of a master pacemaker located in the brain’s suprachiasmatic nuclei (SCN) and self-sustained and cell-autonomous oscillators in nearly all body cells (Partch et al., 2014; Schibler et al., 2015). The SCN, which is synchronized primarily by lightdark cycles, must periodically synchronize peripheral clocks in order to maintain phase coherence within the body. Feeding-fasting rhythms, driven by rest–activity cycles, are the most dominant zeitgebers for the circadian oscillators in peripheral organs (Damiola et al., 2000; Stokkan et al., 2001). This indicates that coordinating the timing of metabolism is a major purpose of peripheral clocks in many organisms.
The liver plays a pivotal role in the diurnal processing of nutrients and detoxification of harmful endo- and xenobiotic components. Accordingly, the accumulation and/or activities of many hepatic regulatory proteins and enzymes participating in these activities display robust daily rhythms. These include transcription factors governing lipid and carbohydrate homeostasis (e.g., members of the REV-ERB, PPAR, SREBP, and PAR bZip families), xenobiotic detoxification (e.g., CAR), and enzymes whose expression is under the control of such transcription factors and/or whose activities are tuned by substrate availability (for review, see Gerhart-Hines and Lazar, 2015). In the course of studies aimed at the identification of signaling pathways involved in the systemic regulation of circadian gene expression, we noticed that the size of hepatocytes seemed to be larger at the end of the activity/feeding phase (ZT0) than at its beginning (ZT12; ZT [zeitgeber time]; ZT0 and ZT12 are the times when lights are switched on and off, respectively; Gerber et al., 2013). If cell size indeed oscillated, the entire liver mass should undergo diurnal changes. Indeed, a few published studies reported on diurnal fluctuations in liver mass in birds (Fisher and Bartlett, 1957; Wilson and McFarland, 1969) and humans (Leung et al., 1986). However, it was not clear from these studies whether these oscillations were accompanied by similar changes in macromolecular content.
The data presented here suggest that hepatocyte size and global RNA and protein levels oscillate in a daily manner in the mouse liver and that these rhythms are driven by both feeding-fasting and light-dark cycles. In liver, ribosomes appear to be rate limiting for protein synthesis, and their number indeed oscillates during the day. Whereas pre-ribosomal RNA (pre-rRNA) is transcribed at nearly constant levels throughout the day, many ribosomal protein mRNAs are translated in a highly rhythmic fashion. We propose that excess rRNAs not assembled into complete ribosomal subunits are polyadenylated in the nucleus by the poly(A) polymerase PAPD5 and degraded by the exosome.
Results
Hepatocyte Sizes Follow Diurnal Oscillations that Depend on the Feeding Regimen
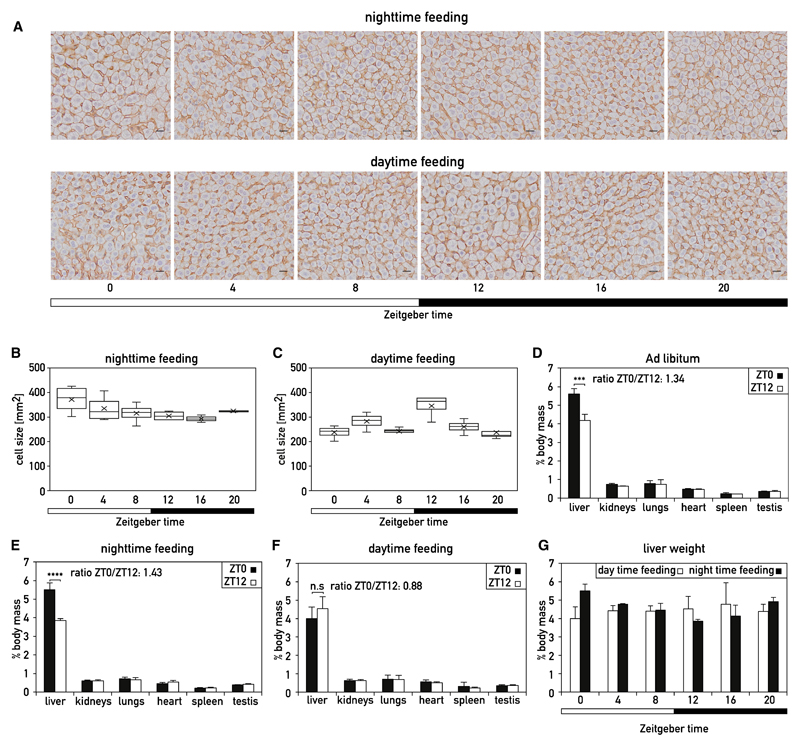
In a previous study, we noticed daytime-dependent changes in hepatocyte sizes in histological mouse liver sections (Gerber et al., 2013; Wang et al., 2017). To extend these observations, we prepared liver sections from mice exposed to different feeding regimens. Mice fed exclusively during the night (night-fed; note that mice are nocturnal and feed primarily during the night) or exclusively during the day (day-fed) were sacrificed at 4-hr intervals around the clock. Liver sections from these animals were stained with β-catenin and Mayer’s hematoxylin to visualize cell contours and nuclei, respectively (Wang et al., 2017; Figure 1A). The average hepatocyte surfaces were determined and are depicted in Figures 1B and 1C. Whereas hepatocyte sizes follow a smooth daily oscillation in night-fed mice, no clear rhythm is observed in day-fed mice, although in the latter, hepatocyte surfaces are somewhat larger at ZT12 than at other time points (ZT0 and ZT12 are the times when the lights are switched on and off, respectively).
Figure 1. Diurnal Changes in Liver Weight Depend on Feeding Cycles.
(A) Representative immunohistochemistry (IHC) images of liver sections from night-fed (top panel) and day-fed (bottom panel) mice sacrificed at 4-hr intervals around the clock. The scale bar represents 20 μm.
(B and C) Diurnal oscillations of hepatocytes areas extracted from IHC images, such as the ones depicted in (A). Each time point shows the median and SEM of four independent biological samples. X, average values. For each sample, more than 1,200 cells were analyzed.
(D–F) Masses of various organs at ZT0 and ZT12, expressed as percentages of total body weight, of male C57BL/6 mice subjected to light-dark cycles and fed ad libitum (D), exclusively during the night (E), and exclusively during the day (F). Data are represented as the mean ± SD for four mice per time point (D) and nine mice per time point (E and F). ***p < 0.001; ****p < 10−6; two-sided Student’s t test; n.s. means “not statistically significant“.
(G) Liver weight determined in night- or day-fed animals at 4-hr intervals around the clock. The Zeitgeber times are indicated below the panel.
See also Figure S1.
Diurnal Changes in Liver Weight Correlate with Oscillations in Cellular RNA and Protein Content
As shown above, cell size oscillates in night-fed animals, and we therefore anticipated daily fluctuation of the total liver mass in these animals. Indeed, we observed a marked difference in liver weight at ZT0 and ZT12 in these mice (Figure 1E), whereas the weight of kidneys, lungs, heart, spleen, and testis remained constant, irrespective of the feeding regimen (Figures 1D–1F, S1A, and S1B). Moreover, the difference in liver weight strongly depended on the feeding regimen. Thus, the ratio of liver weight measured at ZT0 and ZT12 increased from 1.34 in ad-lib-fed mice (Figure 1D) to 1.43 in night-fed mice (NF) (Figure 1E). Yet, in day-fed mice (DF), there was only a statistically insignificant trend of a liver weight increase during the feeding period (Figure 1F). As shown in Figure 1G, the liver mass oscillated in a smooth diurnal cycle in night-fed mice but remained nearly constant in day-fed mice.
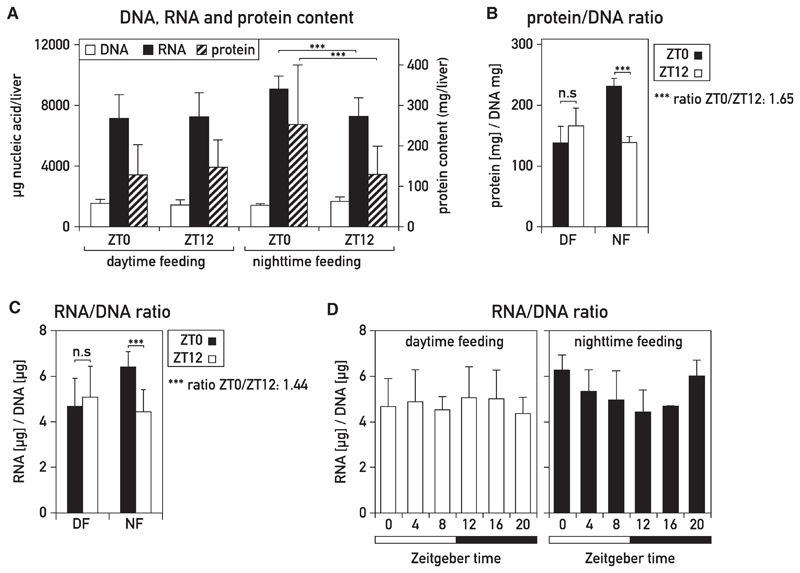
We examined whether the diurnal oscillations in liver mass were also accompanied by corresponding fluctuations in macromolecular content. Glycogen levels vary in a daily fashion, but in ad-lib-fed mice, they only contribute about 4% and 6.5% to liver weight at trough and peak levels, respectively (Udoh et al., 2015). Therefore, they cannot account for the 34% difference in liver mass reported above. As proteins are the most abundant macromolecules in liver and most other organs, we determined their cellular concentrations at ZT0 and ZT12 by employing methods outlined in Figure S2A. Dot-blot experiments (Figure 2B) and standard Bradford assays (Figure S2C) revealed an impressive 1.6-fold difference in protein accumulation at these two time points in night-fed mice but no significant difference in day-fed animals. To determine whether whole-cell RNA mass, which reflects mostly ribosomal 28S and 18S RNA, also oscillates, we used a procedure that measures RNA and DNA in the same sample (Figure S2B). The RNA/DNA ratio of 1.44 for livers harvested at ZT0 and ZT12 from night-fed mice (Figure 2C) was virtually identical to the ratio of liver weight measured at these two time points (Figure 1E). As expected, the RNA/DNA ratio did not change significantly between ZT0 and ZT12 in day-fed animals (Figures 2A, 2C, and 2D). Irrespective of feeding time, DNA levels, reflecting cell number, remained nearly constant throughout the day (Figure 2A). As depicted in Figure 2D, the cellular RNA levels fluctuated with a diurnal rhythm similar to that observed for liver mass (Figure 1G). Thus, both cellular protein and RNA levels underwent substantial daily rhythms in night-fed, but not day-fed, animals. Because food consumption was nearly identical in night- and day-fed mice (Figure S1C), the quantity of ingested nutrients could not explain these changes. Therefore, light-dark cycles and/or the circadian pacemaker must have assisted feeding-fasting cycles to produce the observed oscillations in liver mass and macromolecular content.
Figure 2. Daily Changes in Hepatic RNA and Protein Content in Mouse Liver.
(A) Content in DNA, RNA, and soluble protein determined at ZT0 and ZT12 in livers of mice fed exclusively during the night or exclusively during the day. The methods used are outlined in Figures S2A and S2B and STAR Methods.
(B and C) Protein/DNA ratio (B) and RNA/DNA ratio (C) in the livers of night-fed (NF) or day-fed (DF) mice at ZT0 and ZT12. The data represent the mean ± SD for eight mice per time point. Soluble protein amounts were measured by dot-blot protein assays. See also Figure S2C.
(A–C) The data represent the mean ± SD for eight mice per time point (***p < 0.001; two-sided Student’s t test).
(D) RNA/DNA ratio in nighttime- or day-fed mice around the 24-hr cycle. The Zeitgeber times (ZT), with ZT0, lights on, ZT12, lights off, are indicated below the panel. The data represent the mean ± SD for four to eight mice per time point.
See also Figure S2.
The Diurnal Accumulation of Ribosomes and Proteins Is Regulated by Posttranscriptional Mechanisms
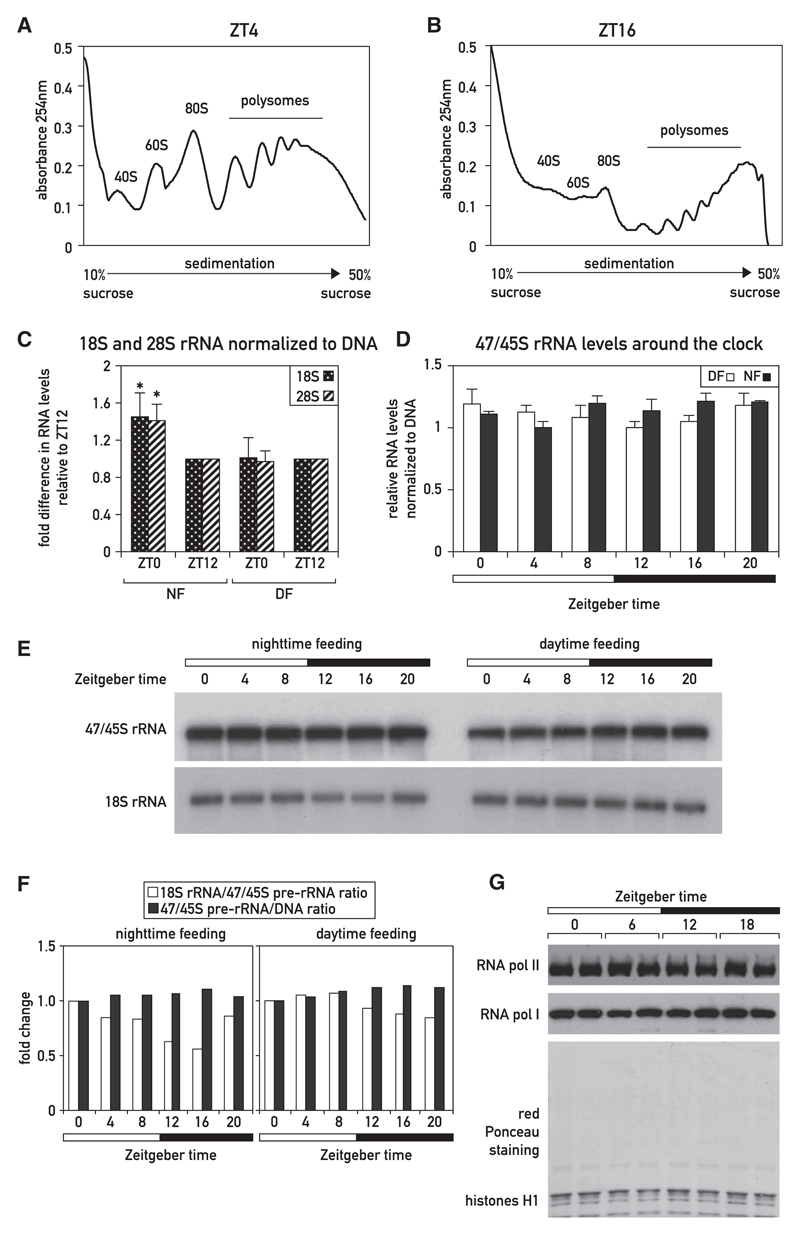
The upsurge in cellular protein accumulation during the activity/feeding phase could have been due to a global increase in mRNA levels, ribosome number, translation efficiency, protein stability, or a combination of these parameters. In agreement with previous reports (Fishman et al., 1969; Jouffe et al., 2013), we observed a higher proportion of ribosomes associated with large polysomes during the activity phase (ZT16) as compared to the resting phase (ZT04; Figures 3A, 3B, S3C, and S3D), suggestive of a global increase in translation efficiency at ZT16. At this time point, free ribosomal 40S and 60S subunit levels were very low (Figures 3B, S3C, and S3D), indicating that ribosomes are rate limiting for protein production. We thus hypothesized that an increase in ribosome number was required for boosting hepatic protein synthesis during the activity/feeding phase. Indeed, because rRNAs represent more than 80% of cellular RNA, the higher RNA/DNA ratio at ZT0 (Figures 2C and 2D) reflects an increased number of ribosomes at this time point. This was confirmed by qRT-PCR assays for 28S and 18S rRNAs (Figures 3C and S3A). As expected, the amounts of 28S and 18S rRNAs showed no such change in day-fed mice (Figures 3C and S3A).
Figure 3. Analysis of Temporal Polysome Distribution, rRNA Transcription, and rRNA Accumulation in Livers of Night-Fed Mice.
(A and B) Representative polysome profiles obtained by sucrose gradient sedimentation of cytoplasmic liver extracts from mice sacrificed at ZT04 (A) and ZT16 (B). For replicates, see Figure S3.
(C) 18S and 28S rRNA levels in night- (NF) or day-fed (DF) mice at ZT0 and ZT12 (see also Figure S3A). Real-time qRT-PCR quantifications of 18S and 28S rRNA from total RNA samples (extracted according to the liver whole-cell RNA protocol) were normalized to DNA content (i.e., cell number), as determined in the experiments displayed in Figure 2A. The fold differences are normalized to ZT12. The data represent the mean ± SD for six mice per time point (*p < 0.05; two-sided Student’s t test).
(D) 47/45S rRNA levels in night- (NF) or day-fed (DF) mice around the clock measured by qRT-PCR and normalized to DNA content, as measured in the data shown in Figure 2A. The data represent the mean ± SD for at least three mice per time point.
(E) 47/45S and 18S rRNA levels in night- (NF) or day-fed (DF) mice around the clock normalized to DNA content and analyzed by northern blot hybridization. Mice were sacrificed at 4-hr intervals (three animals/time point), RNA/DNA ratios were measured, and total RNAs were prepared and pooled.
(F) 18S rRNA/47/45S pre-rRNA ratio and 47/45S pre-rRNA/DNA ratio according to the quantification of the northern blot presented in Figure 3E and the DNA content measured in Figure 2A.
(G) Density of elongating RNA Pol I and Pol II molecules, around the clock. Night-fed mice were sacrificed at 6-hr intervals (two animals/time point), and liver nuclear proteins associated with chromatin were prepared. Protein extracts corresponding to 5 μg of DNA were analyzed by immunoblotting with antibodies recognizing RNA polymerase I (RPA2 subunit) and RNA polymerase II (POLR1B). Red Ponceau staining of the membrane served as loading control.
See also Figure S3.
Surprisingly, rhythmic accumulation of 18S and 28S rRNA was not achieved by cyclic rDNA transcription rates. First, the accumulation of 47/45S pre-rRNA, in contrast to that of 28S and 18S rRNAs, was nearly constant during the day, irrespective of the feeding regimen (Figures 3D, 3E, and S3B). Given the high metabolic stability of ribosomes (Hirsch and Hiatt, 1966; Loeb et al., 1965), high-amplitude oscillations in 47/45S pre-rRNA accumulation would have been required to generate the about 1.4-fold difference in cellular 28S and 18S rRNA (see STAR Methods for mathematical considerations). Second, using previously described methods (Menet et al., 2012; Wuarin and Schibler, 1994), we visualized elongating RNA polymerase complexes, reflecting relative transcription rates, at four time points around the clock in livers of night-fed mice. As depicted in Figure 3G, the levels of chromatin-associated RNA polymerase I and RNA polymerase II, transcribing protein- and rRNA-encoding genes, respectively, remained virtually invariable. We thus concluded that global pre-mRNA and pre-rRNA synthesis remained nearly constant throughout the day and that posttranscriptional, rather than transcriptional, mechanisms are likely to account for the diurnal rRNA accumulation cycles.
Rhythms in Nuclear rRNA Polyadenylation Are Antiphasic to the Oscillations in Total rRNA Accumulation
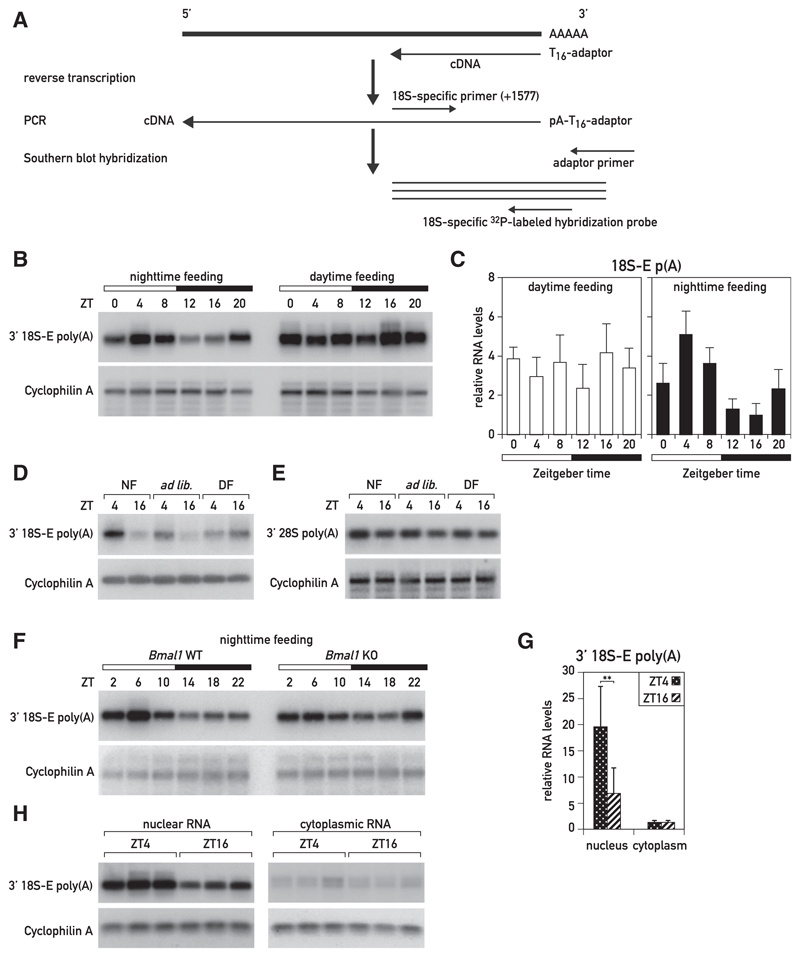
Because the oscillation in rRNA accumulation could not be attributed to diurnal rDNA transcription, rRNAs must have been diurnally degraded. Whereas rRNA transcription and processing have been extensively studied in mammals, the mechanisms involved in rRNA degradation are still poorly understood. In yeast, improperly processed pre-rRNAs are targeted through the addition of short adenylate tails by the TRAMP complex and degraded by the 3′–5′ nuclear exosome (Dez et al., 2006; Houseley and Tollervey, 2006; LaCava et al., 2005; Wery et al., 2009). Polyadenylated rRNA transcripts have also been observed in mouse and human cells, possibly suggesting a similar decay pathway in mammalian cells (Shcherbik et al., 2010; Slomovic et al., 2006). If polyadenylated rRNAs were indeed targeted for degradation, the levels of polyadenylated rRNA in liver should follow a diurnal rhythm with a phase roughly opposite to that of cellular ribosome accumulation. In order to determine the dynamics of polyadenylated rRNA, we synthesized cDNA from total liver RNA collected at 4-hr intervals around the clock by using an oligo(dT)-adaptor primer and amplified the cDNAs of adenylated 18S and 28S 3′ regions with rRNA-specific forward primers and an adaptor-specific reverse primer. In these experiments, we used a low number of PCR cycles in order to keep the amplification rates within the exponential range (Figure 4A). The resulting PCR products were verified by sequencing. Importantly, the single PCR product obtained for polyadenylated 18S rRNA corresponded to the 18S-E precursor RNA, which is confined to the nucleus. This is in keeping with results described below, showing that rRNA polyadenylation occurs in the nuclear compartment. The amounts of polyadenylated rRNAs were quantified by Southern blot hybridization (Figures 4B and S4D) and real-time qPCR (Figure 4C). Whereas nearly constant levels of 3′ polyadenylated 18S-E pre-rRNA were observed around the day in livers of day-fed mice, the accumulation of these transcripts followed a robust diurnal rhythm in night-fed mice, with maximal and minimal levels at around ZT04 and ZT12/ZT16, respectively (Figures 4B and S4D). We also observed a similar temporal polyadenylation pattern for polyadenylated 18S-E rRNA in ad-lib-fed mice (Figure S4A). The accumulation of polyadenylated 28S rRNA was diurnal as well, albeit with a somewhat lower peak-to-trough ratio (Figures 4E and S4F). For both polyadenylated 18S-E and 28S rRNAs, the rhythmic expression profiles were confirmed by another technique, involving the isolation poly(A)+ RNA by adsorption to oligo(dT)-biotin beads, reverse transcription using random primers, and quantification of the resulting cDNAs by real-time qPCR employing rRNA-specific primers (Figures S4B and S4C). The comparison of polyadenylated rRNA accumulation at ZT04 and ZT16 in mice subjected to the three feeding regimens demonstrated that daytime feeding abolished the daily oscillations observed in mice fed during the night or ad libitum (Figures 4D, 4E, S4E, and S4F). Thus, food absorption exclusively or preferentially during the night was required not only for increasing liver mass and protein and ribosome accumulation during the activity/feeding phase but also for the antiphasic increase in the abundance of polyadenylated rRNA species that occurred during the resting/fasting phase.
Figure 4. Polyadenylation of 18S-E rRNA Is Diurnal in the Livers of Night-Fed, but Not Day-Fed, Mice.
(A) RT-PCR protocol used for the semiquantitative analysis of polyadenylated 18S-E rRNA transcripts.
(B) Comparison of polyadenylated 18S-E rRNA levels around the clock in night- and day-fed mice by semiquantitative Southern blot analysis. Hybridization of RT-PCR products using a (32P)-labeled hybridization probe specific for 18S-E rRNA (18S_1773-1802 probe) to detect 3′ polyadenylated 18S-E rRNA in mouse liver. Mice were sacrificed at 4-hr intervals (three animals/time point), and total RNAs were prepared and pooled.
(C) Comparison of polyadenylated 18S-E rRNA levels around the clock in night- and day-fed mice by qRT-PCR. Night- and day-fed mice were sacrificed at 4-hr intervals, and total RNAs were prepared. The values represent mean ± SD for three mice per time point and were normalized to cyclophilin A mRNA levels.
(D and E) Comparison of the levels of polyadenylated 18S-E (D) and 28S (E) rRNA at ZT04 and ZT16 in night-, day-, and ad-lib-fed mice by semiquantitative Southern blot analysis. Southern blot hybridization of RT-PCR products with an 18S rRNA-specific probe (see B; D) and a 28S rRNA-specific probe (28S_4675-4694; E) to detect 3′ polyadenylated rRNAs in mouse liver is shown. Mice (six animals/time point) were sacrificed, and total RNAs were prepared and pooled.
(F) Comparison of polyadenylated 18S-E rRNA levels around the clock in wild-type (WT) and Bmal1 knockout (KO) mice, subjected to a nighttime-restricted feeding regimen, by semiquantitative analysis. Southern blot hybridization of RT-PCR products (using the hybridization probe specified in A) to detect 30 polyadenylated 18S-rRNA in mouse liver. Four to five animals per time point were sacrificed at 4-hr intervals, and total RNAs were prepared and pooled.
(G and H) Comparison of polyadenylated 18S-E rRNA levels in nuclear and cytoplasmic mouse liver RNA at ZT04 and ZT16 by qRT-PCR (G) and semiquantitative Southern blot analysis (H). Night-fed mice were sacrificed at ZT04 and ZT16, liver nuclei and cytoplasmic extracts were prepared, and RNAs were prepared from these subcellular fractions. The specificity of nuclear and cytoplasmic RNA fractions were controlled by qPCR experiments (Figures S4I and S4J). The measurements of polyadenylated rRNA levels determined by qRT-PCR (G) represent means ± SD for six mice per time point and were normalized to Cyclophilin A mRNA levels (**p < 0.01; two-sided Student’s t test). For the hybridization of RT-PCR products by Southern blot hybridization (H), the RNAs from three animals per time point were pooled.
(B–G) Cyclophilin A mRNA was used as a loading control, and the quantifications of the blots are shown in Figures S4D–S4G. See also Figures S4 and S7.
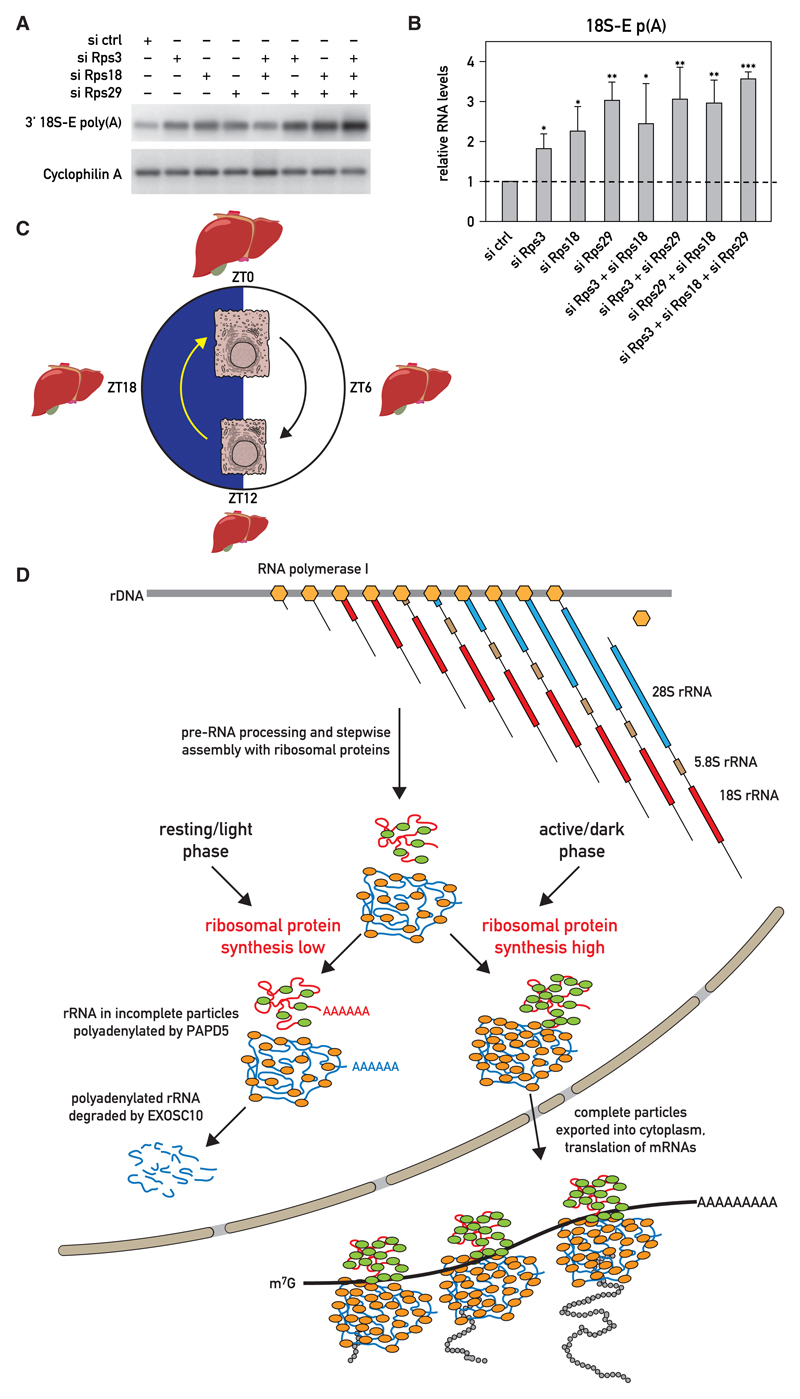
To examine the potential importance of the circadian clock for diurnal rRNA polyadenylation, we performed experiments with livers of clock-deficient Bmal1 knockout (Bmal1 KO) mice exposed to a nighttime feeding schedule. These animals still displayed an oscillation of polyadenylated 18S-E rRNA, albeit with a somewhat blunted peak/trough ratio when compared to wild-type mice (Figures 4F and S4G). Therefore, the circadian oscillator played a less important role than feeding rhythms in governing diurnal 18S-E rRNA polyadenylation.
Given that polyadenylated 18S rRNA was confined to nuclear 18-E rRNA, we expected that rRNA polyadenylation occurred in the nucleus. To examine this more directly, we compared the levels of polyadenylated 18S-E (Figures 4G and 4H) and 28S rRNA (Figure S4H) in liver nuclei and in the cytoplasm of night-fed mice sacrificed at ZT04 and ZT16. The efficacy of our cell fractionation was verified by determining the levels of Lgals1 pre-mRNA and mRNA and MALAT1, an abundant nuclear non-coding RNA, in the nuclear and cytoplasmic fractions (Figures S4I and S4J). As anticipated, the levels of polyadenylated 18S-E and 28S rRNAs were much higher in the nucleus than in the cytoplasm, and the difference of rRNA polyadenylation between ZT04 and ZT16 was statistically significant only in the nucleus (Figures 4G and S4H). Therefore, we hypothesized that poly(A) tails might serve in a nuclear quality control mechanism, in which these posttranscriptionally added sequences were degradation marks for rRNA species not assembled into functionally competent ribosomal subunits. Indeed, according to the rough estimate outlined in STAR Methods, ribosomal proteins (RPs) were unlikely to be produced at amounts sufficient to package all rRNA molecules processed from newly synthesized pre-rRNAs into complete ribosomal subunits during the resting/fasting phase, when nuclear rRNA polyadenylation was maximal. Moreover, our results suggested that RP mRNA translation and not rRNA transcription was rate limiting for the production of ribosomes in liver.
In Cultured NIH 3T3 Fibroblasts, Excess rRNAs Are Polyadenylated in the Nucleus by the Poly(A) Polymerase PAPD5 and Degraded by the Exosome EXOSC10
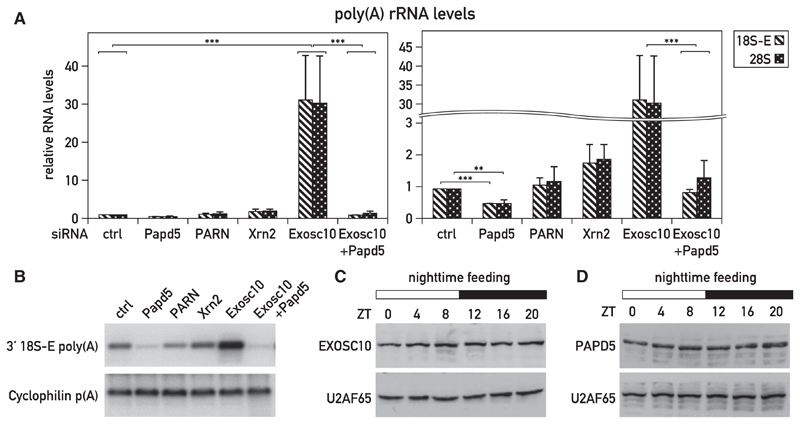
According to the quality surveillance hypothesis proposed above, a nuclear polymerase capable of adding oligo- or poly(A) tails to incompletely assembled rRNAs should exist, polyadenylated rRNAs should be a substrate for degradation, and a reduction in RP synthesis should result in increased rRNA polyadenylation. These conjectures are difficult to study in whole organisms, and we thus decided to explore their validity in cultured NIH 3T3 cells. Knocking down the 3′-to-5′ exoribonuclease EXOSC10 in NIH 3T3 cells by RNAi (Figure S5A) led to an ~30-fold higher accumulation of polyadenylated 18S and 28S rRNA (Figures 5A and 5B). In contrast and as expected, the depletion of the 5′-to-3′ exoribonuclease XRN2 (Figure S5C) only produced a moderate increase in the levels of these transcripts (Figures 5A and 5B). The small interfering RNA (siRNA)-mediated knockdown of the non-canonical poly(A) polymerase PAPD5 (Figure S5B) led to reduced levels of polyadenylated 18S and 28S rRNA transcripts (Figures 5A and 5B), suggesting that this nuclear enzyme (Rammelt et al., 2011) participated in the polyadenylation of rRNAs. It had been proposed previously that the exonucleolytic activity of EXOSC10 is enhanced by the addition of 3′-oligo(A) tails, supposedly by promoting substrate recognition (LaCava et al., 2005; Rammelt et al., 2011; Vanácová et al., 2005; Wyers et al., 2005). If PAPD5 generated the poly(A) tails of rRNAs recognized by EXOSC10, the depletion of both PAPD5 and EXOSC10 (Figures S5A and S5B) would be expected to reduce the polyadenylation of rRNAs observed when only EXOSC10 is depleted. This prediction proved to be accurate (Figures 5A and 5B).
Figure 5. rRNAs Are Polyadenylated in the Nucleus, and Polyadenylated rRNAs Are Recognized by the 3′ -to-5′ Exosome.
(A and B) Effect of RNAi using PAPD5 siRNA (siPapd5), PARN siRNA (siPARN), XRN2 siRNA (siXrn2), and EXOSC10 siRNA (siExosc10) on polyadenylated 18S-E rRNA levels in transfected NIH 3T3 cells. Polyadenylated rRNAs were measured by qRT-PCR (A) or Southern blot hybridization of RT-PCR products with the 18S-E rRNA-specific probe (B). Knockdown efficiencies were controlled by qRT-PCR (Figures S5A–S5D). (A) The data represent mean values ± SD of 18S-poly(A) rRNA levels relative to Cyclophilin A mRNA levels measured in three independent transfection experiments with non-targeting siRNAs (siCtrl) or Papd5/PARN/Xrn2/Exosc10 siRNAs. ***p < 0.001; **p < 0.01; two-sided Student’s t test.
(C and D) EXOSC10 and PAPD5 protein expression around the clock in livers of night-fed animals. Night-fed mice were sacrificed at 4-hr intervals (three animals/time points), and liver nuclear proteins were prepared and pooled. Protein extracts were analyzed by immunoblotting with antibodies recognizing EXOSC10 (C), PAPD5 (D), and U2AF65 (used as a loading control; C and D).
See also Figure S5.
In addition to being channeled to a degradation pathway involving the nuclear exosome, polyadenylated rRNAs may also be subject to deadenylation by nuclear deadenylases. Conceivably, the removal of poly(A) tails by such enzymes would counteract rRNA decay by EXOSC10. Recently, the poly(A)-specific RNase PARN has been shown to remove oligo(A) tails added by PAPD5 to the telomerase RNA component (Shukla et al., 2016). However, because we did not observe increased levels of polyadenylated 18S-E rRNA in PARN-depleted cells (Figures 5A, 5B, and S5D), we concluded that PARN did not play an essential and rate-limiting role in regulating the accumulation of polyadenylated rRNAs.
Given their ubiquitous expression, we considered it likely that PAPD5 and EXOSC10 were also participating in the polyadenylation and degradation, respectively, of rRNAs in the liver. Therefore, we examined whether the accumulation of one or both of these enzymes displayed a diurnal cycle. However, as shown in Figures 5C and 5D, the levels of EXOSC10 and PAPD5 remained nearly constant in liver nuclei. Therefore, the rhythmic accumulation of polyadenylated RNAs could not be explained by a corresponding fluctuation in the levels of these two enzymes.
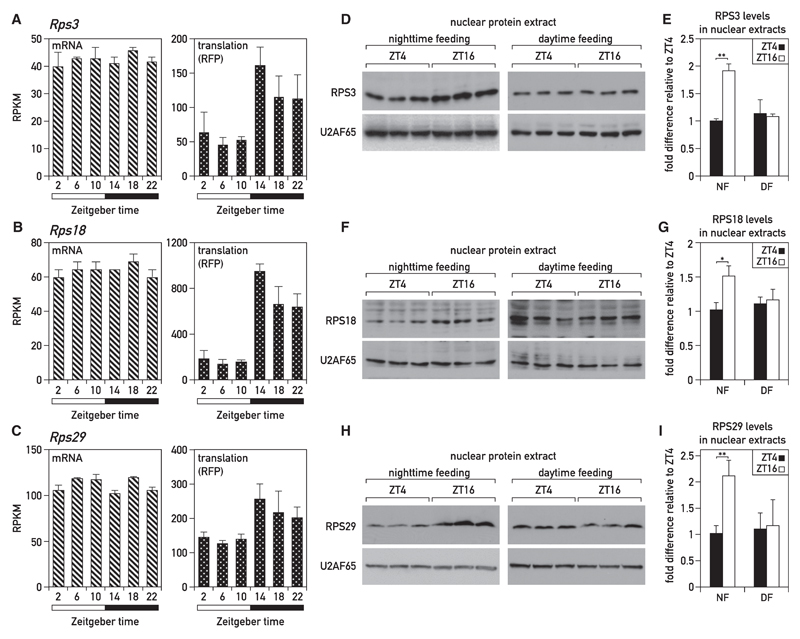
Ribosome biogenesis is a highly complex pathway involving many maturation steps, and the efficient production of functional ribosomal subunits requires the coordination of rRNA and protein expression (Mayer and Grummt, 2006). In the livers of ad-lib-fed and night-fed mice, the translation of most RP-encoding mRNAs has been reported to oscillate in a daily fashion, with highest expression levels reached at the beginning of the dark period (Atger et al., 2015; Janich et al., 2015; Jouffe et al., 2013). RNA accumulation and ribosome profiling data of mice fed either during the night (Figures 6A–6C) or ad libitum (Figures S6J–S6L) confirmed that the translation efficiency of Rps3, Rps18, and Rps29 mRNAs strongly oscillated during the day (right panels). These diurnal translation rates are manifested in a higher nuclear accumulation of RPS3, RPS18, and RPS29 at ZT16 in night-fed mice (Figures 6D–6I). By contrast, the temporal accumulation profiles of these RPs were nearly flat in liver nuclei of day-fed mice. When normalized to cytoplasmic tubulin amounts, the levels of the examined RPs in cytoplasmic extracts were not subject to marked oscillations (Figures S6D–S6I). As global protein concentrations—and therefore tubulin levels—oscillated (Figures 2B and S2C), these results suggested that RP levels also fluctuated during the day, as expected from the quantification of total RNA (of which >80% is rRNA) and 28S and 18S rRNA (Figures 2C, 2D, 3C, and S3A).
Figure 6. Diurnal Synthesis and Nuclear Accumulation of RPs in Livers of Night-Fed Mice.
(A–C) Temporal translation rates (ribosome profiling) and accumulation (RNA sequencing [RNA-seq]) of mRNAs encoding RPS3, RPS18, and RPS29. The data were extracted from an RNA sequencing and ribosome profiling analysis around the 24-hr cycle in livers of mice entrained to a nighttime feeding regimen (Atger et al., 2015). Means of four values per time point ± SD are plotted.
(D, F, and H) RP levels analyzed by immunoblotting with antibodies recognizing RPS3 (D), RPS18 (F), and RPS29 (H) in nuclear liver protein extracts prepared from mice subjected to a nighttime- or daytime-restricted feeding regimen and sacrificed at ZT04 or ZT16. Immunoblotting with antibodies recognizing U2AF65 was used as a loading control.
(E, G, and I) Means ± SD for six mice per time point of representative immunoblots presented in (D, F, and H) normalized to ZT04. The data represent the mean values ± SD (*p < 0.05; **p < 0.01; two-sided Student’s t test).
See also Figure S6.
Because the increase in RP abundance at ZT16 correlated with the lowest level of polyadenylated rRNA, we suspected that the two events were causally related. The sub-stoichiometric production of RPs during the inactivity/light period may thus have triggered the polyadenylation and degradation of nuclear rRNA molecules not assembled into complete ribosomal subunits. To test whether the downregulation of RP expression could indeed provoke the polyadenylation of rRNAs, we determined the levels of polyadenylated 18S-E transcripts in NIH 3T3 cells depleted of ribosomal proteins. Indeed, the knockdown of RPS3, RPS18, and RPS29 expression alone or in various combinations (Figures S6A–S6C) resulted in an augmented accumulation of polyadenylated 18S-E rRNA (Figures 7A and 7B). Our study thus supports a model proposing that RP synthesis is rate limiting for the assembly of mature ribosomal subunits in the liver and that rRNAs produced in excess over RPs are polyadenylated and degraded. As the translational output of most RP mRNAs strongly oscillates during the day, the accumulation of functional ribosomal subunits is also subject to diurnal fluctuations.
Figure 7. 18S-E rRNA Polyadenylation Is Enhanced by the Depletion of Ribosomal Proteins in NIH 3T3 Cells.
(A and B) Effect of Rps3 siRNA (siRps3), Rps18 siRNA (siRps18), and Rps29 siRNA (siRps29) on polyadenylated 18S-E rRNA levels in transfected NIH 3T3 cells, as measured by Southern blot hybridization of RT-PCR products with 18S_1773-1802 probe (A) or by qRT-PCR (B). The knockdown efficiencies were controlled by qRT-PCR experiments (Figures S6A–S6C). (B) Data represent the mean values ± SD of polyadenylated 18S-E rRNA levels relative to Cyclophilin A mRNA levels measured in three independent transfection experiments with non-targeting siRNAs (siCtrl) or Rps3/Rps18/Rps29 siRNAs. *p < 0.05; **p < 0.01; ***p < 0.001; two-sided Student’s t test.
(C) Cartoon displaying diurnal fluctuations in liver volume and hepatocyte cell size. The items in the scheme are not drawn to scale.
(D) Model showing the putative roles of ribosomal protein synthesis and rRNA polyadenylation in the diurnal production of ribosomes. The daily cycle of ribosome accumulation is regulated posttranscriptionally. Whereas the synthesis of 47/45S pre-rRNA and RP mRNAs (not shown in scheme) is constant throughout the day, RP mRNAs are translated more efficiently during the activity/dark phase, during which mice feed. This leads to an imbalance between rRNA and RP production, particularly during the resting/light phase. Excess rRNAs not assembled into complete pre-ribosomal particles are polyadenylated by the TRAMP complex and degraded by the 3′ –5′ nuclear exosome. Complete pre-ribosomal subunits are exported into the cytoplasm, where they mature to functional subunits capable of mRNA translation. RNA polymerase I complexes are symbolized by light brown hexagons, and RPs associated with small and large subunits are represented as green and light brown ovals, respectively.
Discussion
The Entire Liver Oscillates
The processing and detoxification of nutrients in the liver must be coordinated with feeding/fasting rhythms, which are imposed by daily rest/activity cycles. The cyclic expression of genes participating in these processes can be orchestrated by systemic cues, controlled by environmental rhythms and the circadian master pacemaker in the SCN and/or by local circadian hepatocyte oscillators (Kornmann et al., 2007; Partch et al., 2014; Schibler et al., 2015). The underpinning mechanisms can act at the level of transcription (Menet et al., 2012), RNA splicing (Gotic et al., 2016), mRNA polyadenylation and stability (Kojima et al., 2012), and translation (Atger et al., 2015; Janich et al., 2015; Jouffe et al., 2013). Feeding-fasting rhythms play a dominant role in driving oscillations in gene expression and metabolism, either via synchronizing peripheral oscillators in hepatocytes or, more directly, by controlling the activity of regulators and enzymes participating in anabolic and catabolic metabolic functions. Here, we show that the entire liver oscillates with regard to tissue mass, hepatocyte size, and macromolecular content (schematically visualized in Figure 7C). These rhythms are accentuated in mice exclusively fed during the night and dampened in mice exclusively fed during the day. Although the circadian clock contributed to their amplitude, feeding during the dark phase appeared to be the dominant parameter in determining global liver oscillations. Because of the higher amplitudes observed in night-fed mice, we performed most of our experiments with these animals. However, we believe—and demonstrate it for select features—that the same mechanisms are also operative in ad-lib-fed animals.
How general are diurnal oscillations in liver size? Ultrasonic measurements in human subjects conducted at hourly intervals between 10 a.m. and 4 p.m. suggested that liver mass oscillated in a consistent fashion by almost 20% during this time period (Leung et al., 1986). These measurements were, however, not extended to an entire day, and the authors could thus not discriminate between diurnal and ultradian rhythms. Moreover, they could not assess whether the fluctuations in liver volume were accompanied by corresponding changes in macromolecular content. Interestingly, omission of the evening meal eliminated the temporal liver volume changes during the following day, whereas skipping breakfast or lunch had little effect on these fluctuations. Studies published in the 1960s have already reported daily oscillations in liver weight in red-winged black-birds and Japanese quails (Fisher and Bartlett, 1957; Wilson and McFarland, 1969), which showed a difference in the liver-to-body weight ratio of 30% and 20%, respectively. Whereas the authors noticed that changes in liver weight correlated with those measured for lipid and glycogen content, they pointed out that these molecules cannot explain the diurnal differences in liver mass (Wilson and McFarland, 1969). Morphological changes at the cellular level, such as the number and structure of certain organelles, were also reported several decades ago for hepatocytes (for review, see Uchiyama, 1990). As already mentioned, the actin cytoskeleton undergoes dramatic daily changes, with most actin fibers being observed at ZT0, when hepatocytes have reached their maximal size. These studies, together with the data presented here, underscore the highly dynamic structural, morphological, and biochemical changes of hepatocytes during the day, probably reflecting an adaptation of liver physiology to feeding-fasting cycles.
The Number of Ribosomes May Be Rate Limiting for Protein Synthesis
The correlative evidence presented in our study suggests that the number of ribosomes may be rate limiting for protein synthesis in the liver. Indeed, the accumulation of ribosomes follows a diurnal cycle in hepatocytes, with maximal and minimal numbers reached at the beginning and end, respectively, of the activity/feeding phase. Mammalian ribosomes contain 80 RPs, 47 in the large 60S subunits (RPLs) and 33 in the small subunits (RPSs), and four rRNAs, three in the 60S subunit (28S, 5.8S, and 5S) and one (18S) in the small subunit (Khatter et al., 2015). Three of the four RNA species (28S, 18S, and 5.8S) are processed from a polycistronic 47S pre-rRNA specified by about 200–500 tandemly repeated genes residing in the nucleolus, whereas 5S rRNA is encoded by tandemly repeated genes active in the nucleoplasm. Because it is difficult for cells to synthesize 84 components issued from 82 genes in exactly identical numbers, the most straightforward way of producing stoichiometric amounts of large and small subunits would be to synthesize RPs and 5S rRNA in slight excess over 47/45S pre-rRNA and to degrade the surplus of constituents not integrated into complete particles. Yet, our data support an entirely different mechanism, in which pre-rRNA is constitutively synthesized in excess throughout the day, whereas RPLs and RPSs are translated rhythmically from nearly invariable numbers of RP mRNAs. The excess of 28S and 18S rRNAs not incorporated into ribosomal particles is polyadenylated in the nucleus and degraded.
The amplitude of the diurnal RP translation cycle was higher in mice fed exclusively during the night than in ad-lib-fed mice. Conceivably, the global rates of translation and ribosome biogenesis in response to nutrients are controlled by the mTOR (mammalian target of rapamycin) pathway, at least in part (Albert and Hall, 2015; Sengupta et al., 2010). This hypothesis was substantiated by the diurnal phosphorylation profile of RPS6 in night- and day-fed animals (Figures S6M and S6N).
If RP translation cycles were indeed involved in generating the observed oscillations in liver mass, one would expect that RP translation efficiency remains invariable throughout the day in organs with invariable sizes. For one such organ, the kidney, this has recently been confirmed by ribosome profiling (V. Castelo-Szekely, A. Bulak Arpat, P. Janich, and D.G., unpublished data).
rRNA Polyadenylation Cycles May Reflect Rhythmic rRNA Degradation in the Nucleus
Posttranscriptional polyadenylation is not only implicated in the export, stability, and translation of mRNAs (Mitchell and Parker, 2014) but also in the elimination of aberrant and misfolded non-coding RNAs, e.g., tRNAs and rRNAs (Kuai et al., 2004; LaCava et al., 2005; Shcherbik et al., 2010; Wyers et al., 2005), similar to what has been observed for oligo-adenylation in bacteria (Mohanty and Kushner, 2011). Because RNA turnover dynamics are difficult to study in whole organs of living animals, we reasoned that recording polyadenylation kinetics of rRNA might serve as a surrogate for such experiments. Indeed, polyadenylated rRNAs accumulated roughly in antiphase with RP synthesis, in keeping with a scenario in which excess rRNA not assembled into functional subunits are polyadenylated and degraded (Figure 7D). Support for this scenario was further obtained in experiments with cultured NIH 3T3 cells, in which the depletion of RPs through RNAi elicited a higher abundance of polyadenylated rRNA species. In these cells, PAPD5 was implicated in the polyadenylation-mediated degradation of rRNA by the exosome. PAPD5 is a mouse ortholog of the yeast Trf4 and Trf5 proteins, two non-canonical poly(A) polymerases functioning in the TRAMP complex. TRAMP polyadenylates a broad spectrum of nuclear RNAs, including rRNAs and transcribed spacer regions excised from 47S pre-RNA (Egecioglu et al., 2006; Kadaba et al., 2006; Rammelt et al., 2011; Sudo et al., 2016). In mammals, PAPD5 has also been reported to adenylate small nucleolar RNAs and microRNAs during their 3′ end processing (Berndt et al., 2012; Boele et al., 2014), histone mRNAs (Mullen and Marzluff, 2008), and incomplete pre-rRNA transcripts (Shcherbik et al., 2010). Thus, it seems likely that the PAPD5-dependent polyadenylation mechanism in mammalian nuclei is similar to that of the TRAMP complex in yeast nuclei. Non-canonical polyadenylation of rRNA molecules was also reported in human cell lines (Slomovic et al., 2006, 2010). Hence, a mechanism whereby the poly(A) tail added by PAPD5 on its rRNA substrates facilitates access to the exosome complex may be conserved from yeast to humans.
Does Deadenylation Contribute to the Diurnal Accumulation of Polyadenylated rRNA?
In contrast to other examples for poly(A)-mediated degradation, our data showed that PARN, a poly(A)-specific exonuclease, does not remove the poly(A) tail of rRNA. Therefore, the poly(A) tail added to rRNA appears to irreversibly tag rRNA substrates for degradation. We also examined a possible role of Nocturnin (NOC) (also known as CCR4L), another deadenylase, in this process. NOC accumulation exhibits a robustly rhythmic expression pattern, peaking during the night in liver and other organs (Kojima et al., 2010). It would have been conceivable that NOC counteracted rRNA polyadenylation during the night and thus diminished the levels of degradation-prone rRNA substrates. We confirmed the diurnal expression of NOC mRNA and protein and observed that the cyclic expression of NOC was dramatically dampened when the food regimen was inverted (Figures S7A and S7B). However, the liver weights from Nocturnin knockout and wild-type mice did not differ dramatically between the two genotypes (Figures S7D and S7E). Likewise, polyadenylated rRNAs determined at ZT04 and ZT16 were similar in night-fed mice of the two genotypes (Figure S7C). Based on these results, we concluded that NOC did not play a major part in the daily rRNA polyadenylation cycle. Nonetheless, in day-fed mice, the levels of polyadenylated 18S-E rRNA-E were significantly higher at ZT16 than at ZT04 (Figure S7C). However, it is likely that the mechanisms accounting for this difference were of indirect nature and do not involve a NOC-mediated deadenylation of nuclear polyadenylated rRNAs. In fact, NOC is localized primarily in the cytoplasm (Baggs and Green, 2003) and, as indicated by its name, reaches peak levels during the night. Further studies will be required to elucidate the precise role of NOC, if any, in hepatic rRNA metabolism.
Star★Methods
Detailed methods are provided in the online version of this paper and include the following:
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○
Mouse Strains
-
○
Cell Lines and Transfections
-
○
-
-
○
Preparation of Total Nucleic Acid and RNA:DNA Ratio Measurements
-
○
Soluble Protein Measurements
-
○
Western Blotting
-
○
RNA Analysis by Quantitative Real-Time RT-PCR
-
○
Polysome and Ribosome Profiling
-
○
Immunohistochemistry, Image Segmentation, and Hepatocyte Size Estimation
-
○
-
QUANTIFICATION AND STATISTICAL ANALYSES
-
○
Estimation of rRNA Synthesis Necessary to Maintain Cellular Ribosome Numbers
-
○
Quantification of Hepatocyte Surface Areas
-
○
Statistical Analysis
-
○
Star★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Monoclonal anti-RPS3 | Cell Signaling Technology | Cat# 9538 |
| Rabbit Polyclonal anti-RPS29 | Sigma-Aldrich | Cat# AV40329; RRID:AB_1856467 |
| Rabbit Polyclonal anti-RPS18 | Abcam | Cat# ab91293; RRID:AB_2050267 |
| Rabbit Polyclonal anti-EXOSC10 | Abcam | Cat# ab50558; RRID:AB_869937 |
| Goat Polyclonal anti-PAPD5 | Sigma-Aldrich | Cat# SAB2501846 (lot number 10875P1) |
| Mouse Monoclonal anti-RNA PolII F-12 | Santa Cruz Biotechnology | Cat# sc-55492 |
| Rabbit anti-RNA PolI (RPA-116/RPA2) | gifted by Prof. Dr. Ingrid Grummt | N/A |
| Mouse Monoclonal anti-U2AF65 | Sigma-Aldrich | Cat# U4758 |
| Mouse Monoclonal anti-β-Actin | Sigma-Aldrich | Cat# A5441 |
| Rabbit Polyclonal anti-NOCTURNIN | Prepared by C.B. Green | N/A |
| Mouse Monoclonal Anti-α-Tubulin | Sigma-Aldrich | Cat# T9026 |
| B-catenin | BD Biosciences | Cat#610154 |
| Rabbit anti-phospho-S6 Ribosomal Protein (Ser235/236) | Cell Signaling Technology | Cat#2211 |
| Rabbit anti-S6 ribosomal Protein (5G10) | Cell Signaling Technology | Cat#2217 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | Cat# 13778075 |
| Cycloheximide | Sigma-Aldrich | Cat# C7698 |
| TRIZOL | Thermo Fisher Scientific | Cat#15596018 |
| Critical Commercial Assays | ||
| μMACS Streptavidin kit | Miltenyi Biotec | Cat# 130-074-101 |
| SuperScript II reverse transcriptase | Thermo Fisher Scientific | Cat# 18064071 |
| SYBR Green Master Mix | Roche | Cat# 04707516001 |
| Deposited Data | ||
| Ribosome profiling and RNA-seq data (ad lib-fed mice) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GSE67305 |
| Ribosome profiling and RNA-seq data (night-fed mice) | GEO (http://www.ncbi.nlm.nih.gov/geo) | GSE73554 |
| Experimental Models: Cell Lines | ||
| NIH 3T3 cell line | ATCC | Cat# CRL-6442 RRID:CVCL_0594 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Wild-type C57BL/6JRJ | Janvier Labs | RRID:IMSR_JAX:000664 |
| Mouse: WT and Nocturnin KO mice (C57BL/6 background) | Laboratory of C.B. Green | N/A |
| Recombinant DNA | ||
| Sequence-Based Reagents | ||
| ON-TARGET SMART-Pool siRNAs targeting Rps29 sequence | Dharmacon (GE Healthcare) | Cat# L-045910 |
| ON-TARGET SMART-Pool siRNAs targeting Rps18 sequence | Dharmacon (GE Healthcare) | Cat# L-042224 |
| ON-TARGET SMART-Pool siRNAs targeting Rps3 sequence | Dharmacon (GE Healthcare) | Cat# L-047921 |
| ON-TARGET SMART-Pool siRNAs targeting Papd5 sequence | Dharmacon (GE Healthcare) | Cat# L-061333 |
| ON-TARGET SMART-Pool siRNAs targeting Exosc10 sequence | Dharmacon (GE Healthcare) | Cat# L-049286 |
| ON-TARGET SMART-Pool siRNAs targeting PARN sequence | Dharmacon (GE Healthcare) | Cat# L-040664 |
| ON-TARGET SMART-Pool siRNAs targeting Xrn2 sequence | Dharmacon (GE Healthcare) | Cat# L-046490 |
| See Table S1 for primer list | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| MATLAB | MathWorks | https://www.mathworks.com |
| Other | ||
Contact For Reagent And Resource Sharing
Further information and requests for resources and reagents should be directed to the Lead Contact, Ueli Schibler (ueli.schibler@unige.ch).
Experimental Model and Subject Details
All animal procedures were approved by the Veterinary offices of the Cantons of Geneva (U. S.) and Vaud (D. G. and F. G.). The animal experiments (time restricted feeding and tissue collection on WT and NOC KO mice) performed in the laboratory of C.B. Green were approved by the Institutional Animal Care and Use Committee.
Mouse Strains
Male C57BL/6 wild-type mice (13 weeks old) were purchased from Janvier Labs. All animals were housed according to standard. For the phase entrainment to defined LD cycles, the animals were kept in cages during 15 days, placed in homemade, ventilated cabinets in a temperature controlled room (22 ± 1°C), with computer-programmable 12:12 LD cycles (A. Liani and Y-A. Poget, Mechanical Workshop of the Department of Molecular Biology, University of Geneva). When indicated, feeding cycles were imposed by a homemade computer-programmable feeding machine. Immediately after mice were sacrificed, organs were dissected and weighed.
The mice used for the Nocturnin experiments were congenic on a C57BL/6 background and the WT and KO mice were littermates from heterozygous mNoc+/mNoc+/− parents breeding.
Cell Lines and Transfections
The NIH 3T3 cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO) and 1% Penicillin-Streptomycin-Glutamine (PSG, GIBCO). For knockdown experiments, male NIH 3T3 cells were transfected with 15 picomoles of mouse SMART-POOL siRNAs (in a total volume of 2.5 mL) purchased from Dharmacon (GE Healthcare) using Lipofectamine RNAiMAX (Thermo Fisher Scientific), according to the manufacturer’s instructions. Non-targeting siRNAs were used as negative controls at the same concentrations. Forty-eight hours after transfections, total RNAs were extracted.
Method Details
Preparation of Total Nucleic Acid and RNA:DNA Ratio Measurements
For the accurate determination of total RNA and DNA content in the liver we adapted the method described in Schmidt E. E and Schibler U, 1995 (Figure S2A). Livers were homogenized in 9 volumes of HM buffer (10 mM Tris pH7.5, 1mM MgCl2, 50 mM NaCl, 0.1 mM NaF, 0.1 mM Na3VO4, 0.1 mM ZnSO4, 0.5 mM PMSF, 1 mM CP [1 μg/mL pepstatin, 1 μg/mL aprotinin, 1 μg/mL leupeptin]) at 4°C. Two volumes were used to measure the RNA/DNA ratio, and 0.5 volumes for the protein analysis.
For the RNA:DNA ratio, two volumes of liver homogenate were sonicated to reduce viscosity and diluted to a final volume of 20 mL with 1% SDS, 200mM NaCl and 5mM EDTA. An equal volume of Φ/C/I (phenol/chloroform/isoamyl-alcohol-25:24:1 was added, and the emulsion was mixed on a rotating device for at least 1h at room temperature before the phases were separated by centrifugation. The aqueous phase was extracted twice more with (Φ/C/I and total nucleic acids were precipitated with 0.7 volumes of isopropanol. Pellets were resuspended in 5 mL TCS (10 mM Tris, pH 7.5; 0.5 mM CaC12; 1% SDS), proteinase K was added to 40 μg/mL, and samples were incubated at 50°C for 30 min. NaCL was added to 0.2 M, EDTA to 1 mM, and samples were extracted twice with Φ/C/I, once with CHCI3, and nucleic acids were precipitated with 0.7 volumes of isopropanol. Pellets were washed with 5 mL 70% Ethanol, air-dried, and resuspended in one volume of ddH20 (with regard to the liver volume). The final solution contained all polymeric nucleic acids and some glycogen granules. The latter were eliminated by a 10 min centrifugation at full speed in a microcentrifuge at room temperature. The resulting glycogen pellet did not contain co-precipitated nucleic acids (Schmidt and Schibler, 1995).
The following method was developed for measuring RNA and DNA amounts in the same samples. A 100 μL aliquot of the clarified aqueous solution was mixed with an equal volume of 1M NaOH and incubated for 1 hr at 50°C to completely hydrolyze RNA into nucleoside monophosphates (NMPs). This treatment denatures DNA, but does not hydrolyze it. A 50 μL aliquot of the DNA/NMP solution was fractionated into denatured DNA (excluded) and NMPs (included) on a 1.5 mL G-50 Sephadex column prepared in a Pasteur pipet, using NaOH as an eluent (Figure S2B). The OD260nm of small aliquots of the different fractions was measured using a ND-1000 NanoDrop machine (Thermo Scientific). The DNA- and RNA-fractions were pooled, the volumes were measured, and the ODs260nm were measured using a spectrophotometer. To convert the OD260nm measurements to nucleic acid concentrations, we normalized the results to known amounts of RNA or DNA from liver tissue in a mixture treated the same way. Finally, we calculated the RNA/DNA ratio.
Soluble Protein Measurements
For the measurements of soluble proteins 10% of the liver homogenates prepared in HM buffer (see methods for nucleic acid determinations above) were centrifuged at 4,000 rpm in a microfuge and the supernatants were adjusted to 0.1M NaOH. The concentrations of proteins in the alkaline supernatants were determined either by standard Bradford protein assays (Figure S2C) or by a dot-blot protein assays (see below).
For the dot-blot assay, serial dilutions of the alkaline homogenates were blotted on Whatman paper and stained with Coomassie Blue, alongside with serial dilutions of BSA in the same solution. The membranes were scanned, each dot was quantified using the ImageJ software, and the sample concentrations were determined using a standard curve created by plotting the BSA signals versus the signals obtained with liver homogenates.
Western Blotting
Proteins from mouse liver nuclei were prepared according to the NUN procedure (Lavery and Schibler, 1993). Cytoplasmic extracts (post nuclear supernatants) were obtained by homogenizing liver tissue in HM buffer (10 mM Tris pH7.5, 1mM MgCl2, 50 mM NaCl, 0.1 mM NaF, 0.1 mM Na3VO4, 0.1 mM ZnSO4, 0.5 mM PMSF, 1 mM CP [1 μg/mL pepstatin, 1 μg/mL aprotinin, 1 μg/mL leupeptin]) and by centrifuging the homogenate for 10 min at 4°C at 4,000 rpm in a Beckman centrifuge (SW40Ti). Proteins associated with chromatin were extracted from liver cell nuclei according the procedure previously described in Wuarin and Schibler, 1994. SDS-PAGE and immunoblot analysis were performed according to standard protocols. Antibodies used were rabbit RPS29 (Sigma-Aldrich), RPS3 (Cell Signaling Technology), RPS18 (Abcam ab91293), EXOSC10 (Abcam ab50558), goat PAPD5 (Sigma-Aldrich), mouse RNA PolII (Santa Cruz F-12), rabbit RNA PolI (RPA-116/RPA2) generously provided by Prof. Dr. Ingrid Grummt, and mouse U2AF65 (Sigma-Aldrich), ACTIN (Sigma-Aldrich), NOCTURNIN (raised in rabbits by C.B.G), and TUBULIN (Sigma-Aldrich).
RNA Analysis by Quantitative Real-Time RT-PCR
RNAs were extracted from cultured cells using the TRIZOL reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNAs were synthesized from total RNAs using either random hexamers or oligo(dT)-adaptor primers (Table S1), and SuperScript II reverse transcriptase (Thermo Fisher Scientific) following the supplier’s instructions. The cDNAs were quantified by real-time PCR-amplification in a LightCycler 480 (Roche), using the SYBR Green Master Mix (Roche). Mean levels were calculated from triplicate PCR assays for each sample and normalized to the amounts of Cyclophilin A transcripts.
Liver whole-cell RNA was purified essentially as previously described (Schmidt and Schibler, 1995) from mice that were phase-entrained for two weeks by a 12 hr light/12 hr dark regimen and, when indicated, daytime- or nighttime-restricted feeding cycles. Specifically, 250 mg of liver tissue (fresh or kept frozen at −70°C) were ground in 5 mL extraction buffer using a Polytron PT 2500 E homogenizer. The extraction buffer was prepared as follows: 250 g of guanidium thiocyanate were dissolved in 320 mL of H2O and 17.6 mL of 0.75 M sodium citrate, pH 7. Just before use the required volume of extraction buffer was supplemented with 0.1 volumes of 2M ammonium acetate, pH 4, and 0.01 volumes of β-mercaptoethanol. Subsequently, 5 mL of phenol (saturated in H2O) and 2 mL of chloroform-isoamylalcohol (49:1 volume ratio) were added, and the emulsion was vigorously shaken manually, before the aqueous and organic phases were separated by centrifugation at 4,000 rpm for 20 min at 4°C. RNA was precipitated from the aqueous phase by the addition of an equal volume of isopropanol, and the mixture was kept at −20°C during at least 25 min. The precipitated RNA was pelleted by centrifugation at 4,500 rpm during 15 min at 4°C. The pellet was resuspended in 6 mL of 4M LiCl to remove traces of DNA (which, in contrast to RNA, is soluble in 4M LiCl), and the RNA was recovered by sedimentation at 4,500 rpm for 15 min at 4°C. The RNA pellet was washed in 75% ethanol, followed by a centrifugation at 4,500 rpm for 15 min at 4°C, and the pellet was dried at room temperature for at approximately 10 min. In all centrifugation steps a Hettich Rotina 380R tabletop centrifuge was used. The purified RNA was dissolved in 700 μL of diethyl dicarbonate (DEPC)-treated H2O. For the DEPC treatment, double-distilled H2O was vigorously mixed with 0.1% volumes of DEPC, before autoclaving.
For the quantification of transcripts, cDNA synthesis and quantitative RT-PCR were performed as described above for cells. Nuclear and cytoplasmic fractions were prepared exactly as described in (Wuarin and Schibler, 1994) and RNAs from each fraction were prepared as described above for the liver whole-cell RNA. Briefly, liver nuclei were purified by homogenization in a 1.9 M final concentration sucrose solution, followed by pelleting once through a 2 M sucrose cushion. Nuclei were stored in buffer [NSB: 20 mM Tris-C1 (pH 7.9), 75 mM NaC1, 0.5 mM EDTA, 0.85 mM DTT, 0.125 mM PMSF, 50% glycerol] before RNA extraction. Cytoplasmic fractions were precipitated in 5 volumes isopropanol and resuspended in 1 volume H2O before RNA extraction procedure.
Ribosomal RNA levels were normalized to DNA levels by two different methods. Relative rRNA concentrations were first measured by standard real-time RT-qPCR amplification, and the values where normalized to the amounts of DNA determined by DNA:RNA fractionation (see above). The second method was performed by subtraction of the Ct values obtained by real-time PCR-amplification from reverse transcribed total nucleic acids (cDNA/DNA mix) by Ct values obtained from non-reverse transcribed samples (RNA/DNA mix). Primers are listed in Table S1.
RNA Polyadenylation Analysis
RT–PCR assays were performed as previously described (Shcherbik et al., 2010). Briefly, total RNAs were reverse-transcribed using oligo(dT)-adaptor primers (Table S1), and SuperScript II reverse transcriptase following the supplier’s instructions. The 3′ regions of 18S and 28S rRNA, and Cyclophilin A as loading controls, were amplified with either 18S_1577, 28S_4578, or Cyclophilin A_Fw primers, and adaptor reverse primers, by either real-time q-PCR amplification or a standard PCR with a low number of cycles (20 cycles). In the latter case the PCR products were separated on agarose gels and visualized by Southern blot hybridization with the radio-labeled probes 18S_1773-1802, 28S_4675-4694, and Cyclophilin A (Table S1). PCR reactions with primers designed to amplify a fragment of the polyadenylated Cyclophilin A mRNA were used to monitor the efficiency of cDNA synthesis in all samples.
Poly(A)+ RNA isolations were carried out using the μMACS streptavidin kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s protocol. RNAs were recovered from the oligo(dT) microbeads in 200 μL elution buffer and ethanol precipitated. The pellets were then washed with 70% ethanol, dried and dissolved in 10 μL DEPC-H2O. The resulting RNAs were used for reverse transcription using random hexamers, and the resulting cDNAs quantified by real-time PCR, as described above.
Polysome and Ribosome Profiling
Polysome profiles were performed as described in Jouffe et al., 2013 with slight modifications. Briefly, livers were homogenized in 6 volumes of lysis buffer containing 20 mM HEPES (pH 7.6), 250 mM NaCl, 10 mM MgCl2, 10 mM DTT, 20 μg/mL cycloheximide, 10 U/μL RNase inhibitor. The homogenates were centrifuged for 10 min at 9,500 g, and 1 mg/mL heparin, 0.5% Na deoxycholate, and 1% Triton X-100 were added to the supernatant. 200 μL of lysate were deposited on a linear 16 mL sucrose gradient (7% to 47%), prepared in a buffer containing 20 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2, and 10 mM DTT. After a 3 hr centrifugation at 34,000 rpm (SW40Ti rotor) in an ultracentrifuge at 4°C, the optic density of the gradient at 254 nm was recorded using a Teledyne Isco UV detection apparatus to yield the polysomal profile. The accession number for the ribosome profiling and corresponding RNA-seq data are GEO: GSE67305 (http://www.ncbi.nlm.nih.gov/geo/) for mice fed ad libitum (Janich et al., 2015) and GEO: GSE73554 (http://www.ncbi.nlm.nih.gov/geo/) for mice fed exclusively during the night (Atger et al., 2015).
Immunohistochemistry, Image Segmentation, and Hepatocyte Size Estimation
Mouse livers were fixed using Formalin (Sigma Aldrich) for 24h and rinsed twice with PBS before being embedded in paraffin and cut into 4μm thick slices. Tissue sections were deparaffinized and rehydrated in an ethanol series. After washing with phosphate buffered saline PBS, sections were treated 10 min with 3% hydrogen peroxide in PBS to quench endogenous peroxidase. Heat induced epitope retrieval was then performed with 10 mM Tri-Na citrate 20 min at 95°C. After washing, sections were blocked in 1% BSA in PBS for 30 min and incubated with β-catenin antibody overnight at 4°C. After washing, secondary antibody was incubated 40 min at RT. Cell nuclei were stained with Mayer’s hematoxylin solution. Livers slices were imaged using an Olympus slide scanner at 20X magnification.
Quantification and Statistical Analyses
Estimation of rRNA Synthesis Necessary to Maintain Cellular Ribosome Numbers
Liver cells contain ~10 pg of DNA and ~60 pg of RNA at the time (ZT00) of maximal RNA accumulation (Figure 2C and D). Assuming that 28S rRNA, 18S rRNA, and 5.8S rRNA make up about 80% of total RNA, we estimated that a liver cell contains about 1.2x107 ribosomes. If all 47S pre-rRNA molecules were converted into functional ribosomes, and if ribosomes decayed with a half-life of 24 hr following first-order decay, about 6,000 molecules of pre-rRNA would have to be synthesized per minute per cell (1.2 x107x ln2=24×60 min). The ~300 ribosomal rRNA gene copies of a diploid cell, or a fraction thereof, could readily cope with these demands. However, the production of stoichiometric amounts of ribosomal proteins is likely to be more challenging. Liver cells accumulate between ~300 and 600 RP mRNAs (Atger et al., 2015), and the maximal translation initiation rates for mammalian mRNAs have been estimated to approach ~20 proteins/mRNA/min (Schwanhä usser et al., 2011). Hence, those RP mRNAs expressed at ~300 per cell would have to be translated at full speed to ensure a full complement of ribosomal proteins. Ribosome profiling indicates that they are not (examples shown in Figure 6A-C).
Assuming that the half-live of cytoplasmic rRNA is invariable, the accumulation of cytoplasmic rRNA can be calculated as follows:
Where A = fraction of pre-rRNA converted into rRNA (assuming that this fraction remains constant), [rRNA] = concentration of mature rRNA, x = time of mature rRNA accumulation, t = time of 47/45S pre-rRNA accumulation, t1/2 = cytoplasmic rRNA half-life (assuming first order kinetics for rRNA degradation), and f(t)[pre-rRNA] = function of time of 47/45S pre-rRNA accumulation.
Quantification of Hepatocyte Surface Areas
For quantification of hepatocyte size, 15% of whole-liver immunohistochemistry (IHC) images from the centers of the slices were automatically segmented using a custom script in MATLAB, using the same parameters across all time points. Cells and nuclei were first segmented separately using standard functions from Image Processing Toolbox in MATLAB (e.g., imopen, bwareaopen, and watershed). Only cells for which both nuclei and plasma membranes were detected were kept for the calculation of cell areas. For each sample, the surfaces of more than 1,200 cells were measured (minimal number: 1,231; maximal number: 11,542; average: 4024).
Statistical Analysis
All of the data are presented as the mean ± SD or SEM and represent a minimum of three independent experiments. Statistical parameters, including statistical analysis, statistical significance, and n values are reported in the Figure legends. For in vivo experiments, the number of animals (n) is provided.
For statistical comparison of multiple groups, we performed one-way ANOVA. For statistical comparison of two groups, we performed two-tailed Student’s t test. The values of *p < 0.05, **p < 0.01 ***p < 0.001 and ****p < 10−6 were considered significant; n.s. indicates not statistically significant.
Supplemental Information
Supplemental Information includes seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2017.04.015.
Highlights.
Size, ribosome number, and protein content of mouse livers follow a daily rhythm
Feeding-fasting rhythms drive nuclear rRNA polyadenylation cycles
rRNA polyadenylation cycles are antiphasic to ribosomal protein synthesis rhythms
rRNAs in incomplete ribosomal subunits are polyadenylated and degraded
Acknowledgments
We thank André Liani and Yves-Alain Poget for designing and engineering the automated feeding machines; Pascal Gos for help in using them; Dr. Ingrid Grummt, DKFZ, Heidelberg, for providing anti-RNA polymerase I antibodies; and Nicolas Roggli for the artwork. Dr. Charna Dibner, the current employer of F.S., Geneva Medical School, has generously allocated her time for conducting the revisions. We thank Jessica Sordet-Dessimoz, Histology Core Facility, EPFL, for her assistance in the immunohistochemistry experiments. Work in the laboratory of U.S. was supported by the Swiss National Science Foundation (SNF 31-113565 and SNF 31-128656/1), the European Research Council (ERC-2009-AdG-TIMESIGNAL-250117), the State of Geneva, and the Louis Jeantet Foundation of Medicine. F.S. was funded in part by a long-term EMBO fellowship (ALTF 1464-2012). Work in the laboratory of C.B.G. was funded by the NIH (R01GM111387, R01GM112991, and R01AG045795). D.G. acknowledges funding by SNF professorship grant 157528 and the NCCR RNA & Disease. F.G. acknowledges funding by the European Research Council (through individual starting grants ERC-2010-StG-CIRCATRANS-260988) and the Leenaards Foundation (to F.G. and D.G.). F.G. and D.M. are Nestlé Institute of Health Sciences SA employees.
Footnotes
Author Contributions
F.S., A.G., and U.S. designed the research and analyzed the data; F.S. did most of the experiments; D.M., J.W., and F.G. conducted the cell size estimation; the laboratories of F.G. and D.G. conducted the ribosome profiling analysis; J.J.S. and C.B.G. did the studies with Nocturnin KO mice; U.S. and F.G. supervised the research; and F.S. and U.S. wrote the manuscript.
References
- Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci USA. 2015;112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Berndt H, Harnisch C, Rammelt C, Stöhr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez JP, Hüttelmaier S, Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Søkilde R, Hawkins SM, Coarfa C, Ikeda K, Takayama K, et al. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci USA. 2014;111:11467–11472. doi: 10.1073/pnas.1317751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher HI, Bartlett LM. Diurnal cycles in liver weights in birds. Condor. 1957;59:364–372. [Google Scholar]
- Fishman B, Wurtman RJ, Munro HN. Daily rhythms in hepatic polysome profiles and tyrosine transaminase activity: role of dietary protein. Proc Natl Acad Sci USA. 1969;64:677–682. doi: 10.1073/pnas.64.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, Schibler U. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell. 2013;152:492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Lazar MA. Circadian metabolism in the light of evolution. Endocr Rev. 2015;36:289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotic I, Omidi S, Fleury-Olela F, Molina N, Naef F, Schibler U. Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp. Genes Dev. 2016;30:2005–2017. doi: 10.1101/gad.287094.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CA, Hiatt HH. Turnover of liver ribosomes in fed and in fasted rats. J Biol Chem. 1966;241:5936–5940. [PubMed] [Google Scholar]
- Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015;25:1848–1859. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Schibler U. Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- Leung NW, Farrant P, Peters TJ. Liver volume measurement by ultrasonography in normal subjects and alcoholic patients. J Hepatol. 1986;2:157–164. doi: 10.1016/s0168-8278(86)80074-5. [DOI] [PubMed] [Google Scholar]
- Loeb JN, Howell RR, Tomkins GM. Turnover of ribosomal RNA in rat liver. Science. 1965;149:1093–1095. doi: 10.1126/science.149.3688.1093. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascentseq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. Wiley Interdiscip Rev RNA. 2011;2:256–276. doi: 10.1002/wrna.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammelt C, Bilen B, Zavolan M, Keller W. PAPD5, a non-canonical poly(A) polymerase with an unusual RNA-binding motif. RNA. 2011;17:1737–1746. doi: 10.1261/rna.2787011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, Sinturel F, Gosselin P, Gerber A, Fleury-Olela F, et al. Clock-talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb Symp Quant Biol. 2015;80:223–232. doi: 10.1101/sqb.2015.80.027490. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Schmidt JC, Goldfarb KC, Cech TR, Parker R. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat Struct Mol Biol. 2016;23:286–292. doi: 10.1038/nsmb.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Fremder E, Staals RH, Pruijn GJ, Schuster G. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc Natl Acad Sci USA. 2010;107:7407–7412. doi: 10.1073/pnas.0910621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Sudo H, Nozaki A, Uno H, Ishida Y, Nagahama M. Interaction properties of human TRAMP-like proteins and their role in pre-rRNA 5′ ETS turnover. FEBS Lett. 2016;590:2963–2972. doi: 10.1002/1873-3468.12314. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y. Rhythms in morphology and function of hepatocytes. J Gastroenterol Hepatol. 1990;5:321–333. doi: 10.1111/j.1440-1746.1990.tb01634.x. [DOI] [PubMed] [Google Scholar]
- Udoh US, Swain TM, Filiano AN, Gamble KL, Young ME, Bailey SM. Chronic ethanol consumption disrupts diurnal rhythms of hepatic glycogen metabolism in mice. Am J Physiol Gastrointest Liver Physiol. 2015;308:G964–G974. doi: 10.1152/ajpgi.00081.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mauvoisin D, Martin E, Atger F, Galindo AN, Dayon L, Sizzano F, Palini A, Kussmann M, Waridel P, et al. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017;25:102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M, Ruidant S, Schillewaert S, Leporé N, Lafontaine DL. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009;15:406–419. doi: 10.1261/rna.1402709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WO, McFarland LZ. Diurnal changes in livers and digestive systems of coturnix as related to three photoperiodic regimens. Poult Sci. 1969;48:477–482. doi: 10.3382/ps.0480477. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.