Abstract
The use of RNAi to suppress protein synthesis offers a potential way of reducing the level of enzymes or the synthesis of mutant toxic proteins but there are few tools currently available for their delivery. To address this problem, bioconjugated quantum dots (QDs) containing a hydrophobic component (N-palmitate) and a sequence VKIKK designed to traverse across cell membranes and visualize drug delivery were developed and tested on cell lines of brain origin. We used the Zn outer shell of the QD to bind HIS6 in JB577 (W•G•Dap(N-Palmitoyl)•VKIKK•P9•G2•H6) and by a gel-shift assay showed that siRNAs would bind to the positively charged KIKK sequence. By comparing many peptides and QD coatings, we showed that the QD-JB577-siRNA construct was taken up by cells of nervous system origin, distributed throughout the cytosol, and inhibited protein synthesis, implying that JB577 was also promoting endosome egress. By attaching siRNA for luciferase in a cell line over-expressing luciferase, we showed 70% inhibition of mRNA after 24–48 h. To show more specific effects, we synthesized siRNA for neutral (NSMase2), acid (lysosomal ASMase) sphingomyelinase, and sphingosine kinase 1 (SK1), we demonstrated a dose-dependent inhibition of activity. These data suggest that QDs are a useful siRNA delivery tool and QD-siRNA could be a potential theranostic for a variety of diseases.
Keywords: acid sphingomyelinase, neutral sphingomyelinase2, quantum dots, RNA interference
Many neurodegenerative disorders are characterized by the accumulation of abnormal proteins, such as beta-amyloid peptide (Aβ) in Alzheimer’s, or super-oxide dismutase mutations in Amyotrophic Lateral Sclerosis (ALS) (Tiraboschi et al. 2004; Basso et al. 2013). Small RNA interference is believed to suppress the activation of abnormal genes and may control normal neural development and maintenance. Endogenous miRNAs are secreted in extracellular vesicles by cells following sphingomyelinase activation (Trajkovic et al. 2008) and consist of 21–25 nucleotides that negatively regulate transcription, bind to the 3′-untranslated region of target mRNA and facilitate its degradation. miRNA has been found to play an important role in many neurodegenerative disorders. For example, the Aβ, an important factor in Alzheimer’s disease, is controlled in part by miRNA137/181c, implicating miRNA as a potential therapeutic target (Geekiyanage and Chan 2011). Furthermore, miRNAs are important in the development of oligodendrocytes from glial precursor cells and so have been suggested to be viable therapeutic agents (Dugas and Notterpek 2011). Thus, the administration of miRNA-219 to aging rats has been shown to increase myelination, and miRNA-219 could be useful for the treatment of multiple sclerosis (MS) (Pusic and Kraig 2014). Small interfering RNA (siRNA) is an miRNA analogue which is often used in the laboratory setting for gene silencing and is currently being explored as a potential therapeutic agent. siRNA could both treat diseases caused by miRNA misregulation and be therapeutic in instances where disease is caused by abnormal protein activity. However, the delivery remains a problem despite the potential advantages of short nucleic acid based drugs. siRNA can be delivered without a facilitating carrier/agent, such as an aerosolized siRNA, and most other delivery systems have employed direct injection of potentially toxic lipid nanoparticles (DeVincenzo et al. 2008). However, it is hard to deliver siRNA to the central nervous system (CNS) by these approaches and siRNA can be degraded by endogenous RNAses or filtered out by the kidney and removed by phagocytes post-injection (Whitehead et al. 2009). This limits the medical application of siRNA as a small molecule drug. Using bioconjugated quantum dots (QDs) which are florescent nanovectors and probes, we demonstrated the large potential for overcoming these limitations of drug penetration and safe delivery (Walters et al. 2012, 2015; Boeneman et al. 2013; Xu et al. 2013).
We have previously shown that 6 nm CdSe QDs with a ZnS surface and solubilized by a specific dihydrolipoic acid (DHLA)-derived coating will bind a cell-penetrating peptide JB577 (W•G•Dap(N-Palmitoyl)•VKIKK•P9•G2•H6) in which P9G2 acts as a spacer) through (His)6-Zn tight association, and deliver H6-green fluorescent protein to either neurons or glia in the CNS (Walters et al. 2015). The usefulness of QDs for delivery is because of their high quantum yield, large physical cross-section, and their strong one-photon and two-photon absorption over a broad fluorescence range without photobleaching effects (Algar et al. 2011). Their spectral properties make them perfect for long-term imaging and drug/peptide tracking, and QD emissions can be narrowly tuned as a function of their radius, meaning that many different wavelengths are possible (Murray and Kagan 2000).
The large relative surface area of QDs available for conjugation means that we can display > 50 different biomolecules in a controlled manner (Prasuhn et al. 2010a, b). This nanoscaffold could therefore be used to carry various biological materials, such as small siRNAs, peptides (e.g., JB577), or even large proteins (such as H6-green fluorescent protein). Taking advantage of this, Li et al. (2012a,b) synthesized an amino-polyethylene glycol (PEG) complex for the CdSe/ZnS QDs, showed that negatively charged siRNAs were electrostatically adsorbed to the surface of QDs, and demonstrated that these nanocomplexes were taken up by SK-N-SH neuroblastoma cells. When the QD nanocomplex was bound to siRNA for β-secretase (BACE1), there was a 50% reduction in BACE1 expression. To improve delivery efficiency of QDs, we needed to attach a cell-penetrating peptide which promotes egress from the endosomal compartment, and lipopeptide JB577 uniquely provides this function (Delehanty et al. 2010a,b; Boeneman et al. 2013). Further, in order to work with brain tissue (Walters et al. 2012, 2015), we had to overcome two main obstacles to nanoparticle delivery, namely how to selectively target different neural cell types (Walters et al. 2012, 2015) and how to get to subcellular and target organelles without toxicity. We have previously shown JB577 functions as an endosomal release peptide/cytosolic delivery peptide and has neuronal selectivity in rat hippocampal slices (Walters et al. 2012) using negatively charged compact ligands to deliver to neurons and positively charged coats to deliver to glia. We therefore tested the hypothesis that the VKIKK sequence in JB577 could bind and deliver siRNA to intact cells. Our previous studies demonstrated that the coating of the nanoparticle affects the location of delivery, so that a negatively charged compact ligand (CL4) delivered QDs to pyramidal neurons in the hippocampus but not to oligodendrocytes, astrocytes, or microglia. Conversely, if the compact ligand or PEG was positively charged or the extracellular matrix was enzymatically digested with chondroitinases (Walters et al. 2015), the QDs were targeted more to oligodendrocytes. This makes QDs ideal multipurpose delivery vehicles because they not only facilitate specific cell-type delivery (Walters 2015) and are non-toxic, but also allow for real-time tracking of their location in vitro, eliminating the need for the addition of fluorescent tags to track progress. We chose sphingomyelinases to illustrate this delivery because of their importance to the nervous system and their potential role in exosome release (this is blocked by GW4869) (Pusic and Kraig 2014), and our long-term interest in their response to various stress-related neurochemicals (Testai et al. 2004; Jana and Pahan 2007; Qin et al. 2009, 2012; Dawson and Qin 2011).
Material and methods
Quantum dots
CdSe/ZnS core-shell QDs with emission maxima centered at 625 nm were synthesized (Invitrogen, Carlsbad, CA, USA) and made hydrophilic by exchanging the native hydrophobic capping shell with DHLA PEG-based ligands or negatively charged/zwitterionic DHLA-based compact ligands CL1, CL2, and CL4 as described previously (Mei et al. 2008; Algar et al. 2011; Susumu et al. 2011). This renders them soluble in physiological buffers and the resulting particle is about the size of a protein such as the maltose binding protein (Delehanty et al. 2010a,b).
Peptides
The palmitoylated peptide (JB577) sequence used was W•G•Dap(N-Palmitoyl)•VKIKK•P9•G2•H6 where ‘Pal’ corresponds to a palmitate group that is covalently attached to a non-hydrolysable thiolresembling diaminopropionic acid residue functionality synthesized into the peptide backbone (Sapsford et al. 2007). JB578 is AcWG (Pal)P9G2H6 (Boeneman et al. 2013). All peptides were synthesized using Boc (t-butoxycarbonyl)-solid phase peptide synthesis, purified by HPLC, and purity verified by electrospray ionization MS (Dawson et al. 2010). All peptide sequences are written in the conventional N-to-C terminal orientation.
Sphingomyelinase assays
Lysosomal acid sphingomyelinase (ASMase) and neutral sphingomyelinase (NSMase) activities were determined with the fluorimetric substrate hexamethylumbelliferyl (HMU)-phosphorylcholine as previously described (Testai et al. 2004). Briefly, cells were harvested and washed with phosphate-buffered saline (PBS), the pellets were resuspended and lysed in 25 mM Tris-HCl, 150 mM NaCl, and 1% Triton X-100, pH 7.4. For ASMase activity assay, 50 μg proteins were mixed with the fluorogenic substrate HMU-phosphorylcholine, the incubation was carried out at pH 4.5, and 150 mM sodium acetate buffer containing 1 mM EDTA was added to block any NSMase activity. Neutral sphingomyelinase 2 (NSMase2) assay was carried out at pH 7.4 reaction buffer (10 mM MgCl2, 100 mM Tris-HCl, pH 7.4 and 0.1% Triton X-100) included 5 μM fresh dithiothreitol to inhibit any ASMase activity. The HMU released was followed fluorometrically in a 96-well FLX microplate reader. The enzyme activity was calculated from the slope of the graph of intrinsic fluorescence plotted against time and standardized to μg of protein.
RT-PCR for NSMase2 and ASMase mRNA expression in cells
Total RNA was extracted from cultured cells using a Qiagen total RNA extract kit (Valencia, CA, USA). RT-PCR was executed with a RT-PCR one-step kit (Qiagen) and primer pairs specific to mouse Smpd1 using forward: 5′-TGGTTCTGGCTCTGTTTGACTCCA-3′and backward: 5′-TCAGCTGATCTTGG CGAGACTGTT-3′; primer pairs specific to mouse Smpd3 using forward: 5′-ACATCGATT CTCCCACCAACACCT-3′and backward: 5′-AATTCGCACAA TGCAGCTGTCC TC-3′; primer pairs specific to mouse SphK1 using forward: 5′-ATGGTCTGATGCATGAGGTGGTGA-3′and backward: 5′-ATGGTCTGATGCATGAGGTGGTGA-3′and 18s rRNA as control, using forward: 5′-CCAGAGCGA AAGCATTT GCCAAGA-3′and backward: 5′-AATCAACGCAAGCTTATGA CCCG C-3′primers. Briefly, the reaction mixture was prepared in PCR tubes according to the kit menu and put into a Perkin Elmer GeneAMP PCR System 2400 (Perkin Elmer, Waltham, MA, USA). The programming RT-PCR procedure consisted of reverse transcription (50°C for 30 min), initial PCR activation (95°C for 15 min), then 35 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min, annealing temperature may change according to primer melting temperature. The RT-PCR amplified samples were visualized on 1.0% agarose gels using ethidium bromide.
Complex formation and delivery of QD-peptide-siRNA
A quantity of 100 μM solution of JB577 peptide suspended in 10% dimethylsulfoxide, 90% 1×PBS was mixed with 10 μM siRNA for 1 h. siRNA for ASMase and sphingosine kinase 1 (SK1) were purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA and NSMase2 siRNA was purchased from Invitrogen/Life Technologies, Grand Island, NY, USA. Subsequently, a 12–500 μM solution of QD coated with various ligands was added and left to self-assemble for 30 min. QDs were diluted in Dulbecco’s modified Eagle media (DMEM) to a final concentration of 30–50 nM. Human oligodendroglioma (HOG) cells, human cerebral epithelial cells (HCEC) (Dasgupta et al. 2011), mouse fibroblasts (102R), human colorectal carcinoma cells expressing luciferase (HCT-116) and mouse neuroblastoma cells (NB2a) were cultured for 24 h in DMEM supplemented with 10% fetal bovine serum and 1% gentamycin DMEM at 37°C in a humidified atmosphere containing 5% CO2 Cells were treated with QD-JB577-siRNA complex for 24–48 h in serum free DMEM. All experiments with lipofectin were performed according to the manufacturer’s instructions.
Electrophoretic mobility shift assay (EMSA)
For gel mobility shift assays, 0.1 nmol of siRNA was incubated with peptides at 1:X (where X = 0.1, 1, 10, 20, 30 molar ratio of siRNA to peptide) for 30 min, subjected to electrophoresis on 1.5% agarose gels and stained with ethidium bromide. When QDs were incorporated into complexes, JB577 and QD were complexed in a 1: X ratio for 1 h. Subsequently, siRNA was complexed as described above, to produce conjugate with 1: X: 1 molar ratio of QD: JB577: siRNA and subjected to electrophoresis.
Cell lines used in study and cell viability assays
We have previously reported the properties of a cell line derived from an HOG and the characteristics of its sphingomyelinases (Testai et al. 2004). We established stable cell lines from mouse skin fibroblasts which constitutively expressed the SMPD3 gene for neutral sphingomyelinase2 (NSMase2) and the SMPD1 gene for acid sphingomyelinase (Qin and Dawson 2012). HCT-116 cells were a generous gift from Dr. Tong Chuan, University of Chicago, and were designed to express the luciferase protein. HCEC cells were derived from human brain endothelium (Dasgupta et al. 2011) and were a generous gift from Dr. RK Yu. Cells were plated in 24-well culture plates for 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, which was carried out as described previously (Wiesner et al. 1997).
All experiments were approved by the Institutional Biosafety Committee at the University of Chicago (protocol number 829 on 10/09/14).
Cellular luciferase assay for the functional gene silencing effect
Upon 24 or 48 h of incubation after transfecting luciferase siRNA, the HCT-116 luciferase-expressing cells were washed with PBS and lysed in a passive lysis buffer (Promega, Madison, WI) for 15 min. A sample of 20 μL of the lysates were transferred to a black 96-well plate and 100 μL of luciferase assay reagent (Promega) was added per well. After mixing, total luminescence intensity was quantified using a synergy H1 multimode reader, following luciferase assay manufacturer’s instructions. The total protein amount was quantified by a bicinchoninic acid assay to correct the luminescence intensity per milligram of protein.
Cell staining and microscopy image analysis
Monolayer cultures of cells were fixed to gel-coated slides using Prolong Gold Antifade (Invitrogen) containing 4′,6-diamidino-2-phenylindole, a nuclear stain that binds strongly to A–T-rich regions in DNA. siRNA was labeled with Cy3 according to the manufacturer’s instructions (Thermo Fisher Scientific Inc, Skokie, IL), and was transfected as per the previously described protocol. The intracellular distribution of QDs was analyzed by fluorescence microscopy using a Marianas fully automated Yokogawa-type spinning disc confocal microscope equipped with ×40 and ×100 oil immersion lenses.
Statistical analysis
All experiments were repeated at least three times and sample run in triplicate to sufficient to attain statistical significance. The statistical data is expressed as mean ± standard errors and was statistically analyzed by conducting an ANOVA followed by Tukey–Kramer method with a post-hoc analysis (*p < 0.01).
Results
JB577 binds to siRNA with high affinity in vitro
Based on the routine use of polyhistidine-tags on protein purification by tightly binding to Ni or Zn affinity resins, we used an aniline-catalyzed hydrazine coupling to link hexahistidine sequences to peptides, such as JB577 (Prasuhn et al. 2010a,b), so that they would bind to Zn on the surface of our QDs (Fig. 1). We had previously shown that the lipopeptide JB577 (W•G•Dap(N-Palmitoyl)•VKIKK•P9•G2•H6), based on the C-terminal KRas4A structure and designed to be a competitive inhibitor of palmitoyl-protein thioesterase1 (Cho and Dawson 1998), was stable and uniquely able to promote egress from endosomal compartments in cultured cells (Dele-hanty et al. 2010a, b), in effect functioning as a cell-penetrating peptide. Since we demonstrated that JB577 on the surface of CL4-QDs was targetedto neuronalcytosol, we sought to further demonstrate that siRNA could be electrostatically associated/complexed with the VKIKK section of JB577. It has been previously shown that negatively charged nucleic acids can be bound to positively charged amino acids by electrostatic interaction, suggesting that this linkage might possibly hold long enough to facilitate QD-mediated siRNA delivery (Li et al. 2012a,b). Thus, negatively charged siRNA could bind to positively charged lysine residues in the VKIKK sequence of JB577 via such electrostatic interaction. The VKIKK sequence has a similar affinity to the polyarginine motif of RGD-9R (Kumar et al. 2007). However, the RGD-9R sequence did not confer the endosomal escape observed with JB577 (Boeneman et al. 2013) so there must be something unique about JB577. We propose that the VKIKK part of JB577 is required for both the siRNA binding and the endosomal membrane penetration. We observed the complex to be distributed throughout the cytoplasm (Walters et al. 2012; Boeneman et al. 2013), although we do not have data on how long the complex stays associated, the subsequent suppression of specific RNA was very clear.
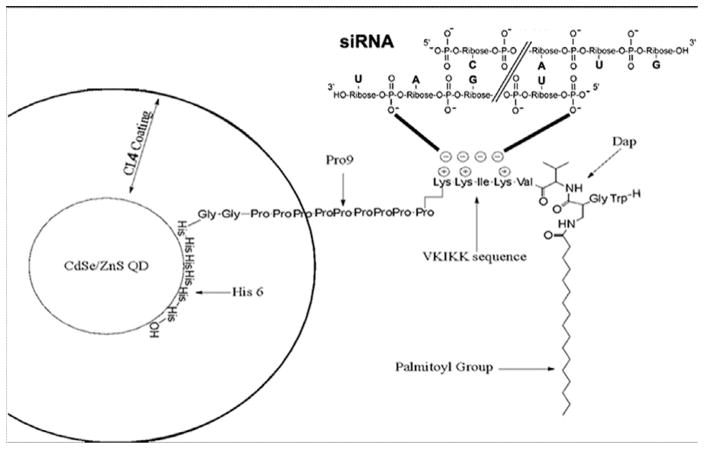
Fig. 1.
Structure of siRNA conjugated JB577 peptide bound to the surface of the QD via a His6 (Histidine) sequence. siRNA is conjugated to the positively charged VKIKK sequence by positive/negative charge interaction. The CdSe/ZnS QD is coated in negatively charged CL4 ligand occupying less space than polyethylene glycol (PEG)-derived ligands used by other investigators. WG is added to monitor the peptide during purification by HPLC and the palmitoyl group is N-linked to a modified cysteine (Dap) to mimic the native thiol linkage. Theoretical calculations for pH7.4 suggest the following properties of the different solubilizing coatings used: CL1 = +0.61−1.00 + 0 = −0.39 charge (one full negative charge) and a partially protonated amine complicated by the piperazine ring. CL2 = −0.04 net charge. Almost a full zwitterion, the amine is partially deprotonated. CL4 = +1.00−2.00 = −1.00 net charge. Two negative charges, one fully protonated amine, positive charge. PEGNH2 = +0.9 net charge. One positive charge mostly fully protonated.
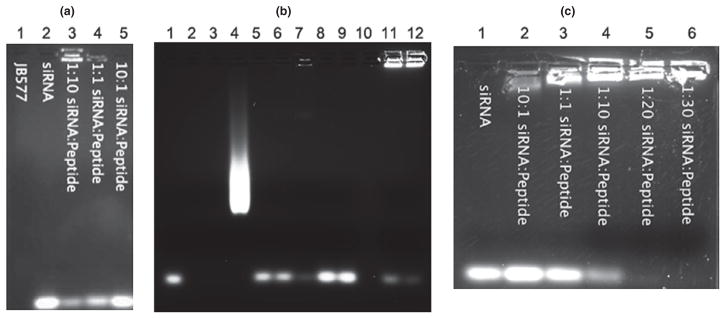
In order to confirm that the lysine residues of JB577 could effectively bind siRNA, we performed an EMSA, in which the amount of unbound siRNA remaining in the front of migration (Fig. 2a) and the siRNA: JB577 complex running close to gel well were visualized by ethidium bromide staining (Fig. 2a, lane 3; Fig. 2b, lane 7). As a control, we showed that JB578 which lacks VKIKK sequence did not bind to the siRNA (Fig. 2b, lanes 8 and 9) (Boeneman et al. 2013). These results suggest the interaction between negatively charged siRNA and positively charged VKIKK on JB577. The ratio of siRNA to peptide was varied with the goal of generating a formulation capable of high cell uptake as well as the ability to carry a significant payload of siRNA. Subsequently, we repeated EMSA with JB577 conjugated with PEG900-QD before siRNA complex assembly over a range of concentrations of JB577 and confirmed that conjugation of JB577 to QD had no effect on the affinity of the VKIKK sequence for siRNA (Fig. 2c). When QD conjugated with JB577: siRNA complex, QD: JB577: siRNA complex bands staying close to gel well (Fig. 2b, lanes 11 and 12) and is much more bright than the band of siRNA: JB577 complex alone (Fig. 2b, lane 7), at the same time free QD smear on agarose gel will be reduced or disappeared (Fig. 2b). The QD concentration used was equimolar to the concentration of siRNA in each group. We determined empirically that 1: 20 was the optimal siRNA to cellpenetrating peptide (CPP) molar ratio, and that there was no benefit to be derived from increasing the peptide concentration beyond this ratio (Fig. 2a and c). Thus, the most advantageous molar ratio of components for our QD conjugates is 1: 20: 1, QD: JB577: siRNA. This is consistent with previous studies in which a transferrin receptor-targeting peptide bound siRNA at a 1: 20 molar ratio, and an RGD peptide with a polyarginine motif bound siRNA at a 1: 10 molar ratio (Kumar et al. 2007; Furgeson et al. 2014). We utilized both CdSe/ZnS core/shell QD’s that were solubilized with PEG-DHLA, which emit at a wavelength of 625 nm, and Cy3 labeled siRNA (Life Technologies) that emitted at a wavelength of 570 nm. Studies have suggested that ~ 50 peptides could be displayed on 530–550 nm QDs functionalized with DHLA-based ligands. Thus, the larger diameter of the 625 nm QDs (~ 10 nm) suggests a capacity to display a greater number of peptides and so easily display the 20 peptides needed for our delivery complex (Delehanty et al. 2009).
Fig. 2.
Gel-shift assays to show siRNA binding to JB577 requires VKIKK. (a) Gel shift mobility assay displaying migration of free or peptide complexed siRNA. Peptide is free and not QD complexed. Three concentrations of siRNA: peptide were tested to determine optimal linkage concentrations on a 2% agarose gel. (b) Gel mobility shift assay showing the relative affinities of JB578, JB577, and QD: JB577 to siRNA. The lane assignments are as follows: 1: siRNA, 2: JB577, 3: JB578, 4: QD, 5: JB577:siRNA/3:1, 6: JB577:siRNA/6:1, 7: JB577:siRNA/9:1, 8: JB578:siRNA/6:1, 9: JB578:siRNA/9:1, 10: Blank, 11: siRNA:JB577:QD/1:6:1, 12: siRNA:JB577:QD/1:9:1. (c) Gel shift mobility assay displaying mobility of free or QD-peptide complexed siRNA. Peptide was assembled to QD prior to treatment with siRNA. Five concentrations of siRNA: peptide: QD were tested to determine optimal linkage concentrations on a 1.5% agarose gel.
QD-JB577 delivers siRNA into cytosol and facilitates endosomal escape in multiple cell lines
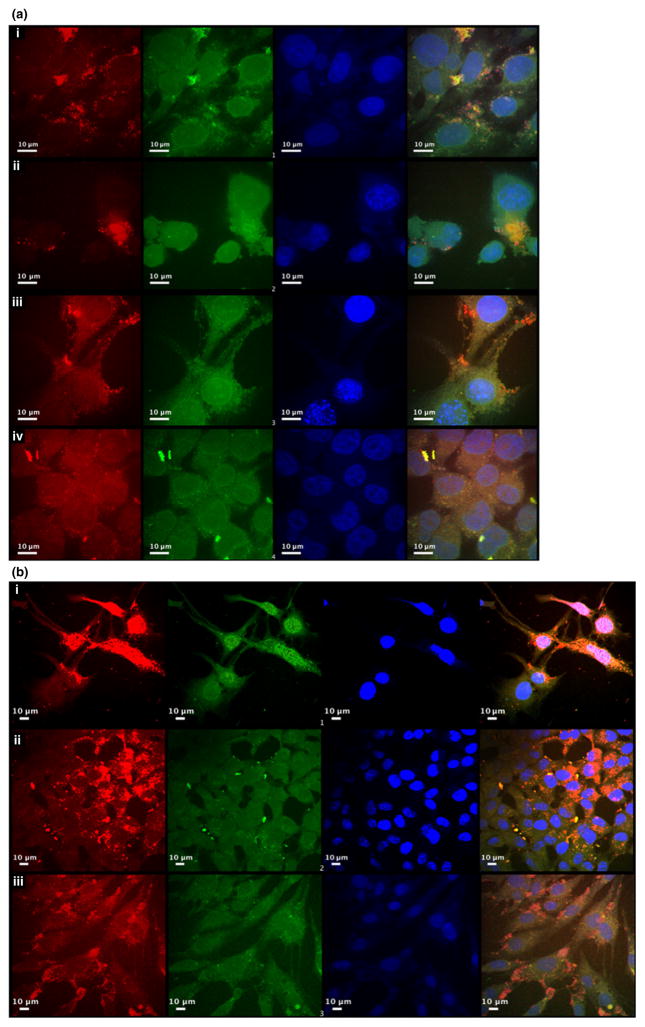
As previously described (Delehanty et al. 2009, 2010a,b; Boeneman et al. 2013), the addition of the CPP, JB577, was able to facilitate cellular uptake and endosomal escape. Moreover, elements, such as charge and lipid content, have been shown to have an effect on the efficiency of delivery (Boeneman et al. 2013). Although QDs have been extensively studied, no one has definitively shown a functional effect by delivering a biological signal with QDs. We confirmed that the QD-PEG900-JB577-siRNA conjugates were able to facilitate endosomal escape in transfecting cells after 24 h. In images taken with a confocal microscope at both 100×(Fig. 3a) and 40×(Fig. 3b) magnification, both siRNA (green) and QD(red) appeared well dispersed throughout the cytosol in variety of cells and occupying the entire cell volume, indicating successful endosomal escape (Fig. 3a, Panels (i–iv) and Fig. 3b Panels (i–iii)). QD-PEG900-JB577-siRNA exhibited delivery efficiency in allowing QDs to access the cytosol of different cell lines representing several tissue types: namely, HOG cells, HCEC, mouse skin fibroblast (102R), and human colorectal carcinoma cells expressing luciferase (HCT-116). QDs appeared evenly distributed in colonic cell lines HCT-116 and HCEC (Fig. 3a and b) and somewhat localized in 102R and HOG cells. This could be because of either aggregation in the media or incomplete escape from endosomal compartments. However, the siRNA (Cy3) does appear to be evenly distributed throughout the cytosol [Fig. 3a panels (i and ii) and 3b, panel (i)], suggesting that siRNA is detaching from JB577 during delivery and that there is robust endosomal escape of siRNA. It is also possible that QD localization is as a result of slower uptake in HOG and 102R cells, as nanoparticle kinetics can vary greatly in different cells lines (Doiron et al. 2011).
Fig. 3.
Confocal microscopy evidence for delivery of siRNA conjugated JB577 QD into cytosol. (a) Cells were transfected with polyethylene glycol (PEG) QD-JB577-siRNA constructs for 24 h, siRNA was labeled with Cy3 (green), quantum dots are shown as red and nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). The cells were then confocal imaged with a 100× oil immersion lens. The cell types were as follows (i) 102R fibroblasts (ii) human oligodendroglioma (iii) human cerebral epithelial cells (HCEC) (iv) HCT-116. (b) Cells were transfected with PEG QD-JB577-siRNA constructs for 24 h, siRNA was labeled with Cy3 (green), quantum dots are shown as red and nuclei stained with DAPI (blue). The cells were then imaged with a 40× oil immersion lens. The cell types were as follows (i) 102R (ii) HCT-116 (iii) HCEC.
QD ligand coatings facilitate cellular siRNA uptake
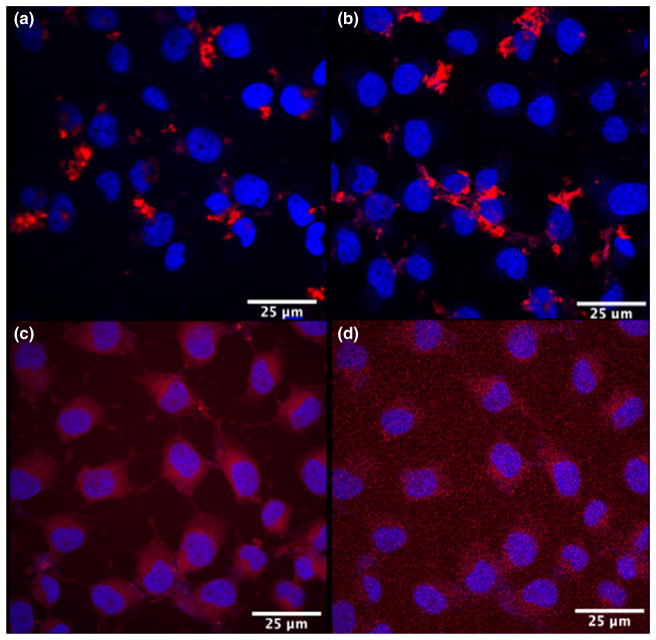
It has been previously demonstrated (Walters et al. 2012, 2015) that the charge of QD coating selectively influences the cell type in which the QD will be delivered. Therefore, negatively charged CL4 ligand has been shown to selectively target neurons over glial cells (Walters et al. 2012). A recent study by us (Walters et al. 2015) shows that the more zwitterionic CL2 and CL1 coating ligands showed declining preferential uptake by neurons, and that QDs coated with a positively charged PEG-600-NH2 was preferentially taken up by oligodendrocytes. We tested four different coatings: the positively charged PEG900, CL1, the zwitterionic CL2, and the negatively charged CL4 to ensure that all QD-polymer coatings could deliver siRNA to the cytosol of HOG cell (Delehanty et al. 2010a,b). Compact ligands CL1, CL2, and CL4 are all based on DHLA with the main difference being charge, whereas PEG900 is less compact (Susumu et al. 2011). Initial results showed robust delivery of QD evenly distributed throughout the cytosol with coatings PEG900 and CL1 (Fig. 4c and d), but not so well with CL2 and CL4 QD (Fig. 4a and b). Subsequently, Cy3 siRNA was attached to QD-JB577 conjugates and used to transfect HCT-116 cells. We showed that PEG, CL1, and CL2 QD appeared well dispersed throughout the cytosol, whereas CL4 appeared to be aggregated (Fig. 5). However, the siRNA was evenly distributed throughout the cytosol, again indicating that siRNA is detaching from JB577 during delivery (Fig. 5a–d). In each case, siRNA was efficiently delivered to the cytosol of cells.
Fig. 4.
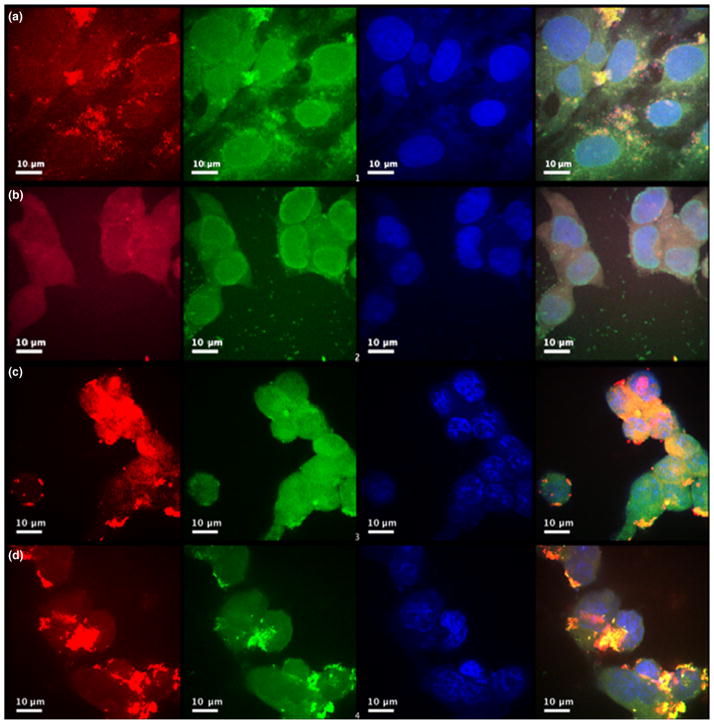
Effect of differently charged compact ligands on QD-JB577 uptake. QDs (red) with different coatings were uptaken by Human oligodendroglioma cells. (a) QDs with CL2 coating. (b) QDs with CL4 coating. (c) QDs with polyethylene glycol coating and (d) QDs with CL1. Nuclei stained with 4′,6-diamidino-2-phenylindole (blue).
Fig. 5.
QDs coated with polyethylene glycol (PEG), CL1, or CL2 and siRNA are evenly distributed throughout the cytosol of HCT-116 cell, indicating that siRNA is efficiently delivered to the cytosol and detached from JB577. Scrambled siRNA labeled with Cy3 (green) was attached to QD-JB577 (red) with coating (a) PEG (b) CL1 (c) CL2 (d) CL4. Cells were transfected for 24, labeled with 4′,6-diamidino-2-phenylindole to visualize the nuclei (blue), then imaged with a 100×oil immersion lens.
siRNA-JB577 QD conjugates facilitate robust enzyme knockdown
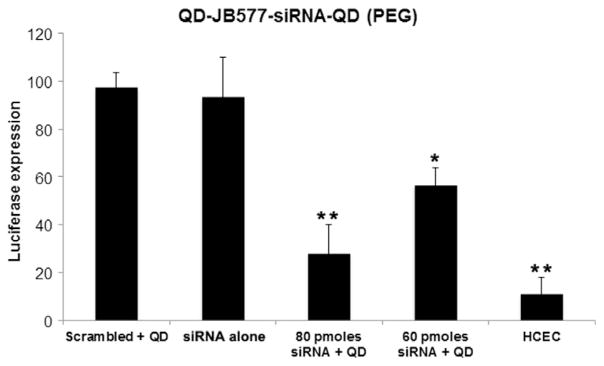
We investigated whether the internalized siRNA leads to efficient enzyme knockdown in HCT-116 cells which constitutively express luciferase. A luciferase reporter assay was previously used to demonstrate functional gene silencing in a human glioma cell line and primary murine neurons/astrocytes (Furgeson et al. 2014). The HCT-116 cells were treated with siRNA targeted to luciferase at various concentrations as well as scrambled siRNA. All transfections were 24–48 h and used 1: 20: 1 molar ratios of QD: JB577: siRNA. QD was coated with PEG-ligand in order to prevent any adherent interactions or interference by QD aggregation (Fig. 3a and b). Luminescence was measured to determine knockdown efficiency (Fig. 6), 80 picomoles of siRNA delivered via the QD-siRNA conjugate reduced luciferase activity to 28.0 ∓ 12.0% of control compared to only 56.5 ∓ 7.5% with 60 picomoles of siRNA. A baseline luminescence (10.9 ∓ 7.2%) was determined by a luminescence assay on HCEC cells which do not express luciferase. Neither of the control groups, the scrambled siRNA-QD-JB577 conjugate or the siRNA without QD or JB577, differed in a statistically significant manner from the untreated HCT-116. The results suggest robust knockdown of luciferase in vitro compared to control groups. The knockdown efficiency reported here exceeds the 50% reported in previous studies (Derfus et al. 2007; Li et al. 2012a,b), indicating an advancement in QD-siRNA delivery technology.
Fig. 6.
Expression of luciferase in HCT cells is reduced by QD-siRNA. QD-siRNA uptake was monitored through the Promega luciferase assay system. All cells were treated with QD-siRNA constructs for 24 h, and luciferase expression was protein normalized. QD-siRNA significantly reduced luciferase activity through a doseresponse manner. Control siRNA (second column) is 80 picomoles. Data is expressed as mean ± standard errors and was statistically analyzed by conducting an ANOVA followed by Tukey–Kramer HSD post-hoc analysis (*p < 0.01, **p = 0.05).
QD-siRNA conjugates facilitate knockdown of NSMase2 and ASMase with similar efficiency to lipofection
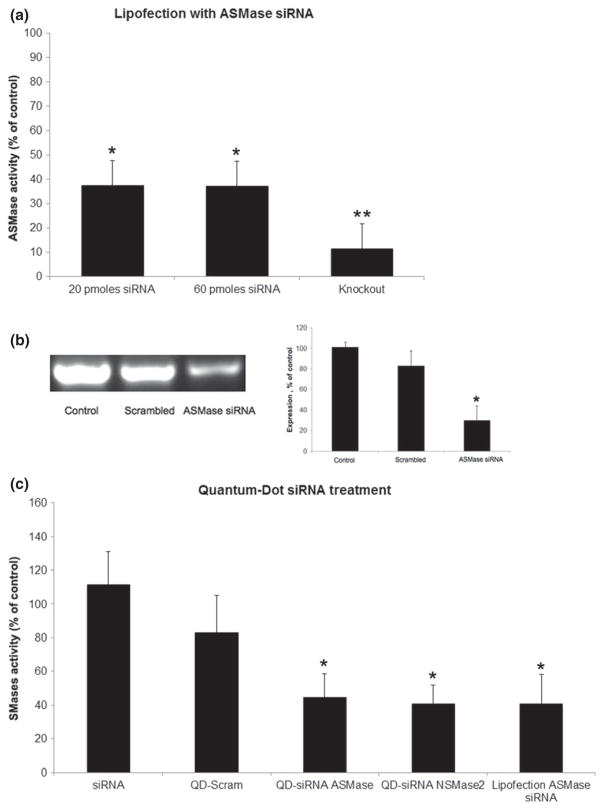
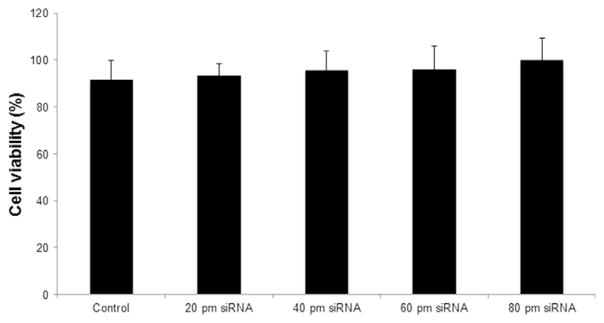
In order to confirm knockdown efficiency, we demonstrated that our QD-siRNA constructs are a viable method of sphingomyelinase knockdown in vitro. We attempted knockdown of ASMase and NSMase2 proteins in 102R and HOG cells, respectively. The high activity of SMases in these cells makes them ideal targets for siRNA-QD knockdown detection. ASMase is abnormal in Niemann-Pick types A and B, and is a target of antidepressive drug treatments. It has been suggested that ASMase knockdown protects against ischemic injury in murine models as it reduces ceramide levels and caspase 3 activation (Llacuna et al. 2006). NSMase2 responds to oxidative and other neurological stresses; more intriguingly, NSMase2 has been shown to induce exosomal secretion and miRNA regulation in many cells (Trajkovic et al. 2008; Kosaka et al. 2013). Additionally, NSMase2 derived ceramide has been implicated in neuronal cell death, and oxidized phosphatidylcholine can activate NSMase2, which converts sphingomyelin into ceramide, causing downstream activation of caspases 3 and 8, which have proapoptotic effects in oligodendrocytes (Qin et al. 2009). In order to demonstrate a practical efficiency, QD transfection results were compared with a standard siRNA lipofection delivery method (Santa Cruz Biotechnologies). After lipofection, RT-PCR indicated that ASMase RNA was reduced to 63.4% of original levels in 102R cells (Fig. 7b), and ASMase activity was 41.0 ∓ 11.2% of the untreated control values (Fig. 7a and b). Unlike QD-siRNA conjugates, the knockdown efficiency was invariant with regard to the amount of siRNA added above 20 picomoles, the minimum according to the manufacturer’s instructions (Fig. 7a). After 60 picomoles of siRNA against either ASMase or NSMase2 siRNA-QD treatment, ASMase activity was to 45.0 ∓ 13.5% and NSMase2 was 41.2 ∓ 12.5% (Fig. 7c). Similarly, lipofection of ASMase siRNA yielded knockdown to 41.0 ∓ 11.2% of control activity, indicating that there is no significant difference between siRNA delivery efficiency between QD and lipofection (Fig. 7c). In contrast to the > 50% significant cell death which occurred during lipofection treatment, there was no statistically significant difference in cell viability at any concentration of QD-siRNA constructs after 24 h of treatment (Fig. 8). In addition, when compared to unaffected cell growth following knocking down of ASMase, we observed an inhibition of cell proliferation in neuroblastoma NB2a cells by a QD-siRNA complex targeting SK1 (Figure S1). SK1 is a key enzyme catalyzing phosphorylation of the metabolite of ceramide sphingosine to form S1P, the cell proliferation stimulator (Ogretmen and Hannun 2004; Pyne and Pyne 2010; Maceyka et al. 2012). Figure 9 is a scheme summarizing how QD delivered siRNA can reduce the expression of SMases.
Fig. 7.
Expression of SMases is reduced by QD-siRNA treatment. (a) Acid sphingomyelinase (ASMase) activity in cells that were transfected with siRNA using a commercial lipofection kit. Column 1 and 2 using 20 and 60 picomoles siRNA to knock down Smpd1 in mouse fibroblasts, and column 3 showing enzyme activity from Smpd1 (ASMase) knock out mouse fibroblasts. (b) Left Panel: RT-PCR electrophoresis showing levels of mRNA in cells treated by lipofectin transfected siRNA. Baseline knockdown efficiency varied between 50% and 63.4% by RT-PCR and fluorometric activity assays. Right Panel: Showing relative percentage of control. (c) Sphingomyelinases activity after 24 h treatment with QD-siRNA constructs. Human oligodendroglioma (HOG) cells and HOG cells over-expressing Neutral sphingomyelinase 2 were treated with QD-JB577-siRNA complexes at a molar ratio of 1 QD: 20 JB577: 1 siRNA, and activity was measured by a fluorometric assay. Lipofection was performed with Santa Cruz commercial lipofection kit using 60 picomoles of siRNA. All results were compared to untreated control. (*p = 0.05).
Fig. 8.
Lack of change in survivability after 24 h of QD-siRNA treatment in this study. Viability was measured via the mitochondrial integrity (3-(4,5-dimethylthiazol-2-yl)-2,5-di phenyltetrazolium bromide) assay.
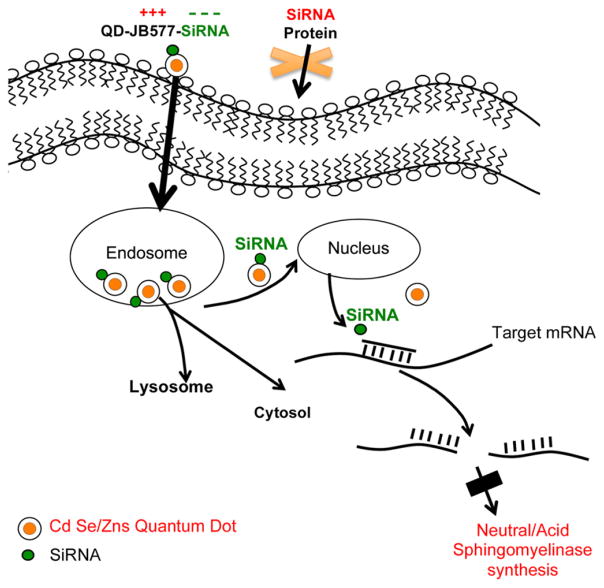
Fig. 9.
A scheme to explain QD delivered siRNA knocking down the synthesis of SMases.
Discussion
A key challenge to the therapeutic potential of siRNA-technology is the need for safe and effective delivery methods. To activate the RNAi pathway, siRNA molecules must be delivered to the interior of target cells and be incorporated into the RNAi machinery. siRNA molecules alone are too hydrophilic to diffuse across cell membranes (Whitehead et al. 2009); meanwhile, the unmodified siRNA is unstable when administered into blood vessels, naked RNA can be degraded by serum nucleases in the bloodstream and does not readily cross membranes to enter cells (Layzer 2004; Jackson and Linsley 2010; Nguyen 2012). Therefore, some protective encapsulation or chemical modification is required to assist the uptake of siRNA by target cells (Whitehead et al. 2009). To date, various delivery platforms for siRNA have been explored in order to address the challenges of in vivo delivery, including lipid nanoparticles, dynamic polyconjugates, triantennary GalNAc-siRNA, and oligonucleotide nanoparticles etc. (Li et al. 2012a,b; Kanasty et al. 2013). Of these, liposomes have been widely used for siRNA delivery because they can carry a relatively large payload, but their usefulness has been limited by their complicated construction and their poor performance in vivo because of uptake by non-specific tissues (Elbayoumi and Torchilin 2009). As a new approach to siRNA delivery, we believe that QDs, with their exclusive characteristics of high, stable intrinsic fluorescence, and low toxicity, allow for both specific delivery and the ability to follow their cellular fate after administration.
The treatment of disease often requires the separate use of both diagnostic tools and therapeutics in order to ensure treatment efficacy. Multipurpose delivery vehicles, such as luminescent nanoparticles could potentially unify these steps, enabling both drug delivery and diagnostic imaging in a single ‘Theranostic’ package. The chemical characteristics of QD-JB577-siRNA complexes make them well suited for the delivery of siRNA. QD-JB577 was found to electrostatically self-assemble with siRNA at an optimal ratio of 1: 20: 1/QD: JB577: siRNA, which is consistent with previous literature values of peptide-siRNA binding affinity (Kumar et al. 2007; Furgeson et al. 2014). Confocal images of four different cell types (HOG, HCT-116, HCEC, and 102R) showed that siRNA was evenly distributed throughout the cytosol of cells. We propose that QD localization is as a result of differences in uptake speed across cells, and nanoparticle kinetics may vary in different cells derived from the nervous and other tissues (Doiron et al. 2011). Endosomal egress is mediated by JB577, and we have previously published optimum conditions for PC12 cells (Boeneman et al. 2013). Our results support the hypothesis that QD-JB577 can effectively bind and deliver siRNA intracellularly to brain-derived cells and that binding depends on the VKIKK sequence. The ability of QD-JB577 to egress endosomes may also depend on the VKIKK sequence since the IKK motif in tree frog Aurein has recently been reported be able to promote endosomal egress (Li et al. 2015).
Likewise, confocal imaging also demonstrated delivery of siRNA utilizing four different QD ligand coatings (PEG, CL1, CL2, and CL4). PEG900, CL1, and CL2 (Fig. 1) all appeared to be distributed throughout the cell volume, while CL4 aggregation could be as a result of effects of negatively charged carboxyl groups on ligand, or lack of endosomal escape and/or clustering in lysosomes. Regardless, siRNA seems to have detached from JB577 and dispersed throughout the cells, indicating that siRNA is able to reach the targeted area. The ligand coating which is needed to solubilize the QD is primarily important because it allows for cell-type specific targeting (Minami et al. 2012; Walters et al. 2012, 2015), and it also might protect siRNA from digestion of intracellular RNase. Furthermore, QD-siRNA constructs demonstrated robust knockdown of 72.0 ∓ 12.0% luciferase, which is similar or better than commercial transfection reagents. The significant knockdown by siRNA-QDs complexes suggests that the siRNA was protected from RNases digestion by the QD coating and multiconjugated JB577 on QDs. This is consistent with other reports (Li et al. 2012a,b). Subsequent treatment with QD-siRNA constructs carrying SMases siRNA resulted in knockdown efficiencies of 45% for ASMase and 41% for NSMase2, respectively. This is comparable to the levels recorded using a commercial lipofection kit. Finally, at the proposed QD and siRNA concentration, there was no observed toxicity in contrast to the toxicity observed with lipofectamines (> 50%). These cationic lipids appear to irreversibly damage the cell surface and cause the DNA to become blobby, stringy, and clump in endosomes (the ‘nasty fuzzballs’ as described by Heuser 2014). The imaging and the transfection results support our claim that these complexes are viable for targeted cellular uptake and functional siRNA release into the cytosol.
Can QDs ever become a viable way of reducing the synthesis of a specific mutant brain protein and thus reversing diseases such as AD, PD, HD, and ALS? Much of the work in this area has been done to image or treat brain tumors. Derfus et al. (2007) previously demonstrated that QDs functionalized with CPP may be conjugated to siRNA via disulfide linkages and targeted to metastatic tumors. However, this study was unable to show targeting properties and a good knockdown efficiency (Derfus et al. 2007). We have recently reported that QDs functionalized and solubilized with CL4 and can specifically target neurons over astrocytes and microglia, and can deliver cargo intracellularly all over the brain when combined with a CPP (Walters et al. 2012, 2015; Boeneman et al. 2013; Agarwal et al. 2015). When injected into the intact developing chick brain, the QD-JB577 conjugates appear in neural precursor cells of developing chick embryo. Migration of QDs are cleared from the choroid plexus of developing chick embryos at days 15–19 with minimal toxicity, suggesting in vivo application could likely be successful (Agarwal et al. 2015). Similar QD studies have shown success when delivering siRNA for matrix-degrading metalloproteinase-9 and siRNA for BACE1 to achieve high transfection efficiency of siRNAs (Bonoiu et al. 2009; Li et al. 2012a,b). In addition, gold nanoparticles have been used to demonstrate transfection agent-free delivery of siRNA (Zheng et al. 2012), and preliminary studies utilizing a TAT-derived CPP claimed successful siRNA delivery to mouse brains (Kumar et al. 2007). The delivery vehicle we used is capable of cellspecific delivery and can be replaced with biodegradable QDs if necessary. Thus, we believe that this study represents a significant neurochemical approach to siRNA delivery, and further suggests that using QD-DHLA PEG900-coated peptide complexes with electrostatically linked siRNA could act as superior delivery vehicles in vivo.
The blood–brain barrier (BBB) is a major obstacle to applying a basic neurochemical approach to clinical situations. It has been previously reported that QD-TAT conjugates intraarterially delivered to a rat brain and were able to pass over the endothelial cell line and reached the brain parenchyma, allowing for successful labeling of some brain tissue regions (Santra et al. 2005). Likewise, some nanoparticles coated with polysorbate 80 could transport a peptide across the BBB after intranasal administration (Ruan et al. 2012). Further, a recent study had also shown that transferrin (Tf) -conjugated QDs traversed across on artificial BBB model (Mahajan et al. 2012). Our previous study has shown QD-JB577 complexes extensively label all parts in the chicken brain when injected into the spinal canal (Agarwal et al. 2015). More promising for the nervous system delivery is our finding that functionalized QD-peptide complexes can track neural cells when they migrate and are cleared from the brain of developing chick embryos with minimal toxicity, and are thus capable of delivery of drugs to brain cells (Agarwal et al. 2015).
In conclusion, the QD-JB577-siRNA complex shows stable self-assembly, and siRNA does not interfere with the ability of the QD-JB577 construct to facilitate either cellular uptake or endosomal escape. Moreover, siRNA was delivered in sufficient quantities as to effect a substantial change in enzyme expression as shown by luciferase and sphingomyelinase assay. We chose SMases knockdown to demonstrate the utility of the QD approach because they are enzymes activated in the CNS following various kinds of stress (Qin et al. 2009, 2012, 2015; Qin and Dawson 2012). Knockdown efficiency by QD-siRNA was roughly comparable to a commercially siRNA delivery lipofection kit but without the cytotoxicity. Our work may ultimately provide a promising strategy for therapeutic siRNA delivery and potential useful for treating variety of diseases in the future.
Acknowledgments
We thank Vitas Bindokas and Christine Labno in the Microscopy Core of University of Chicago for help and advice with the interpretation of images. This work was supported by United States Public Health Service (USPHS), grant numbers NS36866–40 to G.D.; GM098871 to P.D.; I.M. was supported by Naval Research Laboratory (NRL), the Naval Research Laboratory Nanoscience Institute (NRL NSI), Office of Naval Research (ONR), Defense Advanced Research Project Agency (DARPA), Defense Threat Research Agency (DTRA), Joint Science and Technology Office (JSTO), and Military interdepartmental Purchase Request (MIPR) [grant number B112582M].
All experiments were conducted in compliance with the ARRIVE guidelines.
Abbreviations used
- ASMase
acid sphingomyelinase
- CL
compact ligand
- CNS
central nervous system
- CPP
cell-penetrating peptide
- DAPI
4′,6-diamidino-2-phenylindole
- DHLA
dihydrolipoic acid
- EM
electron microscopy
- GFP
green fluorescent protein
- HCEC
human cerebral epithelial cells
- HOG
human oligodendroglioma
- JB577
WG (Dap [Palmitoyl] VKIKK P9 GG His6
- miRNA
microRNA
- NSMase2
neutral sphingomyelinase 2
- PEG
polyethylene glycol
- PPT1
palmitoyl: protein thioesterase 1
- QD
quantum dot
- RNAi
RNA interference
- siRNA
small interfering RNA
- SK1
sphingosine kinase 1
Footnotes
Conflict of interest disclosure:
The authors declare no conflict of interest.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article:
Figure S1. A QD: JB577, and siRNA complex targeted to SphK1, knocks down the expression of SphK1 and inhibits proliferation of neuroblastoma cell NB2a.
References
- Agarwal R, Domowicz M, Schwartz N, et al. Delivery and tracking of quantum dot peptide bioconjugates in an intact developing avian brain. ACS Chem Neurosci. 2015;6:494–504. doi: 10.1021/acschemneuro.5b00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algar W, Susumu K, Delehanty J, Medintz I. Semiconductor quantum dots in bioanalysis: crossing the valley of death. Anal Chem. 2011;83:8826–8837. doi: 10.1021/ac201331r. [DOI] [PubMed] [Google Scholar]
- Basso M, Pozzi S, Tortarolo M, Fiordaliso F, Bisighini C, Pasetto L, Bonetto V. Mutant copper-zinc superoxide dismutase (SOD1) Induces protein secretion pathway alterations and exosome release in astrocytes: implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J Biol Chem. 2013;288:15699–15711. doi: 10.1074/jbc.M112.425066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeneman K, Delehanty JB, Blanco-Canosa JB, et al. Selecting improved peptidyl motifs for cytosolic delivery of disparate protein and nanoparticle materials. ACS Nano. 2013;7:3778–3796. doi: 10.1021/nn400702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonoiu A, Mahajan SD, Ye L, Kumar R, Ding H, Yong K-T, Prasad PN. MMP-9 gene silencing by a Quantum Dot-siRNA nanoplex delivery to maintain the integrity of the blood brain barrier. Brain Res. 2009;1282:142–155. doi: 10.1016/j.brainres.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Dawson G. Enzymatic and molecular biological analysis of palmitoyl protein thioesterase deficiency in infantile neuronal ceroid lipofuscinosis. J Neurochem. 1998;71:323–329. doi: 10.1046/j.1471-4159.1998.71010323.x. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Wang G, Yu RK. Sulfoglucuronosylparagloboside promotes endothelial cell apoptosis in inflammation: elucidation of a novel glycosphingolipid-signaling pathway. J Neurochem. 2011;119:749–759. doi: 10.1111/j.1471-4159.2011.07483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Qin J. Gilenya (FTY720) inhibits acid sphingomyelinase by a mechanism similar to tricyclic antidepressants. Biochem Biophys Res Commun. 2011;404:321–323. doi: 10.1016/j.bbrc.2010.11.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Schroeder C, Dawson PE. Palmitoyl: protein thioesterase (Ppt1) inhibitors can act as pharmacological chaperones in infantile batten disease. Biochem Biophys Res Commun. 2010;395:66–69. doi: 10.1016/j.bbrc.2010.03.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delehanty JB, Mattoussi H, Medintz IL. Delivering quantum dots into cells: strategies, progress and remaining issues. Anal Bioanal Chem. 2009;393:1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- Delehanty JB, Bradburne CE, Boeneman K, et al. Delivering quantum dot-peptide bioconjugates to the cellular cytosol: escaping from the endolysosomal system. Integr Biol (Camb) 2010a;2:265–277. doi: 10.1039/c0ib00002g. [DOI] [PubMed] [Google Scholar]
- Delehanty JB, Medintz IL, Pons T, Brunel FM, Dawson PE, Mattoussi H. Self-assembled quantum dot-peptide bioconjugates for selective intracellular delivery. Bioconjug Chem. 2010b;17:920–927. doi: 10.1021/bc060044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfus A, Chen A, Min D, Ruoslahti Bhatia S. Targeted Quantum dot conjugates for siRNA delivery. Bioconjug Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, Nechev L, Murugaiah V, Van Vliet A, Vaishnaw AK, Meyers R. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral Res. 2008;77:225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Doiron A, Clark B, Rinker K. Endothelial nanoparticle binding kinetics are matrix and size dependent. Biotechnol Bioeng. 2011;8:2988–2998. doi: 10.1002/bit.23253. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Notterpek L. MicroRNAs in oligodendrocyte and Schwann cell differentiation. Dev Neurosci. 2011;33:14–20. doi: 10.1159/000323919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbayoumi TA, Torchilin VP. Cancer-specific anti-nucleosome antibody improves therapeutic efficacy of doxorubicin-loaded long-circulating liposomes against primary and metastatic cancer in mice. Mol Pharm. 2009;6:246–254. doi: 10.1021/mp8001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE. Some personal and historical notes on the utility of “deep-etch” electron microscopy for making cell structure/function correlations. Mol Biol Cell. 2014;25:3273–3276. doi: 10.1091/mbc.E14-05-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- Jana A, Pahan K. Oxidative stress kills human primary oligodendrocytes via neutral sphingomyelinase: implications for multiple sclerosis. J Neuroimmune Pharmacol. 2007;2:184–193. doi: 10.1007/s11481-007-9066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery material for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent Exosomal transfer of angiogenic MicroRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Layzer JM. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JM, Wang YY, Zhao MX, Tan CP, Li YQ, Le XY, Ji LN, Mao ZW. Multifunctional QD-based co-delivery of siRNA and doxorubicin to Hela cells for reversal of multidrug resistanceand real-time tracking. Biomaterials. 2012a;33:2780–2790. doi: 10.1016/j.biomaterials.2011.12.035. [DOI] [PubMed] [Google Scholar]
- Li S, Liu Z, Ji F, Xiao Z, Wang M, Peng Y, Li F. Delivery of quantum dot-siRNA nanoplexes in SK-N-SH cells for BACE1 gene silencing and intracellular imaging. Mol Ther Nucleic Acids. 2012b;1:e20. doi: 10.1038/mtna.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tao Y, Shu Y, LaRochelle JR, Steinauer A, Thompson D, Schepartz A, Chen ZY, Liu DR. Discovery and characterization of a peptide that enhances endosomal escape of delivered proteins in vitro and in vivo. J Am Chem Soc. 2015;137:14084–14093. doi: 10.1021/jacs.5b05694. [DOI] [PubMed] [Google Scholar]
- Llacuna L, Mari M, Garcia-Ruiz C, Fernandez-Checa J, Morales A. Critical role of acidic sphingomyelinase in murine hepatic ischemia-reperfusion injury. Hepatology. 2006;44:561–572. doi: 10.1002/hep.21285. [DOI] [PubMed] [Google Scholar]
- Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50–60. doi: 10.1016/j.tcb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan SD, Law W-C, Aalinkeel R, Reynolds J, Nair BB, Yong K-T, Roy I, Prasad PN, Schwartz SA. Nanoparticle-mediated targeted delivery of antiretrovirals to the brain. Methods Enzymol. 2012;509:41–60. doi: 10.1016/B978-0-12-391858-1.00003-4. [DOI] [PubMed] [Google Scholar]
- Mei BC, Susumu K, Medintz IL, Delehanty JB, Mountziaris TJ, Mattoussi H. Modular poly(ethylene glycol) ligands for biocompatible semiconductor and gold nanocrystals with extended pH and ionic stability. J Mater Chem. 2008;18:4949–4958. [Google Scholar]
- Minami SS, Sun B, Popat K, Kauppinen T, Pleiss M, Zhou Y, Gan L. Selective targeting of microglia by quantum dots. J Neuroinflammation. 2012;9:22. doi: 10.1186/1742-2094-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CB, Kagan CR. Synthesis and characterization of monodisperse nanocrystals and close-packed nanocrystal assemblies. Annu Rev Mater Sci. 2000;30:545–610. [Google Scholar]
- Nguyen DN. Lipid-derived nanoparticles for immunostimulatory RNA adjuvant delivery. Proc Natl Acad Sci USA. 2012;109:E797–E803. doi: 10.1073/pnas.1121423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B, Hannun Y. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- Prasuhn DE, Blanco-Canosa JB, Vora GJ, Delehanty JB, Susumu K, Mei BC, Dawson PE, Medintz IL. Combining chemoselective ligation with polyhistidine-driven self-assembly for the modular display of biomolecules on quantum dots. ACS Nano. 2010a;4:267–278. doi: 10.1021/nn901393v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasuhn DE, Deschamps JR, Susumu K, Stewart MA, Boeneman K, Blanco-Canosa JB, Dawson PE, Medintz IL. Polyvalent display and packing of peptides and proteins on semiconductor quantum dots: predicted versus experimental results. Small. 2010b;6:555–564. doi: 10.1002/smll.200901845. [DOI] [PubMed] [Google Scholar]
- Pusic AD, Kraig RP. Youth and environmental enrichment generate serum exosomes containing miR-219 that promote CNS myelination. Glia. 2014;62:284–299. doi: 10.1002/glia.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne N, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- Qin J, Dawson G. Evidence for coordination of lysosomal (ASMase) and plasma membrane (NSMase2) forms of sphingomyelinase from mutant mice. FEBS Lett. 2012;586:4002–4009. doi: 10.1016/j.febslet.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Testai FD, Dawson S, Kilkus J, Dawson G. Oxidized phosphatidylcholine formation and action in oligodendrocytes. J Neurochem. 2009;110:1388–1399. doi: 10.1111/j.1471-4159.2009.06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Berdyshev E, Poirer C, Schwartz NB, Dawson G. Neutral sphingomyelinase 2 deficiency increases hyaluronan synthesis by up-regulation of Hyaluronan synthase 2 through decreased ceramide production and activation of Akt. J Biol Chem. 2012;287:13620–13632. doi: 10.1074/jbc.M111.304857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JD, Kilkus J, Dawson G. The hyaluronic acid inhibitor 4-methylumbelliforone is an NSMase2 activator – role of Ceramide in MU anti-tumor activity. Biochim Biophys Acta. 2015;1861:78–90. doi: 10.1016/j.bbalip.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Yao L, Zhang B, Zhang S, Guo J. Nanoparticlemediated delivery of neurotoxin-ii to the brain with intranasal administration: an effective strategy to improve antinociceptive activity of neurotoxin. Drug Dev Ind Pharm. 2012;38:123–128. doi: 10.3109/03639045.2011.592533. [DOI] [PubMed] [Google Scholar]
- Santra S, Holloway P, Stanley J, Walter G, Mericle R. Rapid and effective labeling of brain tissue using TAT-conjugated CdS:Mn/ZnS quantum dots. Chem Commun. 2005;25:3144–3146. doi: 10.1039/b503234b. [DOI] [PubMed] [Google Scholar]
- Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. Kinetics of metal-affinity driven self-assembly between proteins or peptides and CdSe–ZnS quantum dots. J Phys Chem C. 2007;111:11528–11538. [Google Scholar]
- Susumu K, Oh E, Delehanty JB, et al. Multifunctional compact zwitterionic ligands for preparing robust biocompatible semiconductor quantum dots and gold nanoparticles. J Am Chem Soc. 2011;133:9480–9496. doi: 10.1021/ja201919s. [DOI] [PubMed] [Google Scholar]
- Testai FD, Landek MA, Dawson G. Regulation of sphingomyelinases in cells of the oligodendrocyte lineage. J Neurosci Res. 2004;75:66–74. doi: 10.1002/jnr.10816. [DOI] [PubMed] [Google Scholar]
- Tiraboschi P, Hansen L, Thal L, Corey-Bloom J. The importance of neurotic plaques and tangles to the development and evolution of AD. Neurology. 2004;62:1984–1989. doi: 10.1212/01.wnl.0000129697.01779.0a. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Weiland F, Schwille P, Brugger B, Simmons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Walters R, Kraig RP, Medintz I, Delehanty JB, Stewart MH, Susumu K, Dawson G. Nanoparticle targeting to neurons in a rat hippocampal slice culture model. ASN Neuro. 2012;4:e00099. doi: 10.1042/AN20120042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters R, Medintz I, Delehanty J, Stewart M, Susumu K, Huston A, Dawson P, Dawson G. The role of negative charge in the delivery of quantum dots to neurons. ASN Neuro. 2015;7:1–12. doi: 10.1177/1759091415592389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner DA, Kilkus JP, Gottschalk AR, Quintans J, Dawson G. Anti-immunoglobulin-induced apoptosis in WEHI 231 cells involves the slow formation of ceramide from sphingomyelin and is blocked by bcl-XL. J Biol Chem. 1997;272:9868–9876. doi: 10.1074/jbc.272.15.9868. [DOI] [PubMed] [Google Scholar]
- Xu G, Mahajan S, Roy I, Yong KT. Theranostic quantum dots for crossing blood-brain barrier in vitro and providing therapy of HIV-associated encephalopathy. Front Pharmacol. 2013;4:140. doi: 10.3389/fphar.2013.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn P, Chen Y, Furgeson DY. A myristoylated cellpenetrating peptide bearing a transferrin receptor-targeting sequence for neuro-targeted siRNA delivery. Mol Pharm. 2014;11:486–495. doi: 10.1021/mp400446v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Giljohann DA, Chen DL, Massich MD, Wang X-Q, Iordanov H, Mirkin CA, Paller AS. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci USA. 2012;109:11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]