Abstract
Noroviruses belong to a genus of genetically diverse viruses within the family Caliciviridae and cause acute gastroenteritis in humans and animals. They are subdivided into genogroups, each of which further segregates into genotypes. Until recently, a new genotype was based on a defined pairwise distance cutoff of complete VP1 sequences, but with the increasing number of available norovirus sequences, this cutoff is no longer accurate, and sequences in the public database have been misclassified. In this paper, we demonstrate that the pairwise distance cutoff method can no longer be used and outline a phylogenetic approach to classify noroviruses. Further-more, we propose a dual nomenclature using both ORF1 and VP1 sequences, as recombination is common and recognizing recombinant viruses may be relevant. With the continuing emergence of new norovirus lineages, we propose to coordinate nomenclature of new norovirus genotypes through an international norovirus working group.
Introduction
Noroviruses are a group of genetically diverse viruses belonging to the genus Norovirus, family Caliciviridae, that cause acute gastroenteritis in humans and animals [14] and are the most common cause of nonbacterial gastroenteritis outbreaks in persons of all ages [8, 17, 21, 25, 42, 46].
Noroviruses are positive-sense, single-stranded, non-enveloped RNA viruses. The linear RNA genome is organized in three open reading frames (ORFs). ORF1 encodes a large polyprotein, which is cleaved by the virus-encoded protease into six non-structural proteins including the viral polymerase. ORF2 and ORF3 encode the major and minor capsid proteins VP1 and VP2, respectively [26].
Human norovirus cannot be grown in cell culture, and to date, only murine norovirus can be cultivated [26]. Norovirus can be segregated genetically into at least five genogroups, I to V, which are further divided into > 30 genotypes [48]. Genotype GII.4 is further divided into strains, or variants [38, 48]. Because different virus types can differ in their ability to cause epidemics, their host range, incidence, virulence, and stability in the environment [11, 24, 28, 40, 43], it is important to correctly identify genetically different noroviruses. The lack of a cell culture system precludes the use of typing into serotypes by neutralization assays to segregate genetically different viruses according to a biologically relevant criterion [9]. With the widespread use of RT-PCR and sequencing, an increasing diversity of noroviruses has been found worldwide. Unfortunately, the lack of standardized typing has led to confusion on the number of genotypes, and to disagreement on what defines a genotype and what is the best genomic region for classification.
Since the mid 1990s, norovirus genotypes have been defined based on the complete VP1. New genotypes have been assigned when VP1 amino acid sequences differed by more than 20 % compared to other genotypes [44]. In recent years, with the rapid accumulation of more norovirus sequence data, the cutoff threshold of 20 % needed adjustment due to increasing within-genotype diversity. The amino acid divergence within a genotype was therefore changed to 14.1 % and a minimum of 15 % pairwise difference between the next-nearest genotype was proposed for classification of a new norovirus genotype [48].
Correct classification is especially important for noroviruses belonging to genogroup II, genotype 4 (GII.4) because of the emergence of new pandemic GII.4 variants [3, 32, 38, 41, 49]. Approximately 5 % aa sequence divergence in VP1 has been reported between GII.4 variants, and up to 2.8 % within variants [3, 49].
The first broadly reactive diagnostic RT-PCR assays targeted highly conserved regions in the polymerase gene, and these are still being used today. These partial polymerase ORF1 sequences are useful for typing of viruses, although they segregate into distinct genotypes clusters that are less robust than those obtained using ORF2 sequences [15]. In addition, analyses comparing partial 3’ ORF1 and ORF2 sequences have shown that many noroviruses have recombinant genomes, with the recombination point located in or around the ORF1/ORF2 junction [6]. Recombination has been recognized as a potentially important means by which these viruses generate diversity [6]. Therefore, criteria were developed for a partial polymerase-based genotype assignment, using 10 and 15 % nucleotide sequence divergence as cutoff values for GI and GII, respectively [44]. As with ORF2, the growing set of available partial polymerase sequences, both in quantity and in time span, began to challenge these genotype cutoff criteria.
Genotyping of recombinant viruses was further complicated when recombinants were identified with novel partial polymerase sequence types combined with an established ORF2 genotype. A partial polymerase genotyping system was proposed by Bull et al. [6] (a to d) for orphan polymerase sequences such as these. However, identical sequences have been published with a different polymerase genotype [10, 39].
Additionally, the lack of an internationally accepted standard for norovirus nomenclature and genotype definition has led to conflicting reports on certain genotypes in the literature [27, 48]. With the increasing understanding of noroviruses having a global distribution, it is critical to establish a standardized nomenclature to allow efficient communication of epidemiologically important lineages. This need for common classification standards was recognized at the 4th International Conference on Caliciviruses in Chile in 2010. An international norovirus working group of researchers was assembled to develop practical standards for a universal nomenclature and typing system. In this paper, the current state of norovirus genotyping is reviewed, and an update of the definition of genotypes is proposed based on phylogeny, as was recently proposed for the families Picornaviridae and Filoviridae [30, 31]. With the continuing emergence of new lineages, we propose to coordinate the nomenclature of new norovirus genotypes and variants through an international norovirus working group, which could be extended to other caliciviruses in the future.
Methods
Phylogeny and distance methods
A set of 197 unique complete ORF2 sequences of GI and GII noroviruses of human or porcine origin was selected from GenBank [2] (download of Feb. 2010), representing the genetic diversity of submitted norovirus sequences. The norovirus sequences were translated into amino acids (aa) and aligned using ClustalW in BioEdit [22], and uncorrected distance matrices were computed from the sequence alignments. To improve the quality of the alignment, the 57 GI sequences and 140 GII sequences were aligned separately. Phylogenetic trees were computed from the aa alignments and nt alignments, using Bayesian analysis with MrBayes [23] and maximum likelihood (ML) with PhyML [19, 20] via the web server at the ATGC Montpellier Bioinformatics platform (http://www.atgc-montpellier.fr/ phyml/). The appropriate substitution model settings were derived using jModeltest [34] for both GI and GII noroviruses. The resulting trees were plotted and edited in FigTree (http://tree.bio.ed.ac.uk/software/figtree/), and genotype clusters were identified using the prototype sequences for all genotypes listed in Fields Virology [16].
Partial ORF1 phylogenetic analysis
A selection was made from GenBank of all unique norovirus GII sequences (n=444) covering at least 1300 nt of the 3′end of ORF1 (pORF1), comprising nearly the complete polymerase gene. Of these, 420 also contained (partial) VP1 sequences, so these could be assigned to a VP1 genotype using the method described above. Twenty-four sequences that did not include a VP1 part could not be genotyped using the method described above. As there was strong overrepresentation of genotype GII.4 (n=360), a subset of GII.4 sequences was randomly deleted to reduce the computation time. The remaining 166 sequences were aligned, and a maximum-likelihood tree was computed for the 1300-nt partial ORF1 (pORF1) part of the sequences using PhyML [19, 20].
Proposed norovirus classification criteria
Phylogenetic distance matrices were computed from the output tree files using Patristic [13]. Average distances within and between phylogenetic clusters, as well as standard deviation (SD) values, were computed in R [35]. These data were used to infer our proposed genotype classification criteria. The average distance between all sequences within a newly identified cluster and its nearest established cluster should not overlap within 2SD of each other (2×SD criterion).
Analysis of sequence data in the public domain using newly proposed criteria for genogroup and genotype assignment
On February 7, 2013, a total of 15,685 nt sequences from viruses classified as members of the genus Norovirus by the submitting scientists were retrieved from GenBank through the NCBI Taxonomy Browser [2, 37]. These sequences included complete genomes as well as partial sequences from different parts of the genome. We analyzed all 15,685 sequences to determine their genogroup using the online norovirus typing tool [29].
Results
Distribution of pairwise similarities of VP1 sequences
The distribution of pairwise aa similarities for VP1 showed two distinct peaks, which represent the similarities within and between genotypes (Supplementary data 1) [48]. The maximum pairwise difference within genotypes was 0.21 for GI and 0.15 for GII, and the minimum difference between genotypes was 0.20 for GI and 0.13 for GII (Table 1).
Table 1.
Overview of sequence analysis results
| Alignments | Capsid (ORF2) |
pORF1 | |||
|---|---|---|---|---|---|
| Genogroup | GI |
GII |
GII | ||
| Amino acids/nucleotides | aa | nt | aa | nt | nt |
| Number of sequences | 57 | 140 | |||
| Length of sequences (nt) | 1593-1641 (complete capsids) | 1300 | |||
| Uncorrected pairwise difference | |||||
| Average within-genotype difference (+ SD) | 0.08 (0.07) | - | 0.06 (0.02) | - | |
| Max. within-genotype difference | 0.21 | - | 0.15 | - | |
| Average between-genotype difference (+ SD) | 0.32 (0.04) | - | 0.35 (0.05) | - | |
| Min. between-genotype difference | 0.20 | - | 0.13 | - | |
| Maximum-likelihood analysis | |||||
| Average a-LRT score of genotype defining branches | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Average within-genotype distance (+ SD) | 0.12 (0.12) | 0.02 (0.09) | 0.11 (0.05) | 0.12 (0.07) | |
| Max. within-genotype distance | 0.38 | 0.82 | 0.25 | 0.60 | |
| Average between-genotype distance (+ SD) | 0.78 (0.22) | 1.50 (0.29) | 1.08 (0.25) | 1.51 (0.33) | 0.72 (0.59) |
| Min. between-genotype distance | 0.36 | 0.75 | 0.27 | 0.60 | |
| Bayesian analysis | |||||
| Average posterior probability score of genotype defining branches | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average within-genotype distance (+ SD) | 0.13 (0.13) | 0.02 (0.09) | 0.17 (0.08) | 0.14 (0.07) | |
| Max. within-genotype distance | 0.42 | 0.81 | 0.40 | 0.65 | |
| Average between-genotype distance (+ SD) | 0.91 (0.19) | 1.44 (0.27) | 1.71 (0.39) | 1.56 (0.31) | |
| Min. between-genotype distance | 0.38 | 0.74 | 0.46 | 0.55 | |
Phylogenetic analysis
All aa and nt ORF2 sequences segregated robustly in defined GI and GII genotypes with branch support values of 1.00 using Bayesian and ML analysis (Table 1, Fig. 1). With the exception of the GII aa data, all pairwise phylogenetic distances within genotypes overlapped with pairwise distances between genotypes both for the Bayesian and ML analysis (Table 1). The distance range where the distances overlapped was different for the two methods: e.g., 0.36-0.38 for the ML analysis and 0.38-0.42 for the Bayesian analysis for GI aa sequences.
Fig. 1.
Phylogenetic trees (MrBayes) of complete capsid aa sequences of norovirus GI (a) and GII (b), showing all genotype clusters as defined in Fields Virology [16] and one new GI cluster GI.9 and three new GII clusters GII.20 – GII.22 (grey italic). The ML trees and the trees based on nucleotides are very similar (data not shown)
Using the newly proposed 2×SD criteria, one new GI (GI.9) and three new GII clusters (GII.20, GII.21, and GII.22) could be identified in addition to the established genotypes [45, 48] (Figs. 1, 2). Two GI genotypes that were identified in the 1990s, GI.4 and GI.5 [1, 33], as well as several well-supported clusters within the established genotypes, i.e. GII.4 variants and sub-clusters within the GI.3 and GI.7 groups [27, 49], did not comply with the 2×SD criterion.
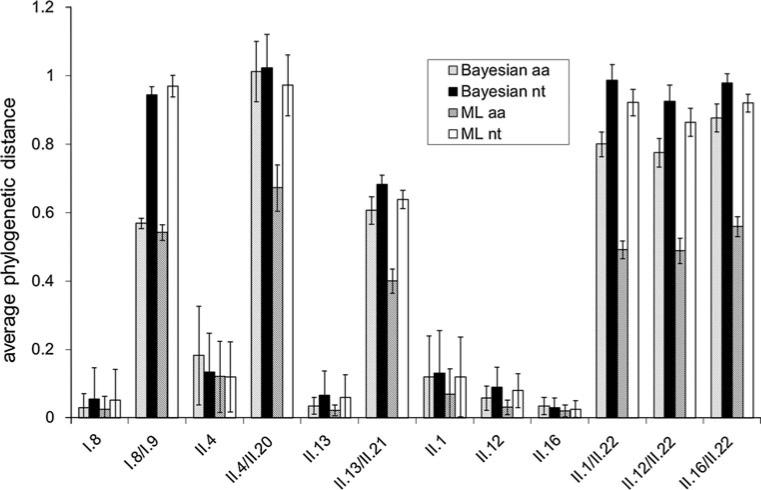
Fig. 2.
Average within-genotype (GI.8, GII.4, GII.13, GII.1. GII.12, GII.16) distances and between-genotype (GI.8/ GI.9 and GII.4/GII.20, GII.13/ GII.21, GII.1/GII.22, GII.12/ G.II.22, GII.12/GII.22) distances of proposed new genotypes and the genotype clusters that are closest to them in the phylogenetic trees, calculated using Baysian and maximum-likelihood methods, and using nt and aa sequence alignments of complete norovirus capsid sequences. Error bars are + 2×SD and – 2×SD
ORF1 analysis
The phylogenetic tree computed from the alignment of 166 representative pORF1 sequences showed 12 clusters of two or more sequences (2-82) with high branch support and 7 branches each, indicating a clearly distinct sequence (Fig. 3). Fifteen of these, nine clusters and six single sequences, could be associated with distinct VP1 types, eight of which (GII.1, GII.2, GII.3, GII.11(GII.19), GII.13(GII.17), GII.16, GII.18 and GII.22) contained the prototype sequence of the associated VP1 type [16] (U07611, X81879, U02030, AB074893(AY823306), AY502009, AY502010, AY823304, AB083780). For a further seven (GII.4, GII.6, GII.7, GII.8, GII.12, GII.20 and GII.21), the pORF1 region has never been sequenced for the VP1 prototype (X76716, AJ277620, AJ277608, AF195848, AJ277618, EU373815, AY675554), so the VP1 type association has been performed using more recent representatives of this VP1 genotype. This leaves three orphan ORF1s, GII.c, GII.e and GII.g, of which GII.c has been described previously [5]. Not all VP1 genotypes are present in the phylogeny, partly because pORF1 sequences of sufficient length were not available in GenBank for some of them. Additionally, two pORF1 types each had two VP1 genotypes associated with them. The latter was the case for VP1 types GII.19 and GII.11, which are of porcine origin, and for the human VP1 types GII.13 and GII.17.
Fig. 3.
Maximum-likelihood tree of partial ORF1 nt sequences (1300 nt) of a set of 166 sequences with 12 clusters of more than sequences – GII.1=5, GII.3=3.G II.4=82, GII.6=2, GII.7=11,G II.12=28, GII.19=2, GII.21=8 (previously GII.b) [4], GII.22=2 (previously GII.d [6]) – and 7 separate single sequences. For nine clusters and six single sequences, the corresponding VP1 genotype is indicated. Two clusters correspond to two VP1 types each, as viruses of the two VP1 types have similar pORF1 sequences. The bracketed
In five pORF1 clusters, sequences with other VP1 types are present besides the VP1 prototype (GII.1, GII.7, GII.12, GII.21 and GII.22). Also, orphan cluster GII.g contains sequences with two different VP1 types, GII.3 and GII.12. Two of the orphan pORF1 clusters described earlier by Bull et al, GII.b and GII.d [5], are now renamed. For the prototype of the VP1 genotype GII.21 (AY675554), only the ORF2 region has been submitted to GenBank, so the ORF1 sequence is unknown. A virus found in India in 2006, Hu/NoV/Ahm PC03/2006/India (EU019230 [7]), is present in the pORF GII.21 cluster, which also contains representatives of the well-described pORF1 type II.b [6, 36]. Thus, the orphan GII.b cluster can be renamed as GII.21. The GII.22 cluster contains one representative of pORF1 orphan type GII.d (DQ366347) and the VP1 type GII.22 prototype Hu/NoV/Yuri/2002/JP (AB083780).
One of the single sequences, ‘r’ (Fig. 3), is a recombinant GII.e /GII.21 virus with a recombination point in the middle of the 1300-nt pORF1 region used in this analysis (Supplementary data 1).
The branch lengths of the pORF1 set are smaller than for the ORF2 set: the average between genotype distances is 0.72 and 1.51, respectively (Table 1). In the calculation of the average between genotype distances for pORF1, the distances with the recombinant virus have been excluded, as these are not true evolutionary distances. As clusters of more than two sequences are required for calculation of the standard deviation of the within-cluster distances, this could only be calculated for eight pPOL clusters, GII.1, GII.3, GII.4, GII.7, GII.12, GII.21, GII.e and GII.g. Of these, seven complied with the 2×SD criterion. GII.1 did not comply with the 2×SD criterion due to high intra-genotype diversity (data not shown).
Analysis of norovirus sequences submitted to GenBank
In all, 15,685 partial and complete norovirus sequences from GenBank were typed using the norovirus online typing tool [29] (Supplementary data 2). Genogroup assignment by the typing tool was compared to the classification in the NCBI Taxonomy Browser, where two groups are defined at the species level: species Norwalk virus and “unclassified norovirus”, with 14 and 8 subgroups, respectively (Table 2). The establishment of the groups and subgroups is a result of the functionality of GenBank's Entrez software, where a new group is automatically established (with label ‘no rank’) for every new name in the field ‘organism’, entered by the scientist who submits the sequence [12].
Table 2.
Classification of all submitted sequences belonging to the genus Norovirus according to the NCBI Taxonomy Browser (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Tree&id=142786&lvl=3&lin=f&keep=1&srchmode=1&unlock), compared to the classification using the phylogenetic typing tool
| Grouping in NCBI Taxonomy Browser |
Grouping according to phylogenetic typing |
|||||||
|---|---|---|---|---|---|---|---|---|
| Species level | Subdivision (no ranking) | N1 | GI | GII | GIII | GIV | GV | Other |
| Species Norwalk virus | ||||||||
| Norovirus genogroup I | 1478 | 1469 | 7 | 1 sapovirus 1 unknown | ||||
| Norovirus genogroup II | 9026 | 48 | 8974 | 1 | 2 | 1 sapovirus | ||
| Norovirus isolates | 3445 | 461 | 2782 | 134 | 16 | 23 | 4 neboviruses | |
| 23 sapoviruses | ||||||||
| 2 valoviruses2 | ||||||||
| Norwalk-like virus | 361 | 48 | 256 | 39 | 5 | 13 short primer sequences3 | ||
| Other small groups (n=10) | 772 | 176 | 470 | 27 | 59 | 2 | 7 sapoviruses 1 unknown 30 short primer sequences3 | |
| Unclassified norovirus (no rank, 8 subgroups) | 603 | 62 | 202 | 112 | 6 | 195 | 26 neboviruses | |
| Total | 15685 | 2264 | 12691 | 313 | 88 | 220 | 109 | |
G genogroup
Number of sequences in GenBank
“Valovirus” is a proposed genus
Thirty sequences were not classified, as these were short primer sequences
A total of 10.504 (67 %) of all sequences fall in two of the subgroups in the taxonomy browser called ‘genogroup I’ and ‘genogroup II’. Fifty-eight (0.5 %) of these belong to another genogroup according to the phylogenetic genogroup assignment by the typing tool. An additional 4457 (86 %) sequences from the other groups and subgroups were classified as GI and GII viruses by the typing tool (747 and 3710 respectively). The remaining 724 sequences from the other groups were classified by the typing tool as belonging to other norovirus genogroups GIII, GIV or GV, or to other genera within the family Caliciviridae, and two sequences were not recognized by the typing tool.
Discussion
To provide a basis for a uniform genotype assignment for noroviruses, we analyzed 197 complete norovirus capsid sequences, representing the genetic diversity of noroviruses in the field. The previously used pairwise uncorrected distance method [44, 48] resulted in an overlap in pairwise distances from within and between genotypes. In contrast, phylogeny-based clustering using both ML and Bayesian methods segregated all currently recognized VP1 ones will not be used for typing (GII.17 and GII.19). For three clusters and one single sequence, there is no corresponding VP1 sequence: GII.c, G II.e, GII.g and r. GII.c is a previously described orphan pORF1 type ([6]). GII.e and GII.g [10] are two additional orphan types. ‘r’ is a recombinant GII.e /G II.b virus (EU921388) with a recombination point in the middle of the 1300-nt region used in this analysis. On the most relevant branches, the branch support values are indicated genotypes at the aa and nt level with robust branch support. We proposed to use 2×SD criteria to group sequences into a particular genotype and identified four novel VP1 genotypes (one GI and three GII genotypes) in addition to the published genotypes. Using this proposed criterion, GI.4 and GI.5, would not be considered a separate genotype due to overlapping standard deviations. The clustering of viruses based on 1300-nt pORF1 sequences is clear, and although the branch lengths are shorter, the 2×SD criterion could be applied except for in the case of GII.1 [44]. The GII.1 cluster is a very diverse cluster, with five pORF1 sequences, which are associated with three different VP1 types, including the GII.1 prototype (U07611), dating back to 1971.
Recombination of noroviruses in the ORF1-ORF2 junction region is common [6], and some genotypes seem to be more prone to recombination than others. The pORF1 sequences of VP1 genotypes such as GII.2 and GII.12, for example, can be found in several clusters, some of which are orphan clusters. Bull et al. [6] recognized the need to name both the ORF1 and ORF2 genotype of a virus, because recombination is common in NoV. Besides the four orphan pORF1 types recognized by Bull et al. (GII.a, GII.b, GIIc and GII.d), new orphan clusters were found in the pORF1 sequence set, which were identified by successive letters, GII.e and II.g (Fig. 3).
Two former pORF1 orphan clusters, GII.b and GII.d [6], can now be associated with a new and unique VP1 type, so these have been renamed GII.21 and GII. 22 (Fig. 3). It should be noted that the fact that a virus has an identical VP1 and pORF2 type does not necessarily mean that it is not a recombinant.
Our analysis of the norovirus (partial) ORF1 and (partial) ORF2 sequences deposited in GenBank further shows the need to standardize the norovirus nomenclature, as heterogeneous classification has evolved which is inconsistent with phylogenetic classification.
All sequences from members of the genus Norovirus that were submitted to GenBank could be assigned as norovirus GI to GV using the typing tool, except for a small number, which were reclassified as belonging to other calicivirus genera. This includes the 603 sequences that, according to the NCBI classification, do not belong to the species Norwalk virus but are assigned to a separate group, “unclassified norovirus”. Thus, the species Norwalk virus is the only species within the genus Norovirus and is, as such, not an informative taxonomic classification. As suggested previously by Zheng et al. [48], the genogroup classification should be used to define the species within the genus Norovirus.
We propose to coordinate the assignment of new genotypes and variants via an international group of norovirus experts, thus securing a consensus nomenclature, analogous to the procedure for influenza virus, Picornaviridae and Filoviridae [30, 31, 47], and use a dual typing system based on complete capsid (VP1) and partial polymerase (1300 nt).
VP1 genotype assignment
We propose to build on the norovirus genotypes described in Fields Virology [16], including two GI (GI.4, GI.5) genotypes that do not satisfy the newly defined 2×SD criterion, since re-naming of these genotypes would lead to confusion.
At least two complete capsid sequences from geographically diverse locations should be available for these sequences to be considered to represent a new genotype. We propose to name the virus with the first full-length capsid sequence available in the public-domain databases (GenBank, EMBL, DDBJ) as the prototype for that genotype. The numbering of a genotype will be designated by the norovirus working group. Single complete capsid sequences forming distinct branches will be preliminarily labelled NA (not assigned) until additional information is available (e.g., GII. NA1, GII NA2).
Partial polymerase typing
The pORF1 nucleotide sequences are named according to their phylogenetic clustering. For clusters containing the pORF1 sequence of the prototype virus of an established VP1 genotype, this VP1 name is used. pORF1 genotypes are designated by a capital P (for ‘polymerase’), followed by the genotype designation, i.e., GII.P4, in order to differentiate them from the VP1 type nomenclature. If only one sequence of a corresponding new VP1 is available, the preliminary NA+number nomenclature of the VP1 is also applied to the pORF1. Orphan pORF1 sequences are given a preliminary letter-based name, following the previously proposed nomenclature [6]. Such a pORF1 genotype is likely derived from a recombination event (especially when found with multiple VP1 genotypes), but the potential exists for the identification of a lineage with this new pORF1 type, combined with a new capsid genotype. Thus, a pORF1 genotype may change from a letter to a number in some cases. One pORF1 sequence of at least 1300 nt covering the 3’ end of ORF1 is sufficient for assignment of orphan pORF1 types. The prototype for each pORF1 type is the first sequence of the required minimal genomic region that is available in the public domain. For novel pORF1 types, the full capsid sequence should also be available.
Variants within genotypes
In the absence of objective criteria such as the 2×SD criterion, further subtyping into variants will be based on phylogenetic analysis, coupled with a relevance criterion established by consensus of the norovirus working group. For example, new GII.4 variants are recognized only after evidence is provided that they have become the epidemic lineages in at least two geographically diverse locations. New GII.4 variants will be named according to year and location of the first full-length capsid sequence in the public domain, e.g., NewOrleans_2009, For some older variants, reference viruses that have been used extensively in publications do not comply with this rule. For example, Farmington Hills and Hunter are not the first complete capsid sequences, but as these names are broadly used, we propose that they should not be altered. A list of proposed GII.4 epidemic variants is presented in Table 3 (further details on GII.4 variants in Eden et al., J Virol, In Press).
Table 3.
Proposed epidemic norovirus GII.4 variants
| Proposed epidemic variant name | GenBank no.1 |
|---|---|
| US95_96 | AJ0048642 |
| Farmington_Hills_2002 | AY4856423 |
| Asia_2003 | AB2209213 |
| Hunter_2004 | AY8830962 |
| Yerseke_2006a | EF1269632 |
| Den Haag_2006b | EF1269652 |
| NewOrleans_2009 | GU4453253 |
| Sydney_2012 | JX4599083 |
GenBank accession number of the first submitted capsid sequence of this variant
Capsid sequence
Complete genome
Variants within other established genotypes, e.g., GI.3, GII.2 and GII.3, are foreseen in the future, and relevance criteria will be discussed by the norovirus working group on a case-by-case basis.
Communication
The norovirus working group will communicate the agreed nomenclature, prototypes of genotypes and preliminary types, and other reference sequences via a publicly accessible typing tool (http://www.rivm.nl/mpf/norovirus/typingtool) [29].
When a submitted sequence cannot be assigned using this typing tool, the originator of the sequence will have the option of providing sequences for additional analysis within the norovirus working group, possibly resulting in an update of the reference set of the typing tool. The working group members will periodically monitor the norovirus sequences submitted to the public domain for possible new genotypes.
Norovirus designation
An adaptation of the previously described cryptogram for noroviruses [18] comparable to the proposed standard described by Kuhn et al. [30] would further facilitate communication by inclusion of the genogroup, genotype and variant assignment. GenBank records should have the species name in the ”Organism” field: norovirus GI, norovirus GII, etc.
The strain name should be written as follows:
| host/Hu (human) | Bo (bovine), Mu (murine), Po (porcine), Ca (canine). A list of the host name abbreviations is publised on the norovirus typing tool website |
| country code (ISO)/year of sampling/genogroup and genotype/(ORF1 and ORF2) | FR, DE, US, JP, etc. GII.P4_GII.4, or if only the ORF2 sequence is known:GII.4 |
| variant name | city, if necessary followed by a serial number |
For example: norovirus GII/Hu/FR/2004/GII.P12-GII.3/ Paris23, norovirus GII/Hu/GB/2010/GII.P4_GII.4_New-Orleans2009/London48, or if only the capsid sequence is known: norovirus GII/Hu/FR/2004/GII.12/Paris25.
Supplementary Material
Acknowledgements
We thank FBVE and Noronet network for collecting and sharing sequences, and Maarten Schippers (RIVM) for development of the R script.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00705-013-1708-5) contains supplementary material, which is available to authorized users.
Disclaimer The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the US Centers for Disease Control and Prevention (CDC). This article received clearance through the appropriate CDC channels prior to submission.
Contributor Information
Annelies Kroneman, National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Everardo Vega, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Harry Vennema, National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Jan Vinjé, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Peter A. White, School of Biotechnology and Biomolecular Sciences, Faculty of Science, University of New South Wales, Sydney, Australia
Grant Hansman, CHS Foundation, University of Heidelberg, Heidelberg, Germany.
Kim Green, Laboratory of Infectious Diseases, NIAID, NIH, Bethesda, MD 20892, USA; Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD 20742, USA.
Vito Martella, Department of Veterinary Public Health, University of Bari Aldo Moro, Valenzano, Italy.
Kazuhiko Katayama, Department of Virology II, National Institute of Infectious Diseases, Tokyo, Japan.
Marion Koopmans, National Institute for Public Health and the Environment, Bilthoven, The Netherlands; Virology Department, Erasmus Medical Center, Rotterdam, The Netherlands.
References
- 1.Ando T, Noel JS, Fankhauser RL. Genetic classification of “Norwalk-like viruses. J Infect Dis. 2000;181(Suppl 2):S336–S348. doi: 10.1086/315589. [DOI] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bok K, Abente EJ, Realpe-Quintero M, Mitra T, Sosnovtsev SV, Kapikian AZ, Green KY. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J Virol. 2009;83:11890–11901. doi: 10.1128/JVI.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buesa J, Collado B, Lopez-Andujar P, Abu-Mallouh R, Rodriguez Diaz J, Garcia Diaz A, Prat J, Guix S, Llovet T, Prats G, Bosch A. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J Clin Microbiol. 2002;40:2854–2859. doi: 10.1128/JCM.40.8.2854-2859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis. 2005;11:1079–1085. doi: 10.3201/eid1107.041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull RA, Tanaka MM, White PA. Norovirus recombination. J Gen Virol. 2007;88:3347–3359. doi: 10.1099/vir.0.83321-0. [DOI] [PubMed] [Google Scholar]
- 7.Chhabra P, Walimbe AM, Chitambar SD. Molecular characterization of three novel intergenotype norovirus GII recombinant strains from western India. Virus Res. 2010;147:242–246. doi: 10.1016/j.virusres.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.de Wit MA, Koopmans MP, Kortbeek LM, Wannet WJ, Vinje J, van Leusden F, Bartelds AI, van Duynhoven YT. Sensor, a population-based cohort study on gastroenteritis in the Netherlands: incidence and etiology. Am J Epidemiol. 2001;154:666–674. doi: 10.1093/aje/154.7.666. [DOI] [PubMed] [Google Scholar]
- 9.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. J Gen Virol. 2004;85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- 10.Eden JS, Bull RA, Tu E, McIver CJ, Lyon MJ, Marshall JA, Smith DW, Musto J, Rawlinson WD, White PA. Norovirus GII.4 variant 2006b caused epidemics of acute gastroenteritis in Australia during 2007 and 2008. J Clin Virol. 2010;49:265–271. doi: 10.1016/j.jcv.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J Infect Dis. 2002;186:1–7. doi: 10.1086/341085. [DOI] [PubMed] [Google Scholar]
- 12.Fauquet CM, Fargette D. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J. 2005;2:64. doi: 10.1186/1743-422X-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fourment M, Gibbs MJ. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol. 2006;6:1. doi: 10.1186/1471-2148-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass RI, Noel J, Ando T, Fankhauser R, Belliot G, Mounts A, Parashar UD, Bresee JS, Monroe SS. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J Infect Dis. 2000;181(Suppl 2):S254–S261. doi: 10.1086/315588. [DOI] [PubMed] [Google Scholar]
- 15.Green J, Norcott JP, Lewis D, Arnold C, Brown DW. Norwalk-like viruses: demonstration of genomic diversity by polymerase chain reaction. J Clin Microbiol. 1993;31:3007–3012. doi: 10.1128/jcm.31.11.3007-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green K. The noroviruses. In: Knipe DM, Howley PM, et al., editors. Fields virology. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 949–980. [Google Scholar]
- 17.Green KY. The role of human caliciviruses in epidemic gastroenteritis. Arch Virol Suppl. 1997;13:153–165. doi: 10.1007/978-3-7091-6534-8_15. [DOI] [PubMed] [Google Scholar]
- 18.Green KY, Ando T, Balayan MS, Berke T, Clarke IN, Estes MK, Matson DO, Nakata S, Neill JD, Studdert MJ, Thiel HJ. Taxonomy of the caliciviruses. J Infect Dis. 2000;181(Suppl 2):S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- 19.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 20.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 21.Hale A, Mattick K, Lewis D, Estes M, Jiang X, Green J, Eglin R, Brown D. Distinct epidemiological patterns of Norwalk-like virus infection. J Med Virol. 2000;62:99–103. doi: 10.1002/1096-9071(200009)62:1<99::aid-jmv15>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 22.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 23.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 24.Huhti L, Szakal ED, Puustinen L, Salminen M, Huhtala H, Valve O, Blazevic V, Vesikari T. Norovirus GII-4 causes a more severe gastroenteritis than other noroviruses in young children. J Infect Dis. 2011;203:1442–1444. doi: 10.1093/infdis/jir039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iturriza-Gomara M, Elliot AJ, Dockery C, Fleming DM, Gray JJ. Structured surveillance of infectious intestinal disease in pre-school children in the community: ‘The Nappy Study’. Epidemiol Infect. 2009;137:922–931. doi: 10.1017/S0950268808001556. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 27.Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Kojima S, Takai R, Oka T, Takeda N, Katayama K. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to Norovirus in Japan. J Clin Microbiol. 2004;42:2988–2995. doi: 10.1128/JCM.42.7.2988-2995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, van Duynhoven Y, Gray J, Iturriza M, Bottiger B, Falkenhorst G, Johnsen C, von Bonsdorff CH, Maunula L, Kuusi M, Pothier P, Gallay A, Schreier E, Hohne M, Koch J, Szucs G, Reuter G, Krisztalovics K, Lynch M, McKeown P, Foley B, Coughlan S, Ruggeri FM, Di Bartolo I, Vainio K, Isakbaeva E, Poljsak-Prijatelj M, Grom AH, Mijovski JZ, Bosch A, Buesa J, Fauquier AS, Hernandez-Pezzi G, Hedlund KO, Koopmans M. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J Clin Microbiol. 2008;46:2959–2965. doi: 10.1128/JCM.00499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroneman A, Vennema H, Deforche K, Avoort HV, Penaranda S, Oberste MS, Vinje J, Koopmans M. An automated geno-typing tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn JH, Bao Y, Bavari S, Becker S, Bradfute S, Brister JR, Bukreyev AA, Chandran K, Davey RA, Dolnik O, Dye JM, Enterlein S, Hensley LE, Honko AN, Jahrling PB, Johnson KM, Kobinger G, Leroy EM, Lever MS, Muhlberger E, Netesov SV, Olinger GG, Palacios G, Patterson JL, Paweska JT, Pitt L, Radoshitzky SR, Saphire EO, Smither SJ, Swanepoel R, Towner JS, van der Groen G, Volchkov VE, Wahl-Jensen V, Warren TK, Weid-mann M, Nichol ST. Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch Virol. 2013;158:301–311. doi: 10.1007/s00705-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauber C, Gorbalenya AE. Toward genetics-based virus taxonomy: comparative analysis of a genetics-based classification and the taxonomy of picornaviruses. J Virol. 2012;86:3905–3915. doi: 10.1128/JVI.07174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, Vinje J, Baric RS. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:e31. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noel JS, Ando T, Leite JP, Green KY, Dingle KE, Estes MK, Seto Y, Monroe SS, Glass RI. Correlation of patient immune responses with genetically characterized small round-structured viruses involved in outbreaks of nonbacterial acute gastroenteritis in the United States, 1990 to 1995. J Med Virol. 1997;53:372–383. doi: 10.1002/(sici)1096-9071(199712)53:4<372::aid-jmv10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 34.Posada D. Selection of models of DNA evolution with jModelTest. Methods Mol Biol. 2009;537:93–112. doi: 10.1007/978-1-59745-251-9_5. [DOI] [PubMed] [Google Scholar]
- 35.R_Development_Core_Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2008. [Google Scholar]
- 36.Reuter G, Vennema H, Koopmans M, Szucs G. Epidemic spread of recombinant noroviruses with four capsid types in Hungary. J Clin Virol. 2006;35:84–88. doi: 10.1016/j.jcv.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Krasnov S, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Karsch-Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. Database resources of the National Center for Bio-technology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J Virol. 2007;81:9932–9941. doi: 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takanashi S, Wang Q, Chen N, Shen Q, Jung K, Zhang Z, Yokoyama M, Lindesmith LC, Baric RS, Saif LJ. Characterization of emerging GII.g/GII.12 noroviruses from a gastroenteritis outbreak in the United States in 2010. J Clin Microbiol. 2011;49:3234–3244. doi: 10.1128/JCM.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuladhar E, Hazeleger WC, Koopmans M, Zwietering MH, Beumer RR, Duizer E. Residual viral and bacterial contamination of surfaces after cleaning and disinfection. Appl Environ Microbiol. 2012;78:7769–7775. doi: 10.1128/AEM.02144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, Iritani N, Kroneman A, Vennema H, Vinje J, White PA, Koopmans M. Indications for worldwide increased noro-virus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 2013;18:8–9. [PubMed] [Google Scholar]
- 42.van Duynhoven YT, de Jager CM, Kortbeek LM, Vennema H, Koopmans MP, van Leusden F, van der Poel WH, van den Broek MJ. A one-year intensified study of outbreaks of gastroenteritis in The Netherlands. Epidemiol Infect. 2005;133:9–21. doi: 10.1017/s0950268804002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoef LP, Kroneman A, van Duynhoven Y, Boshuizen H, van Pelt W, Koopmans M. Selection tool for foodborne norovirus outbreaks. Emerg Infect Dis. 2009;15:31–38. doi: 10.3201/eid1501.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vinje J, Green J, Lewis DC, Gallimore CI, Brown DW, Koop-mans MP. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses”. Arch Virol. 2000;145:223–241. doi: 10.1007/s007050050020. [DOI] [PubMed] [Google Scholar]
- 45.Wang QH, Costantini V, Saif LJ. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine. 2007;25:5453–5466. doi: 10.1016/j.vaccine.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler JG, Sethi D, Cowden JM, Wall PG, Rodrigues LC, Tompkins DS, Hudson MJ, Roderick PJ. The Infectious Intestinal Disease Study Executive. Vol. 318. BMJ; 1999. Study of infectious intestinal disease in England: rates in the community, presenting to general practice, and reported to national surveillance. pp. 1046–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.WHO A revision of the system of nomenclature for influenza viruses: a WHO Memorandum. Bulletin of the World Health Organization. 1980;58(4):585–591. [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Zheng DP, Widdowson MA, Glass RI, Vinje J. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J Clin Microbiol. 2010;48:168–177. doi: 10.1128/JCM.01622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.