ABSTRACT
Bacterial translocation is defined as the passage of live bacteria from the gut lumen to distant sites. Gut commensal bacteria translocation has been attributed to ‘leakiness’, or ‘barrier breach’ of the intestinal epithelium, allowing live bacteria to cross an inappropriately permeable barrier and disseminate to distant sites. Alternatively, studies suggest dendritic cells directly capture luminal commensal bacteria and transport them to distant sites in the steady-state by extending dendrites between epithelial cells into the lumen. Recently we identified translocation of commensal gut bacteria following antibiotics was associated with the formation of goblet cell associated antigen passages (GAPs) in the colon and dependent upon goblet cells (GCs). The translocation of native gut commensal bacteria resulted in low-level inflammatory responses and potentiated mucosal damage in response to concurrent epithelial injury. Here we extend these observations and demonstrate properties of colonic GAPs and observations supporting their priority in the translocation of colonic commensal bacteria.
KEYWORDS: antibiotics, bacterial translocation, goblet cells
At the time of their introduction, antibiotics were viewed as miracle drugs, allowing patients to survive diseases that were previously thought to be untreatable. However widespread exposure to antibiotics is now associated with multiple immune mediated disorders, which in turn have been correlated with antibiotic induced alterations of the gut microbiota.1-6 Yet how dysbiosis of the gut microbiota confers a risk for inflammatory immune responses at local and distant sites is poorly understood. We identified an unappreciated effect of antibiotics inducing the translocation of commensal gut bacteria and inducing inflammatory responses by allowing colonic goblet cells (GCs) to form goblet cell associated antigen passages (GAPs) and deliver luminal substances, including live bacteria, to antigen presenting cells (APCs) in the colon lamina propria (LP).7
The gut LP contains a spectrum of myeloid APCs, including CD11b+ CD103- CX3CR1+ APCs with features of macrophages and CD11b+ CD103+ CX3CR1- APCs with features of dendritic cells,8-11 which will be referred to collectively as mononuclear phagocytes (MNPs). GAPs were observed to be a major pathway delivering luminal antigens to LP-MNPs in the steady-state, as deletion of GCs or manipulation of GAPs abrogates antigen delivery to LP-MNPs as evidenced by their inability to induce T cell responses to luminal antigen.12,13 GAPs form in response to acetylcholine (ACh) acting upon the muscarinic ACh receptor 4 (mAChR4) expressed by GCs.13 The formation of GAPs occurs in the steady-state in the small intestine (SI), but conversely GAP formation is inhibited in the colon of SPF housed mice by GC intrinsic Myd88 dependent sensing of the microbiota.13 Myd88 dependent microbial sensing in GCs trans-activates the epidermal growth factor receptor (EGFR) activating p42/p44 mitogen activated protein kinase (MAPK), which suppresses responses to ACh via the mAChR413. Consistent with this we observed that treatment with oral antibiotics induced the formation of colonic GAPs, and this was followed by spontaneous translocation of native colonic commensal bacteria to the colon draining mesenteric lymph node (MLN) as the gut microbiota was being restored following antibiotic cessation.7
This effect of antibiotics occurs following a single dose of antibiotics, during continuous sub-therapeutic antibiotic therapy, or during a period of time after cessation of therapeutic antibiotic therapy in which colonic GAPs are formed due to the loss of suppressive effects of the intact gut microbiota on GAP formation, but when the gut bacterial load is sufficient to translocate.7 The translocation of commensal bacteria after antibiotics or in mice with reduced or absent microbiota, such as germfree mice, is a well-documented phenomena,14-17 correlating to our finding that the presence of the normal gut flora inhibits colonic GAPs and prevents bacteria translocation. Further we observed that most but not all antibiotics evaluated induced colonic GAP formation and bacterial translocation despite equivalent alterations in the overall colonic bacterial load,7 suggesting that some bacteria may have an enhanced capacity to inhibit GAPs and subsequent translocation and giving hope that antibiotic therapies could be tailored to mitigate this untoward effect. However the considerable questions as to which bacterial species inhibit colonic GAP formation and translocation in a given individual's gut microbiota and how antibiotics could be tailored for an individual's gut microbiota to avoid translocation will require significant more work to be answered.
Regional location and properties of colonic GAPs and translocation of commensal gut bacteria in the presence and absence of antibiotic treatment
Translocation of gut commensal bacteria to the MLN draining the SI was not observed in the absence of antibiotic therapy, and was rare in the presence of antibiotic therapy,7 suggesting that SI GAPs, which are present independent of antibiotic treatment do not have the property of commensal bacterial translocation. The small amount of bacteria seen in a few instances following antibiotic therapy in the MLN draining the SI may have translocated via colonic GAPs as the lymph drainage patterns between the SI and portions of colon can overlap. We expanded these observations by evaluating the presence of GAPs in various regions of the colon using in vivo 2-photon (2P) imaging approaches and by imaging fixed tissue sections following the administration of luminal fluorescent dextran.12,13 The latter approach allows evaluation of GAP density in regions of the GI tract that are difficult to image using the in vivo approach due to the abundant luminal contents, such as in the cecum, or areas that are not easily accessible for in vivo imaging due to anatomy, such as the sigmoid colon. Consistent with our prior observations, we found that GAPs were rare, or nearly absent, in the cecum, ascending colon, and transverse colon in the steady-state (Fig. 1A, B, D, and E). However in contrast to the proximal colonic regions, GAPs were observed in the crypts of the descending colon and sigmoid colon in the absence of antibiotic treatment (Fig. 1A, B, D, and E). The presence of GAPs in the distal colon and their absence in the proximal colon in the steady-state, may provide the basis for the ability of SPF housed mice to be tolerized to high doses of antigen in the colonic lumen in the iliac and caudal lymph nodes, which drain the distal colon segments, but not in the MLNs draining the proximal colon in the steady-state.18
Figure 1.
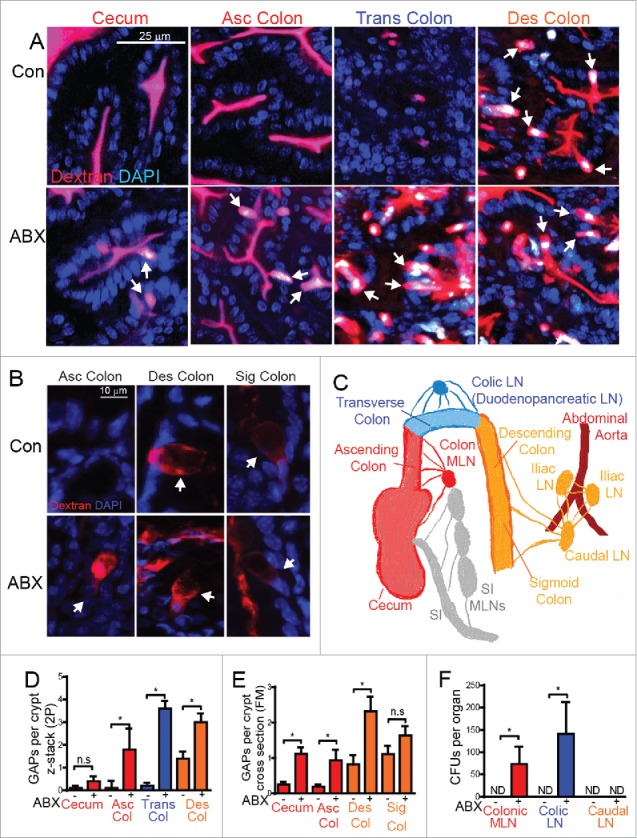
Regional location of colonic GAPs and translocation of commensal gut bacteria in the presence and absence of antibiotic treatment. (A) Mice were given regular water (top row; Con) or Ampicillin (1 g/L), metronidazole (1 g/L), neomycin(1 g/L), and vancomycin(500 mg/L) (second row; ABX) in drinking water for 1 week, and then placed on regular drinking water for 3 days.7 For two-photon (2P) imaging, mice were injected intraluminally with 2 mg 10 kDa rhodamine-labeled dextran and 1 mg DAPI. 20 minutes later mice were imaged as described previously.7,12,13 White arrow denotes GAPs; GCs filled with dextran. (B) After 30 minutes of fixation in 10% buffered formalin, colonic tissue with intraluminal dextran was sectioned and imaged by routine fluorescent microscopy to visualize structure of GAPs. White arrows denote GAPs. (C) Schematic summarizing the lymphatic draining pattern of the intestine. (D-E) Graphical representation of GAPs per crypt in the regions of the colon in control mice or mice following antibiotic treatment by D) 2P imaging E) or fluorescent microscopy (FM) of fixed tissue sections. Color of the bars in graphs correlates with the segment of colon draining to the corresponding LN in panel C and F. (F) Graphical representation of CFUs per LN after plating LN homogenates7 on LB agar overnight in control mice or mice following antibiotic treatment. n = 4 mice per group, Asc colon = ascending colon, Trans colon = transverse colon, Des colon = descending colon, Sig colon = sigmoid colon, ND = not detected, ns = not significant, statistical analysis performed using a Student's t-test, * = p < 0.05.
The colonic lymphatics drain into 3 anatomically distinct lymph node (LN) populations19,20 (Fig. 1C). The cecum and ascending colon lymphatics drain into the colonic MLN, the transverse colon lymphatics drain to a duodendopancreatic LN found behind the colon, referred to here as the colic LN, and the descending and sigmoid colon drain into the caudal LN and iliac LNs19-21 (Fig. 1C). This lymphatic drainage pattern allows evaluation of live bacteria in the various LN populations to inform where bacteria have traversed the colonic epithelium21,22 Despite the presence of colonic GAPs in the descending and sigmoid colon, we did not observe live commensal bacteria in the caudal or iliac LNs at steady-state (Fig. 1F). The presence of GAPs and the absence of bacterial translocation to the LN draining the distal colon may be due to the more dense and less penetrable mucus layer in the distal colon,23 which would reduce the exposure of GCs to bacterial products and allow GAPs to form while at the same time preventing luminal bacteria from accessing the GAPs.
Following antibiotic therapy, which disrupts the luminal commensal bacteria, we observed a significant increase in GAP formation in all segments of the colon with the exception of the sigmoid colon, which showed a trend towards an increase (Fig. 1A, B, D, and E). Imaging of fixed tissues revealed that GAPs forming in the sigmoid colon in the steady-state and those forming after antibiotic therapy appeared structurally similar (Fig. 1B). The level of GAP induction following antibiotic treatment correlated with the level of translocation of live commensal bacteria to the LNs, with the exception of the LN draining the distal colon, which did not contain bacteria in the steady-state or following antibiotic treatment (Fig. 1D-F), further supporting that a more dense mucus layer may protect the caudal LN from translocation of bacteria through the distal colon. Colonic GAPs in the proximal colon that were induced by antibiotic therapy transported a variety of luminal substances including proteins, lectins, and sugars (Fig. 2A, C); however unlike their SI counterparts, colonic GAPs did not transport LPS (Fig. 2B, C) as GC sensing of luminal LPS rapidly shut off GAPs in the colon, but not in the SI.13
Figure 2.
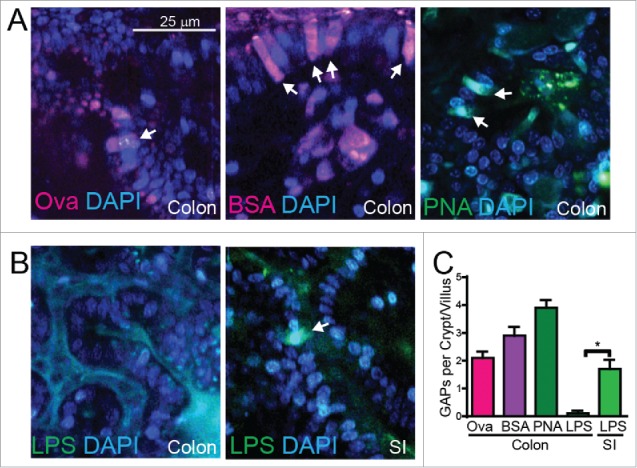
Colonic GAPs acquire a variety of substances following antibiotic treatment. (A) Representative 2-photon images of transverse colonic GAPs after antibiotic treatment filled with (left) luminal alexa-647 labeled ovalbumin (Ova), (middle) luminal alexa-647 labeled bovine serum albumin (BSA), or (right) luminal fluorescein isothiocyanate (FITC) labeled peanut agglutinin (PNA), white arrows denote GAPs. (B) Representative 2-photon images of (left) transverse colon or (right) distal ileum showing colonic GCs do not, but SI GC do fill with FITC labeled lipopolysaccharide (LPS), white arrows denote GAPs. (C) Graphical representation of GAPs per colonic crypt or SI villus filled with various proteins or lectins as assessed by 2P imaging. n = 3 mice per group, statistical analysis was performed using a t-test, * = p < 0.05.
CX3CR1+ LP-MNPs interact with GAPs in the SI and colon
CX3CR1- (CD103+) LP-MNPs were observed to interact with SI GAPs more often that CX3CR1+ (CD103-) LP-MNPs in the steady-state, however luminal antigen was acquired in a manner capable of inducing T cell responses by both SI LP-MNP subtypes when GAPs were robustly induced by exogenous administration of the ACh analog carbamylcholine.12 This suggests that CX3CR1+ LP-MNPs also acquire antigen from GAPs, but consistent with other reports are less efficient at stimulating T cell responses.8 Furthermore CX3CR1+ LP-MNPs interact with and acquire luminal antigen from colonic GAPs,7,13 and are necessary for the dissemination of translocated bacteria to the MLN,7,24 suggesting that CX3CR1+ LP-MNPs interact with colonic GAPs to acquire luminal commensal bacteria following antibiotic therapy. We therefore evaluated if CX3CR1+ LP-MNPs associated with GAPs in the various regions of the intestine in the steady-state and following antibiotic therapy. We observed CX3CR1+ LP-MNPs making multiple contacts with GAPs in the SI, spanning several z-stacks (Fig. 3A) and forming contacts with 20% of the GAPs during in vivo imaging (Fig. 3C). We found that CX3CR1+ LP-MNPs were removed from the epithelium in the ascending and transverse colon in the steady-state (Fig. 3D), but associated with the epithelium and were found interacting with GAPs, wrapping dendrites around GAPs, and forming contacts with >60% of GAPs in the ascending and transverse colon following antibiotic therapy (Fig. 3D, E, F). CX3CR1+ LP-MNPs made contacts with 40% of the GAPs in the distal colon in the steady-state, but did not significantly increase after antibiotic treatment (Fig. 3D, F). Furthermore we found that following antibiotic treatment, CD103+ LP-MNPs and CD103- LP-MNP, which are CX3CR1+,7,11 acquired luminal ovalbumin in a manner able to induce naïve T cell proliferation (Fig. 3G), further supporting that CX3CR1+ LP-MNPs can acquire luminal substances, including soluble antigens and bacteria from GAPs.7
Figure 3.
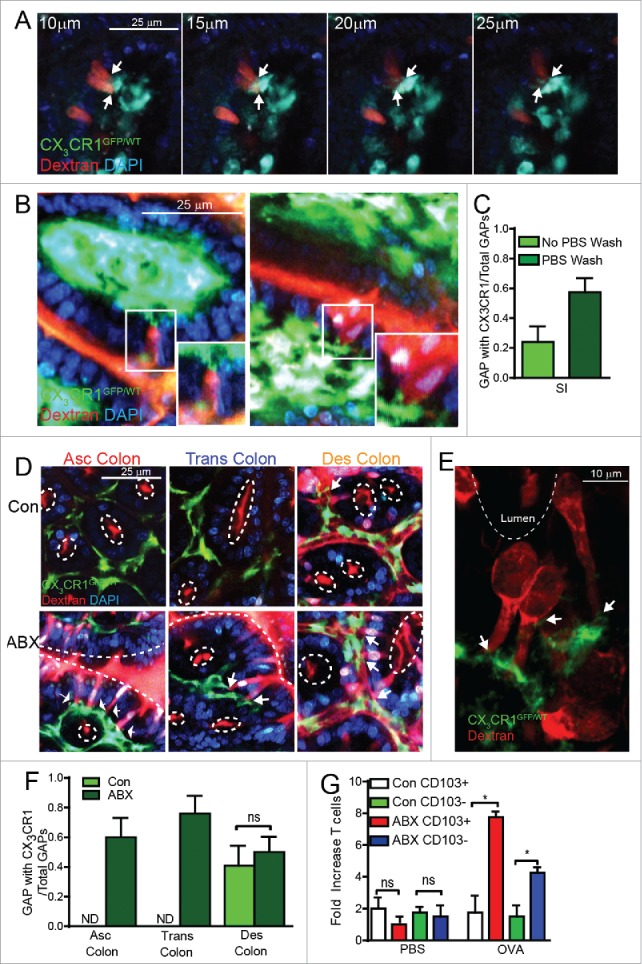
CX3CR1+ LP-MNPs interact with GAPs and acquire luminal antigen in the colon following antibiotic treatment. (A) Serial 5µm z-stacks of a villus from a CX3CR1GFP/WT reporter mouse41 starting 10µm from the lumen acquired by in vivo 2P imaging after intraluminal injection with 10 kDa fluorescent dextran and DAPI; white arrows denotes CX3CR1+ cell contacting a GAP. (B) 2P images of a villus from a CX3CR1GFP/WT mouse after intraluminal injection with dextran and DAPI, inset shows higher magnification of CX3CR1+ cell contacting a GAP. (C) Graphical representation of the ratio of GAPs being contacted by a CX3CR1+ cell to the number of total GAPs in the SI in control mice or after washing the lumen with 5 ml of 37°C PBS to remove the luminal contents and mucus, as described in imaging approaches used by others.27-29,33,34 (D) 2P images of the colon of a control CX3CR1GFP/WT mouse (Con) or following antibiotic treatment (ABX) after intraluminal injection of dextran and DAPI, white arrows denotes CX3CR1+ cell contact with a GAP, white dotted line indicates lumen. (E) Confocal microscopy image of CX3CR1GFP/WT mouse following antibiotic treatment showing CX3CR1+ cell contacting a dextran filled GAP (white arrow) in the transverse colon; white dotted line indicates lumen. (F) Graphical representation of the ratio of GAPs being contacted by a CX3CR1+ cell to the number of total GAPs in the colon of control mice or after antibiotic treatment. (G) Control mice (Con) or antibiotic treated mice (ABX) were injected intraluminally with PBS or 10 mg ovalbumin (Ova); 2 hours later LP-MNPs were isolated from the colon, sorted as CD45+CD11c+MHCII+CD103+ cells or CD45+CD11c+MHCII+CD103− cells, and cultured with sorted CD45+CD62L+CD3+CD48+Vα2+ Ova specific splenocytes from OTI T cell receptor transgenic mice42 at a 1:10 ratio. Graphical representation of fold increase OTI T cells 3 d after co-culture with LP-MNPs. n = 3 mice per group, statistical analysis was performed using a Student's t-test, * = p < 0.05, ns = not significant, ND = not detected.
Priority of GCs/GAPs in the translocation of native commensal bacteria following antibiotic therapy
Translocation of non-pathogenic luminal bacteria after antibiotic treatment is not associated with barrier breach or changes in tight junction permeability,25 making alterations in barrier function an unlikely mediator of this event. Villous M-cells could translocate luminal bacteria to the underlying LP without altering barrier function, however M-cells have not been observed on the non-follicle bearing epithelium of the colon26 where the translocation of commensal bacteria occurs following antibiotics,7,15 making them unlikely participants in this process. The discovery of the ability of LP-MNPs to extend trans-epithelial dendrites (TEDs) to sample the lumen provided an explanation for how bacterial translocation might occur in the absence of epithelial damage or loss of barrier function. However several inconsistencies, including the density and regional location of TED extension27-32 and the ability of microbial stimuli to induce TED extension while suppressing bacterial translocation,7,24,28 suggest that TEDs may not be the mechanism for translocation of luminal commensal bacteria following antibiotic therapy. We evaluated the ability of CX3CR1+ colonic LP-MNPs to extend TEDs in the presence and absence of antibiotic therapy. Consistent with other reports,30 we did not observe TED extension by CX3CR1+ colonic LP-MNPs in the steady-state or following antibiotic treatment (Fig. 4A). While CX3CR1+ colonic LP-MNPs increased contact with the colonic epithelium and GAPs after antibiotic treatment, CX3CR1+ dendrites were never seen crossing the epithelial barrier to contact the lumen. Thus colonic TED formation was unlikely to mediate translocation of colonic luminal bacteria.
Figure 4.
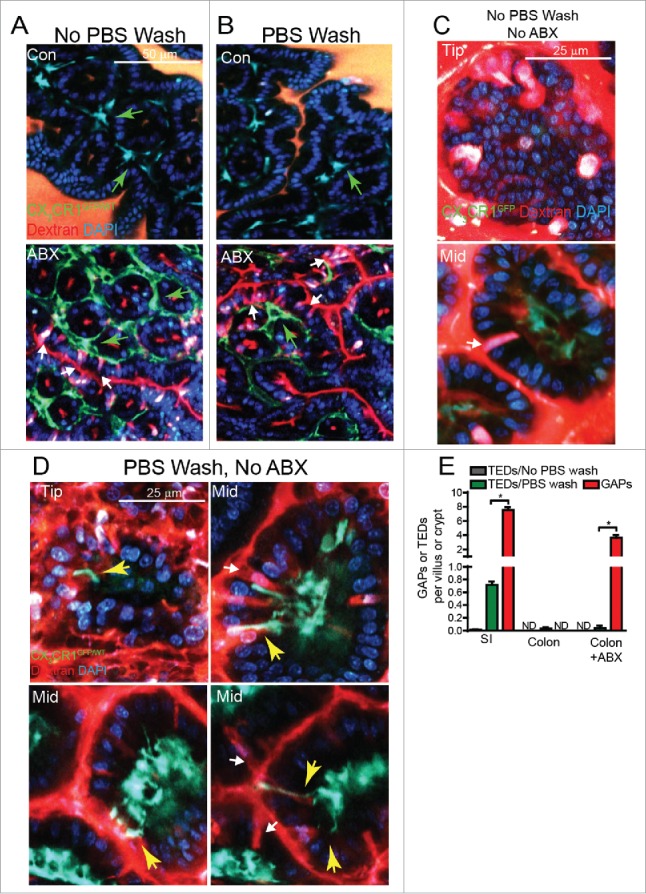
GAPs are significantly more common than the extension of trans-epithelial dendrites (TEDs) by CX3CR1+ LP-MNPs in the steady-state or following antibiotic treatment. 2P images of the transverse colon (A) without PBS wash or (B) with PBS wash to remove luminal contents and mucus from CX3CR1GFP/WT mice given drinking water alone (Con) or antibiotic treatment (ABX) and intraluminal injection of dextran and DAPI; green arrows denote CX3CR1+ LP-MNPs, white arrows denote GAPs. (C) 2P images of the tip and mid small intestinal villus of an untreated CX3CR1GFP/WT mouse after intraluminal injection of dextran and DAPI, white arrows denote GAPs. (D) 2P images of the tip and mid small intestinal villus of an untreated CX3CR1GFP/WT mouse after intraluminal injection of dextran and DAPI following PBS rinsing, white arrows denote GAPs, yellow arrow denotes a TED. (E) Graphical representation of the number of TEDs, counted as CX3CR1GFP/WT dendrites extending into the lumen, as delineated by the DAPI+ epithelial border, per villus or ascending colonic crypt and the number of GAPs per villus or ascending colonic crypt to show frequency of TEDs compared the frequency of GAPs. n = 3 mice per group in panel E, statistical analysis was performed using a Student's t-test, * = p < 0.05, ND = not detected.
Furthermore we found the formation of TEDs to be an exceedingly rare even in the SI of untreated mice (Fig. 4C); we observed only 2 TEDs while evaluating over 500 entire villi, from tip to base (Fig. 4E). We noted that studies of LP-MNP TED extension in the SI, rinsed the lumen of the intestine with PBS to remove the luminal contents and mucus,27-29,33,34 a practice we did not use as mucus is a vital component of the barrier protecting the epithelium from the luminal contents.23,35 When we removed the luminal contents and mucus by rinsing with PBS, TED extension by LP-MNPs became apparent on the tip and in the middle of SI villi (Fig. 4D) at a rate of approximately 1.5 TEDs per villus in the ileum or ∼0.7 TEDs per villus combining all regions of the SI when examining over 300 villi (Fig. 4E), which is highly consistent with prior studies reporting the density and regional location of TED extension.27,28 Often TEDs could be seen near or extending past GAPs (Fig. 4D right top, right bottom) but TEDs also formed independently of GAPs (Fig. 4D left bottom), indicating that TED formation was not dependent upon GAPs. Even after stringent washing, TEDs failed to form in the colon (Fig. 4B), corroborating the lack of TED extension in the colon in other models.30,31
Evaluating the density of TED extension and GAPs simultaneously within the same mouse we observed that GAPs were significantly more common than the extension of TEDs in the SI in the steady-state (∼1000 fold) and in the colon following antibiotic treatment (∼1000 fold; Fig. 4E). Additionally, removal of the protective mucus barrier from the epithelium before imaging increased the contacts CX3CR1+ LP-MNPs made with SI GAPs to 60% of the GAPs (Fig. 3C), suggesting CX3CR1+ SI LP-MNPs increase both TED extension and sampling from GAPs when the mucus barrier is disturbed. Where examined, all studies evaluating the extension of TEDs by LP-MNPs agree that they are induced by enteric pathogens.27-29,34 Our observation that TED extension by LP-MNPs becomes more common after removal of the mucus layer and observations that epithelial Myd88 signals induce SI LP-MNPs to extend TEDs,28 together suggest that TED extension occurs in response to epithelial stress signals. Conversely GAPs are inhibited by epithelial sensing of the microbiota in the colon.13 While SI GAPs were not inhibited by the commensal microbiota, SI GCs expressed TLRs, MyD88, and the EGFR, and activation of this pathway by EGFR ligands can inhibit GAPS in the SI. The ability of the commensal microbiota to inhibit GAPs in the colon, but not in the SI, could be due to lower levels of expression of TLRs and/or higher expression of negative regulators of TLR signaling in SI GCs when compared with colon GCs.13 However the preservation of this pathway in SI GCs raises the possibility that SI GAPs could be inhibited in some situations, such as enteric infection. These observations suggest that GAPs and TEDs may play opposite roles as routes for the immune system's exposure to luminal substances, with GAPs having a higher priority in homeostatic responses and TED extension by SI LP-MNPs having a higher priority during stress and infection.
Dissemination of luminal bacteria to the MLN requires sphingosine 1 phosphate receptor 1 (S1PR1)
The gut commensal microbiota restricts translocation and dissemination of commensal bacteria.7,24 Translocation of bacteria in the colon is independent of Myd88 signals, however the carriage of bacteria to the MLN was induced by the presence of the microbiota7,24,30 Migration of leukocytes to and from LNs is dependent on sphingosine 1 phosphate gradients and inactivation of the sphingosine 1 phosphate receptor (S1PR) inhibits circulation of lymphocytes.36 Intestinal APCs express the S1PR,37 and inhibition of S1PR in clinical trials is an effective treatment of IBD and colitis,38,39 though the mechanism of action is incompletely understood. Additionally, it has been shown CX3CR1+ MNPs require S1PR expression for trafficking of cells carrying pathogenic bacteria to secondary LNs in a model of Yersinia infection.40 Pan S1PR inactivation with FTY720 or selective S1PR1 inactivation with SEW2871 inhibited bacterial translocation to the colon draining MLN (Fig. 5A), suggesting that S1PR blockade might prevent inflammatory responses in the MLN.
Figure 5.
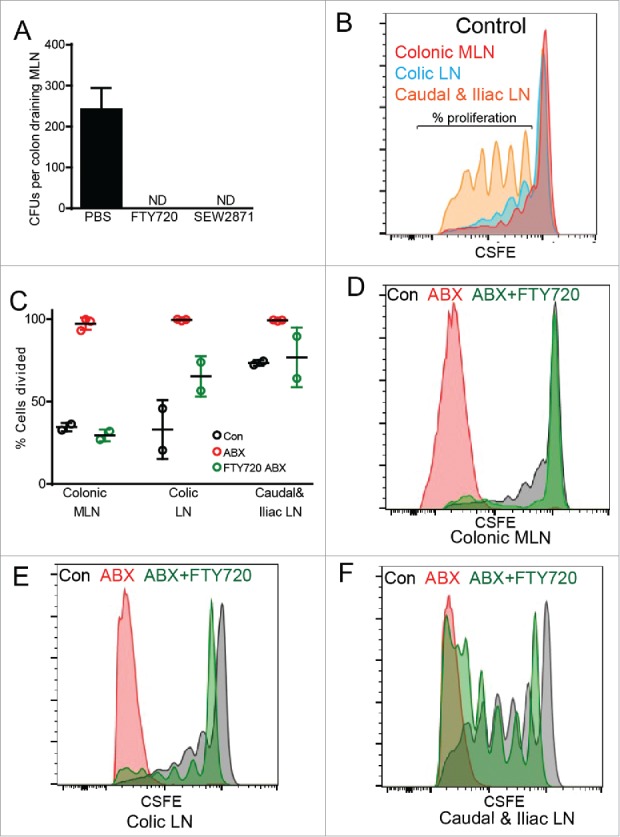
Dissemination of translocated bacteria and induction of T cell responses in the LN draining the proximal colon following antibiotic therapy are dependent upon S1PR. (A) Graphical representation of CFUs per colon draining MLN in mice following antibiotic treatment and i.p. injection with 5 mg/kg FTY72040 or 20 mg/kg SEW287143 48 hours before MLN harvest. (B-F) Mice were given normal drinking water (Control; Con) or antibiotic treatment (ABX), i.v. injected with 4.2 × 106 CFSE labeled ovalbumin (Ova) specific CD4+ splenic T cells from OTII T cell receptor transgenic mice, and 24 hours later given 50 mg Ova via enema and i.p. injection of vehicle or 5 mg/kg FTY720. Three days following ovalbumin administration, colon draining LN homogenates were analyzed by flow cytometry for CFSE dilution. (B) Representation flow plot of colonic draining MLN, colic LN, and caudal and iliac LN in mice not receiving antibiotics (Control). (C) Graphical representation of percent of OTII cells from the various LN and treatments that had diluted CFSE past the initial fluorescent intensity. (D-F) Representation flow plot of (D) colonic draining MLN, (E) colic LN, and (F) caudal and iliac LN in control mice and mice following antibiotic treatment with or without FTY720 injection. n = 2 or 3 mice per group, ND = not detected.
Antibiotic therapy induces T cell responses to luminal antigens in the colon draining LN
We previously observed that following antibiotic therapy, inflammatory cytokine responses were seen in the colon draining MLN. These inflammatory responses could be due to innate immune responses to bacteria and bacterial products as well as T cell responses to luminal antigens delivered by colonic GAPs. We evaluated the ability to induce antigen specific immune responses in the colon draining LNs following antibiotic therapy. In the absence of antibiotic therapy, little to no antigen specific T cell proliferation in response to luminal antigen was observed in LN draining the cecum, ascending colon (colonic MLN) and transverse colon (colic LN), while proliferation was observed in the LNs draining the descending colon and sigmoid colon (caudal and iliac LN; Fig. 5B), correlating with presence of GAPs in the descending colon and sigmoid colon and their absence in proximal regions of the colon in the steady-state (Fig. 1A, C, D and E). This also is consistent with GAPs as a mechanism delivering antigen to the caudal and iliac LNs to induce tolerance to rectally administered antigens.18 In contrast, following antibiotic therapy, robust T cell proliferation to luminal antigen was observed in the LNs draining all segments of the colon (Fig. 5C-F), correlating with the presence of GAPs in all segments of the colon after antibiotic treatment (Fig. 1A, C, D and E). This suggests that the inflammatory cytokines we observed to be produced in the colon draining LN7 may influence naïve T cell differentiation to promote inflammatory responses to otherwise harmless luminal antigens encountered in the setting of bacterial translocation following antibiotic therapy.
Treatment with FTY720 to inhibit S1PR dependent migration at the time of the luminal OVA administration almost completely abrogated antigen specific T cell proliferation in the LNs draining the cecum, ascending colon and transverse colon (colonic MLN and colic LN), while only modestly affecting antigen specific T cell proliferation in the LNs draining the descending colon and sigmoid colon (caudal and iliac LN), which resembled proliferation to luminal antigen untreated mice (Fig. 5C-F). This suggests that the colonic LP-MNPs acquiring luminal antigen and inducing robust T cell responses in the MLNs following antibiotic therapy require S1PR to migrate to the MLN. This also suggests that a second LP-MNP population may acquire luminal antigens in the distal colon and carry them to the LN to induce more moderate proliferative responses in an S1PR independent manner in the absence of antibiotic therapy. Our observations might also indicate that the beneficial effect S1PR inhibitors have on colitis are in part due to inhibition of LP-MNP trafficking to the LN and subsequent inhibition of T cell responses.
Conclusion
The translocation of bacteria across the intestinal epithelium following antibiotic treatment is a well recognized and, until now, enigmatic process. Here we show a window of time following antibiotics where the colonic microbiota is permissive to the formation of colonic GAPs. Once formed, colonic GAPs can deliver a variety of substances, including protein antigens and live commensal bacteria. CD103+ and CX3CR1+ LP-MNPs interact with GAPs to access luminal antigen, while commensal bacteria are primarily acquired by CX3CR1+ LP-MNPs.7 After acquiring commensal bacteria and luminal antigens LP-MNPs traffic to the draining LNs promoting an inflammatory environment, which could result in inflammatory T cell responses to otherwise innocuous antigens. It is not known if inflammatory responses are induced in the LN draining the distal colonic segments following antibiotic therapy, which may be an exception to this sequence of events, as the distal colon GAPs are present in the steady-state and bacterial translocation to the LNs draining the distal colon was not observed in the steady-state or following antibiotic treatment. Alterations in the gut microbiota and antibiotic use are increasingly being associated with inflammatory disorders. The observations presented here demonstrate that inflammatory responses following antibiotic use occur in part due to translocation of gut bacteria via GAPs and suggest that strategies such as manipulation of the gut microbiota following antibiotic therapy to prevent translocation or blockade of LP-MNP trafficking to prevent bacterial dissemination may be approaches to mitigate antibiotic associated inflammatory responses.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Mark J. Miller for insightful critiques and assistance with 2P imaging and analysis. The 2P imaging was performed at the In Vivo Imaging Core at Washington University School of Medicine. The High Speed Cell Sorter Core at the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO. provided flow cytometric cell sorting services. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant P30 CA91842. Studies were supported by the Washington University Digestive Diseases Research Center Core P30 DK052574, DK097317-RDN, Children's Discovery Institute Grant MD-II-2015–481–RDN, AI112626–RDN, AGA Providence Food Intolerance Award–RDN, and DK097893-KAK.
Funding
Work supported by NIH grants P30 CA91842, P30 DK052574, DK097317-RDN, AI112626–RDN AI131342-RDN, Children's Discovery Institute Grant MD-II-2015–481–RDN, AGA Providence Food Intolerance Award–RDN, and DK097893-KAK
References
- [1].Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al.. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012; 488:621-6; PMID:22914093; http://dx.doi.org/ 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Norgaard M, Nielsen RB, Jacobsen JB, Gradus JL, Stenager E, Koch-Henriksen N, Lash TL, Sørensen HT. Use of penicillin and other antibiotics and risk of multiple sclerosis: a population-based case-control study. Am J Epidemiol 2011; 174:945-8; PMID:21920946; http://dx.doi.org/ 10.1093/aje/kwr201 [DOI] [PubMed] [Google Scholar]
- [3].Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics and new diagnoses of Crohn's disease and ulcerative colitis. Am J Gastroenterol 2011; 106:2133-42; PMID:21912437; http://dx.doi.org/ 10.1038/ajg.2011.304 [DOI] [PubMed] [Google Scholar]
- [4].Scribano ML, Prantera C. Antibiotics and inflammatory bowel diseases. Dig Dis 2013; 31:379-84; PMID:24246992; http://dx.doi.org/ 10.1159/000354704 [DOI] [PubMed] [Google Scholar]
- [5].Canova C, Zabeo V, Pitter G, Romor P, Baldovin T, Zanotti R, Simonato L. Association of maternal education, early infections, and antibiotic use with celiac disease: a population-based birth cohort study in northeastern Italy. Am J Epidemiol 2014; 180:76-85; PMID:24853109; http://dx.doi.org/ 10.1093/aje/kwu101 [DOI] [PubMed] [Google Scholar]
- [6].Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother's and offspring's use of antibiotics and infant allergy to cow's milk. Epidemiology 2013; 24:303-9; PMID:23348066; http://dx.doi.org/ 10.1097/EDE.0b013e31827f520f [DOI] [PubMed] [Google Scholar]
- [7].Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 2016; 65:1100-9; PMID:26045138; http://dx.doi.org/ 10.1136/gutjnl-2014-309059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med 2009; 206:3101-14; PMID:20008524; http://dx.doi.org/ 10.1084/jem.20091925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, Kalchenko V, Geissmann F, Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med 2007; 204:171-80; PMID:17190836; http://dx.doi.org/ 10.1084/jem.20061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity 2009; 31:502-12; PMID:19733097; http://dx.doi.org/ 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- [11].Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, et al.. Origin of the lamina propria dendritic cell network. Immunity 2009; 31:513-25; PMID:19733489; http://dx.doi.org/ 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012; 483:345-9; PMID:22422267; http://dx.doi.org/ 10.1038/nature10863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol 2015; 8:198-210; PMID:25005358; http://dx.doi.org/ 10.1038/mi.2014.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berg RD, Garlington AW. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun 1979; 23:403-11; PMID:154474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Berg RD. Promotion of the translocation of enteric bacteria from the gastrointestinal tracts of mice by oral treatment with penicillin, clindamycin, or metronidazole. Infect Immun 1981; 33:854-61; PMID:6456996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berg RD, Owens WE. Inhibition of translocation of viable Escherichia coli from the gastrointestinal tract of mice by bacterial antagonism. Infect Immun 1979; 25:820-27; PMID:159260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berg RD. Inhibition of Escherichia coli translocation from the gastrointestinal tract by normal cecal flora in gnotobiotic or antibiotic-decontaminated mice. Infect Immun 1980; 29:1073-81; PMID:6448820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Veenbergen S, van Berkel LA, du Pre MF, He J, Karrich JJ, Costes LM, Luk F, Simons-Oosterhuis Y, Raatgeep HC, Cerovic V, et al.. Colonic tolerance develops in the iliac lymph nodes and can be established independent of CD103(+) dendritic cells. Mucosal Immunol 2016; 9:894-906; PMID:26577569; http://dx.doi.org/ 10.1038/mi.2015.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014; 14:667-85; PMID:25234148; http://dx.doi.org/ 10.1038/nri3738 [DOI] [PubMed] [Google Scholar]
- [20].Van den Broeck W, Derore A, Simoens P. Anatomy and nomenclature of murine lymph nodes: Descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. J Immunol Methods 2006; 312:12-9; PMID:16624319; http://dx.doi.org/ 10.1016/j.jim.2006.01.022 [DOI] [PubMed] [Google Scholar]
- [21].Carter PB, Collins FM. The route of enteric infection in normal mice. J Exp Med 1974; 139:1189-203; PMID:4596512; http://dx.doi.org/ 10.1084/jem.139.5.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gautreaux MD, Deitch EA, Berg RD. Bacterial translocation from the gastrointestinal tract to various segments of the mesenteric lymph node complex. Infect Immun 1994; 62:2132-4; PMID:8168984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastrointest Liver Physiol 2013; 305:G341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 2013; 494:116-20; PMID:23334413; http://dx.doi.org/ 10.1038/nature11809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu LC, Shih YA, Wu LL, Lin YD, Kuo WT, Peng WH, Lu KS, Wei SC, Turner JR, Ni YH. Enteric dysbiosis promotes antibiotic-resistant bacterial infection: systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. Am J Physiol Gastrointest Liver Physiol 2014; 307:G824-35; PMID:25059827; http://dx.doi.org/ 10.1152/ajpgi.00070.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jacob E, Baker SJ, Swaminathan SP. 'M' cells in the follicle-associated epithelium of the human colon. Histopathology 1987; 11:941-52; PMID:3666678; http://dx.doi.org/ 10.1111/j.1365-2559.1987.tb01900.x [DOI] [PubMed] [Google Scholar]
- [27].Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al.. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 2005; 307:254-8; PMID:15653504; http://dx.doi.org/ 10.1126/science.1102901 [DOI] [PubMed] [Google Scholar]
- [28].Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med 2006; 203:2841-52; PMID:17145958; http://dx.doi.org/ 10.1084/jem.20061884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vallon-Eberhard A, Landsman L, Yogev N, Verrier B, Jung S. Transepithelial pathogen uptake into the small intestinal lamina propria. J Immunol 2006; 176:2465-9; PMID:16456006; http://dx.doi.org/ 10.4049/jimmunol.176.4.2465 [DOI] [PubMed] [Google Scholar]
- [30].Hapfelmeier S, Muller AJ, Stecher B, Kaiser P, Barthel M, Endt K, Eberhard M, Robbiani R, Jacobi CA, Heikenwalder M, et al.. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent stepin invG S. Typhimurium colitis. J Exp Med 2008; 205:437-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cruickshank SM, Deschoolmeester ML, Svensson M, Howell G, Bazakou A, Logunova L, Little MC, English N, Mack M, Grencis RK, et al.. Rapid dendritic cell mobilization to the large intestinal epithelium is associated with resistance to Trichuris muris infection. J Immunol 2009; 182:3055-62 http://dx.doi.org/ 10.4049/jimmunol.0802749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol 2010; 184:2026-37; PMID:20089703; http://dx.doi.org/ 10.4049/jimmunol.0901936 [DOI] [PubMed] [Google Scholar]
- [33].Kim KW, Vallon-Eberhard A, Zigmond E, Farache J, Shezen E, Shakhar G, Ludwig A, Lira SA, Jung S. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood 2011; 118:e156-67; PMID:21951685; http://dx.doi.org/ 10.1182/blood-2011-04-348946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, Furtado GC, Lira SA, Shakhar G. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 2013; 38:581-95; PMID:23395676; http://dx.doi.org/ 10.1016/j.immuni.2013.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 2008; 105:15064-9; PMID:18806221; http://dx.doi.org/ 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004; 427:355-60; PMID:14737169; http://dx.doi.org/ 10.1038/nature02284 [DOI] [PubMed] [Google Scholar]
- [37].Karuppuchamy T, Behrens Eh, Gonzalez-Cabrera P, Sarkisyan G, Gima L, Boyer JD, Bamias G, Jedlicka P, Veny M, Clark D, et al.. Sphingosine-1-phosphate receptor-1 (S1P1) is expressed by lymphocytes, dendritic cells, and endothelium and modulated during inflammatory bowel disease. Mucosal Immunol 2016; 10(1):162-71; PMID:27049060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sandborn WJ, Feagan BG, Wolf DC, D'Haens G, Vermeire S, Hanauer SB, Ghosh S, Smith H, Cravets M, Frohna PA, et al.. Ozanimod induction and maintenance treatment for ulcerative colitis. N Engl J Med 2016; 374:1754-62; PMID:27144850; http://dx.doi.org/ 10.1056/NEJMoa1513248 [DOI] [PubMed] [Google Scholar]
- [39].Bamias G, Rivera-Nieves J. Targeting S1P receptors, a new mechanism of action for inflammatory bowel disease therapy. Gastroenterology 2016; 151(5):1025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].St John AL, Ang WX, Huang MN, Kunder CA, Chan EW, Gunn MD, Abraham SN. S1P-Dependent trafficking of intracellular yersinia pestis through lymph nodes establishes Buboes and systemic infection. Immunity 2014; 41:440-50; PMID:25238098; http://dx.doi.org/ 10.1016/j.immuni.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 2000; 20:4106-14; PMID:10805752; http://dx.doi.org/ 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell 1994; 76:17-27; PMID:8287475; http://dx.doi.org/ 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- [43].Sanna MG, Liao J, Jo E, Alfonso C, Ahn M-Y, Peterson MS, Webb B, Lefebvre S, Chun J, Gray N, et al.. Sphingosine 1-phosphate (S1P) receptor subtypes S1P1 and S1P3, respectively, regulate lymphocyte recirculation and heart rate. J Biol Chem 2004; 279:13839-48; PMID:14732717; http://dx.doi.org/ 10.1074/jbc.M311743200 [DOI] [PubMed] [Google Scholar]


