Abstract
Ligand-dependent activation of the Ah receptor (AhR) can result in an extremely diverse spectrum of biological and toxic effects that occur in a ligand-, species- and tissue-specific manner. While the classical mechanism of AhR-dependent signal transduction is directly related to its ability to modulate gene expression, the dramatic diversity in responses observed following AhR activation or inhibition is inconsistent with a single molecular mechanism of AhR action. Recent studies have revealed that key molecular events underlying the AhR signaling pathway are significantly more varied and complex than previously established, and the specificity and diversity in AhR response can be selectively modulated by a variety of factors. Here we describe new insights into the mechanistic diversity in AhR signal transduction that can contribute to ligand-, species- and tissue-specific differences in AhR reponse.
Keywords: Ah Receptor, AhR, TCDD, Ligand Binding, Ligand Specificity, Toxicity
Graphical abstract
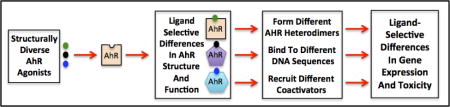
1. Introduction
The Ah receptor (AhR) is a ligand-dependent basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS)-containing transcription factor that responds to exogenous and endogenous chemicals by inducing or repressing the expression of a number of genes and mediating a diverse spectrum of biological and toxic effects in a wide range of species and tissues [1–7]. Additionally, the AhR has been shown to pay a key modulatory role in the regulation of a variety of physiological responses including developmental and immune processes [6–9]. While the AhR signal transduction pathway has similarities to that of nuclear receptors (e.g. steroid hormone receptors), the AhR is unique in that it can differentially respond to structurally diverse chemicals to produce a variety of ligand-selective toxic and/or biological effects, which in turn can be mediated by several different AhR-dependent mechanisms. This review highlights our current understanding of the diversity in ligand-dependent mechanisms of AhR signal transduction and response.
2. Diversity in AhR-Dependent Mechanisms of Gene Expression
Initiation of the classical or cannonical AhR signal transduction mechanism requires binding of the inducing ligand to the PASB domain of the AhR, which is part of a cytosolic multiprotein complex containing heat shock protein 90 (hsp90), XAP2 and p23 [3,4,6], and a subsequent ligand-dependent conformational change in the AhR leading to its nuclear translocation [3,4,6]. Dimerization of the AhR with ARNT (AhR nuclear translocator), a structurally and functionally related bHLH-PAS protein, displaces the AhR from its associated proteins and transforms the AhR into its high affinity DNA binding form [4,10,11]. Binding of the transformed ligand:AhR:ARNT complex to its specific DNA recognition site, the dioxin responsive element (DRE; also referred to as a xenobiotic responsive element (XRE) or Ah responsive element (AHRE)), present in or adjacent to AhR-responsive genes, leads to coactivator recruitment, chromatin rearrangement, and increased gene transcription [4,6,10,12].
While numerous gene products have been identified that are consistently altered in different species and tissues (e.g. CYP1A1) in response to a given AhR ligand, gene expression array and chromatin immunoprecipitation (ChIP) analysis has also revealed significant differences in gene expression profiles [13–15]. The diversity in AhR-dependent gene expression responses observed between cell types can be attributed to a variety of factors, including, but not limited to: AhR/ARNT expression, the presence/absence of specific co-activators/co-repressors and/or transcription factors that can compete with the AhR for ARNT (e.g. Hypoxia Inducible Factor 1α (HIF1α) or AhR repressor (AHRR)), and differences in chromatin structure and epigenetic modifications of AhR target genes [4,12,16–19]. AhR-dependent alterations in the expression of genes that lack an apparent AhR DNA (DRE) binding site, coupled with established cross-talk between the AhR and cellular signaling pathways and other transcription factors, suggests that the AhR participates in several novel noncannonical pathways by which the AhR can stimulate gene expression [6,20–23]. Ligand-activated AhR can dimerize with nuclear proteins other than ARNT (e.g., Kruppel-like factor 6 (KLF6) and RelB), and these unique heterodimers stimulate gene expression via their interaction with DNA binding sites that are significantly different from that of a DRE [24–28] to regulate a unique set of genes (Figure 1). While little is known about the specific protein:protein interactions that occur between the AhR and RelB [24], deletion and functional analysis studies revealed that the mode of AhR:KLF6 dimerization is distinctly different from that of the AhR:ARNT dimer [27]. In addition to these unique AhR heterodimers, ligand-activated AhR can enhance gene expression via its ability to function as a coactivator for other nuclear transcription factors such as the estrogen receptor and E2F1 [29,30]. More recently, it has been observed that binding of the AhR by selective AhR modulator ligands can repress the expression of a unique battery of genes and although the mechanism remains to be determined, it does not appear to require the AhR DNA binding domain [31,32]. While the canonical AhR:ARNT:DRE-dependent mechanism appears to be the principal AhR signaling pathway, ligand-dependent activation and nuclear localization of the AhR can regulate expression of diverse genes via multiple mechanisms, and others may still be identified (Figure 1).
Figure 1.
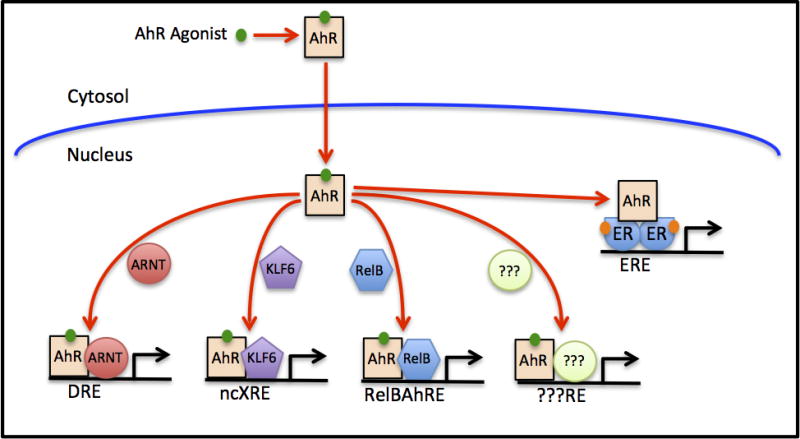
Multiple mechanisms by which a specific ligand-activated AhR can stimulate gene expression. In the classical mechanism of AhR action, ligand binding stimulates AhR nuclear translocation, dimerization with the ARNT protein and the binding of the ligand:AhR:ARNT complex to its DNA binding site (the DRE) stimulates gene expression. However, the dimerization of liganded AhR with other proteins (e.g., KLF6 or RelB) results in the formation of unique protein complexes that bind to distinctly different DNA recognition sites (e.g., a ncXRE or RelBAhRE, respectively) to regulate subsets of genes not regulated by the AhR:ARNT complex. Whether ligand bound AhRs can interact with additional DNA binding partners remains to be determined, but is a possibility. In addition to multiple heterodimers, the AhR has also been observed to bind to other nuclear protein complexes (e.g., estrogen receptor (ER) dimers) and function as a coactivator, enhancing gene expression by these transcription factors.
3. Diversity in AhR Ligand Structure
The best-characterized high affinity ligands for the AhR include a variety of toxic halogenated aromatic hydrocarbons (HAHs), such as the polychlorinated dibenzo-p-dioxins (e.g., 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin)), dibenzofurans, and biphenyls, and numerous polycyclic aromatic hydrocarbons (PAHs) and PAH-like chemicals, such as 3-methylcholanthrene (3MC) and beta-naphthoflavone (BNF) [2,6,33]. It is now well established, by our laboratory and others, that the AhR can bind and be activated/inhibited by a relatively large number of natural, endogenous and synthetic AhR agonists/antagonists whose structure and physicochemical characteristics are dramatically different from the prototypical HAH and PAH ligands [6,33–38]. The lack of a 3D crystal/NMR structure of the AhR ligand binding domain (LBD) has hampered a detailed mechanistic understanding of AhR binding by structurally diverse ligands. However, site-directed mutagenesis and structure-function analysis, including those based on a homology model of the AhR LBD originally developed in collaboration with Dr. Laura Bonati [39–41], have not only provided further evidence for differential binding of structurally diverse ligands within the ligand binding pocket of a single AhR or between AhRs from different species [39–46]. These studies have also provided new insights into the mechanisms contributing to significant differences in the binding affinity of structurally different AhR ligands.
The extreme structural diversity of ligands for the AhR is very similar to the well-established ligand promiscuity reported for the pregnane X receptor (PXR), a member of the nuclear receptor superfamily [47–49]. Crystal and NMR structural analysis of PXR revealed that it has a very large and flexible ligand binding pocket and ligands reportedly can bind in different orientations and with different residues within the pocket [47,48]. The structural diversity of ligands and promiscuity of ligand binding demonstrated by both AhR and PXR ligands suggested some similarities in the ligand binding pockets and ligand-selective activation by these two different receptors. Gene expression analysis of a chemical library of >300,000 compounds allowed direct comparison of ligand-dependent activation of both AhR and PXR reporter gene responses by structurally diverse chemicals [50]. In these studies, a collection of 2281 structurally diverse chemicals, selected from 7790 AhR active compounds identified in the first screen of the library, were tested for their ability to simulate both AhR- and PXR-dependent induction of gene expression in stably transfected human hepatoma (HepG2) cells. These analyses revealed for the first time a striking overlap of AhR and PXR agonists, with 1982 of the 2281 structurally diverse schemicals stimulating AhR-dependent gene expression and 2017 of the 2281 chemicals stimulating PXR-dependent gene expression; 126 chemicals were shown to be selective for the AhR. The ability of the most potent AhR-selective agonists to competitively bind to and/or stimulate AhR DNA binding in vitro was demonstrated in subsequent analysis. The identification of potent and high affinity structurally diverse AhR-selective ligands/agonists that did not activate PXR indicated that although these receptors demonstrate innate similarities in ligand promiscuity, the molecular mechanism(s) of AhR and PXR activation by structurally diverse ligands is not identical [50].
AhR ligand diversity is commonly determined by measuring the ability of a chemical(s) to simulate AhR-dependent gene expression in cells in culture. However, Rannug and coworkers recently challenged this concept and proposed that the apparent structural diversity observed for AhR ligands is actually an artifact of cell culture-based gene expression assay systems [8,9,51,52]. These authors suggest that the AhR has a limited range of acceptable ligands, and that the apparent AhR-dependent gene induction observed with structurally diverse chemicals was actually an indirect response resulting from the ability of these diverse chemicals to inhibit cytochrome P4501-dependent degradation of 6-formylindolo[3,2-b]carbazole (FICZ), a high affinity AhR agonist reportedly present in cell growth media, and that FICZ was the actual inducing chemical [8,9,51,52]. While FICZ could play a role in AhR-dependent gene induction in certain experimental conditions with certain chemicals as these authors proposed, this “indirect activation” hypothesis failed to consider the extensive amount of in vitro binding results available in the published literature that provide strong support for AhR ligand promiscuity. Such studies have already clearly demonstrated the ability of structurally diverse chemicals to not only directly bind to the AhR in vitro (using cytosolic and/or in vitro expressed AhR in competitive radiolabeled ligand binding assays), but also to stimulate AhR DNA binding in vitro (using cytsolic and/or in vitro expressed AhR/ARNT in gel retardation assays) [6,33–37,44,46,53]. In these experimental methods, FICZ, even if present, would not affect the ability of the test chemical to bind to the available unliganded AhR and/or to stimulate AhR DNA binding. Thus, the extensive amount of published literature provides strong support the conclusion that the AhR can directly bind and be activated by structurally diverse chemicals.
4. Diversity in AhR Ligand-Dependent Gene Induction
Given that it has already been established that the AhR can heterodimerize with factors other than ARNT to produce distinct complexes that can bind to distinctly different DNA sequences [24,26–28], the idea that diverse ligands simply activate the AhR to produce an identical AhR:ARNT complex that binds to the same DNA binding sites (i.e. the DRE) and yet produce diverse gene expression responses is clearly not correct. Studies by our laboratory and others have shown that the structural promiscuity of AhR ligands results from differences in the interactions of these diverse ligands with residues within the AhR binding pocket, however, whether these ligand-selective interactions produce AhRs with altered structure and/or functional activity remains an open question. The idea that the structure and functional activity of the AhR may be differentially altered depending on the specific ligand to which it is bound has been suggested by gene expression studies, where equipotent concentrations of diverse AhR ligands not only produce distinctly different magnitudes of induction of the same gene, but also induce a ligand-specific set of AhR-dependent gene products in the same cells [6,13,14,54–56].
It is possible that specific AhR heterodimers or heterodimer combinations can be formed in a ligand-selective manner, and this could contribute to ligand diversity in response, but this has not been examined. Alternatively, it has been suggested that ligand-specific differences produced in the overall structure of the AhR and/or ARNT could alter the nucleotide specificity of AhR:ARNT DNA binding (or perhaps DNA binding of other AhR heterodimers), leading to ligand-specific differences in gene induction responses. While this was an attractive hypothesis and several novel ligand-selective DNA binding sites were proposed [54–57], subsequent PCR-based binding site analysis revealed that AhR:ARNT complexes activated by structurally diverse agonists only bound to DRE-containing DNA [53] and the proposed novel ligand-selective DNA binding sites could not be confirmed [58].
Alternatively, by analogy with steroid hormone receptor mechanisms [59–62], ligand-selective modulation of AhR signaling pathways and the magnitude of response in a given cell may result from ligand-specific changes in the structure of the AhR, AhR:ARNT, and/or other AhR:protein complexes, which could allow interactions with different subsets of transcriptional modulators (e.g. coactivators, corepressors), thereby producing different gene expression responses. This mechanism (Figure 2) is consistent with results from a two-hybrid analysis study that demonstrated that the binding of different HAH ligands to the AhR resulted in distinct differences in coactivator recruitment and were suggestive of ligand-selective differences in AhR structure [63]. However, since that study used only a small fragment of the AhR as part of a protein chimera and did not include ARNT, it remains to be determined whether the ligand-selective differences in coactivator recruitment occurs with the full-length AhR and/or ARNT proteins. To date, although ligand (TCDD)-dependent alterations in AhR structure have been observed using in vitro synthesized [35S]-labeled AhR and proteolysis approaches [64], ligand-specific differences in overall AhR and/or ARNT structure have not yet been reported. However, a recent study was one of the first to clearly demonstrate AhR ligand-specific differential gene induction in a single cell type [65]. Stannocalcin 2 (Stc2) is a gene whose promoter contains numerous DREs [56,65], however, while classical AhR agonists (TCDD, 3MC and BNF) stimulate AhR binding to DREs upstream of CYP1A1 (measured using ChIP analysis) and induced CYP1A1 gene expression in primary hepatocytes, they failed to stimulate Stc2 gene expression or ligand-dependent binding of AhR to Stc2 promoter DREs [56,65]. In contrast, cinnabarinic acid, a newly identified tryptophan-derived AhR agonist [66], stimulated both AhR binding to the Stc2 promoter DREs and Stc2 gene expression, but failed to stimulate AhR binding to CYP1A1 DREs or induce CYP1A1 gene expression [65]. While details of the mechanism(s) responsible for the differential ligand responses of CYP1A1 and Stc2 genes remain to be elucidated, these results are consistent with ligand-selective differences in AhR gene expression and are suggestive of ligand-selective differences in AhR structure/function.
Figure 2.
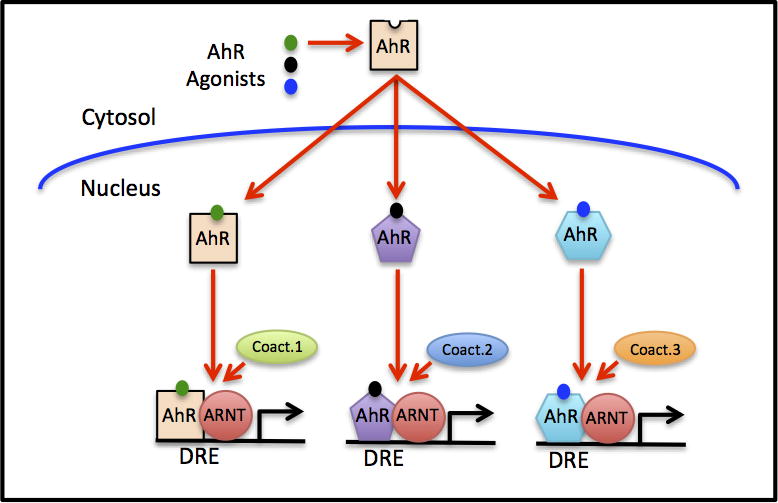
Ligand-selective differences in AhR-dependent gene expression. Binding and activation of the AhR by structurally diverse AhR ligands could produce significant differences in the overall structure of the AhR and/or its dimerization partner that result in recruitment of distinctly different coactivators to the DNA bound AhR complex and differential gene expression. Although this figure only depicts an alteration in the structure of the AhR, ligand-selective structural changes could also occur in the ARNT protein, or in any other protein(s) to which the AhR is bound (such as KLF6 or RelB (see Figure 1)), facilitating differential coactivator recruitment by a greater diversity of ligand-activated AhR complexes and even a greater diversity in gene expression responses.
5. Diversity in AhR Ligand-Dependent Toxicity
While structurally diverse ligands can stimulate AhR-dependent gene expression and produce biological responses like TCDD and TCDD-like HAHs, they do not produce the major toxic effects observed with these compounds (i.e., lethality, wasting, birth defects, chloracne, etc) [1,2,6,33,37,67]. This suggests differences in the overall mechanism of action of “toxic” and “nontoxic” AhR ligands. Current evidence suggests that the persistence of AhR-dependent gene expression produced by metabolically stable TCDD-like HAHs is responsible for the prototypical spectrum of AhR-dependent toxicity [1,2,28,67]. In contrast, metabolically labile AhR ligands (e.g. BNF, 3MC and most structurally diverse ligands) only transiently activate AhR-dependent gene expression, which is suggested to be insufficient to produce the prototypical spectrum of dioxin-like toxicity. If persistent AhR activation is responsible for the observed toxic effects of TCDD-like HAHs, it could be postulated that chronic daily exposure to high doses of a “nontoxic” AhR agonists would be expected to result in persistent AhR activation and produce AhR-dependent dioxin-like toxicity. This has only been indirectly examined in one study in which C57 mice were chronically fed high doses (150 mg/kg) of the relatively potent AhR agonist BNF, 5 days a week for 6 weeks [68]. The lack of any reported AhR-dependent toxic effects suggests that additional factors may contribute to toxicity beyond simply persistence of AhR activation. One hypothesis is that there is an additional molecular target that is selectively affected by toxic TCDD-like HAHs and not by nontoxic AhR ligands, and the combined activation of these distinct targets is required for the observed AhR-dependent toxicity to be manifested. While several AhR-independent effects of TCDD have been previously reported [6,15,69–73], their role in the prototypical spectrum of TCDD toxic responses is unknown. Alternatively, ligand-selective modulation of AhR:ARNT structure and function has also been proposed to at least partially explain the differential ability of ligands to produce the prototypical spectrum of AhR-dependent toxic effects (i.e., lethality, wasting, birth defects, chloracne, etc) and selective TCDD-like HAH gene expression responses [1,2,33,46,67]. Site-directed mutagenesis and functional analysis has revealed that the binding of TCDD-like HAHs within the AhR LBD was distinctly different from that of structurally diverse nontoxic AhR ligands [46], suggesting that AhRs bound by TCDD-like HAHs could have a distinctly different structure/function. Whether HAH-specific structural/functional differences in the AhR in combination with the metabolic persistence of HAHs contribute to AhR-dependent toxicity remains to be examined.
6. Concluding Remarks
Early insights into the molecular mechanism of AhR signal transduction were primarily the result of research into the effects of TCDD and TCDD-like HAHs on CYP1A1 gene expression, and these studies provided new avenues to understand the mechanism by which these widespread environmental contaminants produced toxicity. However, these studies provided few insights into how this relatively simple mechanism could produce the diverse spectrum of toxic and biological effects of these compounds. What was apparent was that a wide variety of structurally diverse dioxin-like and non-dioxin-like ligands (agonists) could stimulate expression of the same spectrum of classical AhR-dependent genes that are induced by TCDD (e.g. CYP1A1, CYP1B1, etc.). Thus, simply demonstrating the ability of a chemical or mixture to stimulate expression of a given AhR-dependent gene provides no useful information as to its ability or potential to produce dioxin-like toxicity, only that it is an AhR agonist. For an AhR ligand to actually be considered dioxin-like, it must be able to produce TCDD-like toxicity in vivo. Understanding the diversity in AhR signaling and response is a complex process. The AhR is known to be a key regulatory factor in a wide variety of endogenous physiological processes, adaptive gene responses and adverse health effects, and that the specificity and magnitude of individual AhR responses vary in a ligand-, cell-, tissue- and species-specific manner. Recent demonstration that the AhR can form heterodimers with nuclear factors other than ARNT to stimulate expression of a distinctly different subset of genes, coupled with the ability of the AhR to bind and be activated by structurally diverse ligands and produce differential gene responses, has expanded the complexity of the AhR signaling pathway and provided new avenues in which to begin to understand AhR diversity in response (Figure 3). The recent identification of a role of the AhR in human disease has not only made it a new and significant target for the development of human therapeutic drugs, but demonstration of both the structural diversity and species selectivity of AhR ligands has provided pharmaceutical companies with numerous lead compounds for AhR drug development. However, an increased understanding of the biochemical and molecular mechanisms by which toxic and nontoxic ligands can differentially regulate AhR functionality and downstream responses is now even more important for the continued development and ultimate approval of such drugs for human use. Overall, even though a significant amount of information has been generated on the AhR signaling pathway, it remains an exciting area for continued research and many significant open questions still remain.
Figure 3.
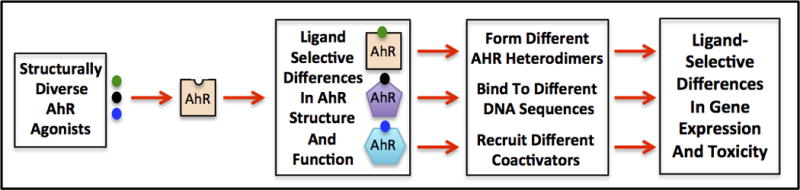
Overall mechanisms by which structurally diverse AhR ligands can contribute to ligand-selective differences in AhR-dependent gene expression and toxicity. See text for details.
Highlights.
The AhR can bind and be activated by structurally diverse ligands and species differences in ligand selectivity have been observed
The AhR can stimulate gene expression by a combination of cannonical and noncannonical mechanisms
AhR-dependent gene expression can vary in a ligand-, mechanism-, cell-, species-, and tissue-specific manner
Mechanisms responsible for the toxicity of select AhR ligands still remains to be determined
Acknowledgments
We apologize to our colleagues whose research contributions could not be included due to space limitations. The authors gratefully acknowledge support from the National Institute of Environmental Health Sciences [R01ES007685 and P42ES004699 (MSD) and a predoctoral fellowship T32ES007059 (SCF)], the California Agriculture Experiment Station, and the American taxpayers.
Abbreviations
- AhR
Aryl hydrocarbon receptor
- AhRE
Ah responsive element
- AHRR
Ah receptor repressor
- ARNT
Ah receptor nuclear translocator
- bHLH-PAS
Basic Helix-Loop-Helix-Per-ARNT-Sim
- BNF
β-Naphthoflavone
- ChIP
Chromatin immunoprecipitation
- DRE
Dioxin responsive element
- FICZ
6-formylindolo[3,2-b]carbazole
- HAHs
Halogenated aromatic hydrocarbons
- HIF1a
Hypoxia inducible factor 1α
- hsp90
Heat shock protein 90
- KLF6
Kruppel-Like Factor 6
- LBD
Ligand binding domain
- 3MC
3-Methylcholanthrene
- PAHs
Polycyclic aromatic hydrocarbons
- PXR
Pregnane X receptor
- Stc2
Stannocalcin 2
- TCDD
2,3,7,8-Tetrachlorodibenzo-p-dioxin
- XRE
Xenobiotic responsive element
References and Recommended Reading
Papers of particular interest have been highlighted as follows:
• of special interest
•• of significant interest
- 1.Poland A, Glover E. 2,3,7,8,-Tetrachlorodibenzo-p-dioxin: segregation of toxocity with the Ah locus. Mol Pharmacol. 1980;17:86–94. [PubMed] [Google Scholar]
- 2.Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit Rev Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 4•.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. Comprehensive review of the molecular mechanisms of AhR-dependent gene expression and coactivators important in AhR signal transduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White SS, Birnbaum LS. An overview of the effects of dioxins and dioxin-like compounds on vertebrates, as documented in human and ecological epidemiology. J Environ Sci Health Part C, Environ Carcin Ecotoxicol Rev. 2009;27:197–211. doi: 10.1080/10590500903310047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. First comprehensive review of the classical and nonclassical/noncannonical mechanisms of AhR action that contribute to its diversity in response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67:259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 9.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 10•.Denison MS, Fisher JM, Whitlock JP., Jr The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J Biol Chem. 1988;263:17221–17224. This was the first paper to identify the DNA binding sequence (i.e., the DRE) for the ligand-activated AhR:ARNT complex. [PubMed] [Google Scholar]
- 11.Soshilov A, Denison MS. Ligand displaces heat shock protein 90 from overlapping binding sites within the aryl hydrocarbon receptor ligand-binding domain. J Biol Chem. 2011;286:35275–35282. doi: 10.1074/jbc.M111.246439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hankinson O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch Biochem Biophy. 2005;433:379–386. doi: 10.1016/j.abb.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 13••.Nault R, Forgacs AL, Dere E, Zacharewski TR. Comparisons of differential gene expression elicited by TCDD, PCB126, betaNF, or ICZ in mouse hepatoma Hepa1c1c7 cells and C57BL/6 mouse liver. Toxicol Lett. 2013;223:52–59. doi: 10.1016/j.toxlet.2013.08.013. Describes comparative gene expression analysis induced different AhR ligands in cells in culture and in vivo and provided evidence for ligand-selective AhR gene activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodale BC, Tilton SC, Corvi MM, Wilson GR, Janszen DB, Anderson KA, Waters KM, Tanguay RL. Structurally distinct polycyclic aromatic hydrocarbons induce differential transcriptional responses in developing zebrafish. Toxicol Appl Pharmacol. 2013;272:656–670. doi: 10.1016/j.taap.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. This paper describes microarray analysis and AhR knockout mice to characterize genes that are regulated in a TCDD- and AhR-dependent and -independent mechanisms. [DOI] [PubMed] [Google Scholar]
- 16.Gradin K, McGuire J, Wenger RH, Kvietikova I, fhitelaw ML, Toftgard R, Tora L, Gassmann M, Poellinger L. Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol Cell Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, Nelson WE, Probst MR, Rosenfeld MG, Hankinson O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol. 2002;22:4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans BR, Karchner SI, Allan LL, Pollenz RS, Tanguay RL, Jenny MJ, Sherr DH, Hahn ME. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol Pharmacol. 2008;73:387–398. doi: 10.1124/mol.107.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beedanagari SR, Taylor RT, Bui P, Wang F, Nickerson DW, Hankinson O. Role of epigenetic mechanisms in differential regulation of the dioxin-inducible human CYP1A1 and CYP1B1 genes. Mol Pharmacol. 2010;78:608–616. doi: 10.1124/mol.110.064899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinehara M, Fukuda I, Yoshida K, Ashida H. High-throughput evaluation of aryl hydrocarbon receptor-binding sites selected via chromatin immunoprecipitation-based screening in Hepa-1c1c7 cells stimulated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Genes Genet Syst. 2008;83:455–468. doi: 10.1266/ggs.83.455. [DOI] [PubMed] [Google Scholar]
- 21••.Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011;12:365. doi: 10.1186/1471-2164-12-365. This paper identified genes apparently regulated in a DRE-independent manner suggestive of alternative mechnisms of AhR action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo R, Matthews J. High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol Sci. 2012;130:349–361. doi: 10.1093/toxsci/kfs253. [DOI] [PubMed] [Google Scholar]
- 23.Tanos R, Patel RD, Murray IA, Smith PB, Patterson AD, Perdew GH. Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology. 2012;55:1994–2004. doi: 10.1002/hep.25571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. This paper identified a novel heterodimer between AhR and RelB that stimulates gene expression via a novel DNA binding element (the RelBAhRE) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel CF, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang G, Elferink CJ. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol. 2012;81:338–347. doi: 10.1124/mol.111.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exper Ther. 2013;345:419–429. doi: 10.1124/jpet.113.203786. The first report describing the dimerization of the AhR with KLF6 and stimulating gen expression via a novel DNA binding sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Jackson DP, Joshi AD, Elferink CJ. Ah Receptor Pathway Intricacies; Signaling Through Diverse Protein Partners and DNA-Motifs. Toxicol Res (Camb) 2015;4:1143–1158. doi: 10.1039/c4tx00236a. Comprehensive review of classical and novel noncannonical AhR signaling pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruegg J, Swedenborg E, Wahlstrom D, Escande A, Balaguer P, Pettersson K, Pongratz I. The transcription factor aryl hydrocarbon receptor nuclear translocator functions as an estrogen receptor beta-selective coactivator, and its recruitment to alternative pathways mediates antiestrogenic effects of dioxin. Mol Endocrinol. 2008;22:304–316. doi: 10.1210/me.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watabe Y, Nazuka N, Tezuka M, Shimba S. Aryl hydrocarbon receptor functions as a potent coactivator of E2F1-dependent trascription activity. Biol Pharm Bull. 2010;33:389–397. doi: 10.1248/bpb.33.389. [DOI] [PubMed] [Google Scholar]
- 31•.Murray IA, Morales JL, Flaveny CA, Dinatale BC, Chiaro C, Gowdahalli K, Amin S, Perdew GH. Evidence for ligand-mediated selective modulation of aryl hydrocarbon receptor activity. Mol Pharmacol. 2010;77:247–254. doi: 10.1124/mol.109.061788. This paper describes a novel mechanism by which activation of the AhR by a selective AhR modulator (SAhRM) can repress gene expression in a non-DRE-dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray IA, Flaveny CA, Chiaro CR, Sharma AK, Tanos RS, Schroeder JC, Amin SG, Bisson WH, Kolluri SK, Perdew GH. Suppression of cytokine- mediated complement factor gene expression through selective activation of the Ah receptor with 3′,4′-dimethoxy-α-naphthoflavone. Mol Pharmacol. 2011;79:508–19. doi: 10.1124/mol.110.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denison MS, Seidel SD, Rogers WJ, Ziccardi M, Winter GM, Heath-Pagliuso S. Natural and synthetic ligands for the Ah receptor. In: Puga A, Wallace KB, editors. Molecular Biology Approaches to Toxicology, Taylor and Francis, Pennsylvania. 1998. pp. 393–410. [Google Scholar]
- 34••.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. This paper provides an overview of the structurally diversity of exopgenous and endogenous AhR ligands. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence BP, Denison MS, Novak H, Vorderstrasse BA, Harrer N, Neruda W, Reichel C, Woisetschlager M. Activation of the aryl hydrocarbon receptor is essential for mediating the anti-inflammatory effects of a novel low-molecular-weight compound. Blood. 2008;112:1158–1165. doi: 10.1182/blood-2007-08-109645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeGroot DE, He G, Fraccalvieri D, Bonati L, Pandini L, Denison MS. AhR ligands: promiscuity in binding and diversity in response. In: Pohjanvirta R, editor. The AH Receptor in Biology and Toxicology. Wiley Hoboken; New Jersey: 2011. pp. 63–79. [Google Scholar]
- 38.Stejskalova L, Dvorak Z, Pavek P. Endogenous and exogenous ligands of aryl hydrocarbon receptor: current state of art. Curr Drug Metab. 2011;12:198–212. doi: 10.2174/138920011795016818. [DOI] [PubMed] [Google Scholar]
- 39••.Pandini A, Denison MS, Song Y, Soshilov AA, Bonati L. Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochem. 2007;46:696–708. doi: 10.1021/bi061460t. The paper describes the first homology model of the AhR LBD based on the template structures of related bHLH-PAS proteins and validated aspects of the model by site-directed mutagenesis and AhR functional analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Pandini A, Soshilov AA, Song Y, Zhao J, Bonati L, Denison MS. Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochem. 2009;48:5972–5983. doi: 10.1021/bi900259z. This paper was the first to identify the amino acid residues in the AhR LBD responsible for binding TCDD with high affinity using amino acid conservation from AhR sequence alignment and functional confirmation using site-directed mutagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraccalvieri D, Soshilov AA, Karchner SI, Franks DG, Pandini A, Bonati L, Hahn ME, Denison MS. Comparative analysis of homology models of the AH receptor ligand binding domain: verification of structure-function predictions by site-directed mutagenesis of a nonfunctional receptor. Biochem. 2013;52:714–725. doi: 10.1021/bi301457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, Schultz PG, Cooke MP. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science (New York, NY) 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whelan F, Hao N, Furness SG, Whitelaw ML, Chapman-Smith A. Amino acid substitutions in the aryl hydrocarbon receptor ligand binding domain reveal YH439 as an atypical AhR activator. Mole Pharmacol. 2010;77:1037–1046. doi: 10.1124/mol.109.062927. [DOI] [PubMed] [Google Scholar]
- 44.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiizaki K, Ohsako S, Kawanishi M, Yagi T. Identification of amino acid residues in the ligand-binding domain of the aryl hydrocarbon receptor causing the species-specific response to omeprazole: possible determinants for binding putative endogenous ligands. Mol Pharmacol. 2014;85:279–289. doi: 10.1124/mol.113.088856. [DOI] [PubMed] [Google Scholar]
- 46••.Soshilov AA, Denison MS. Ligand promiscuity of aryl hydrocarbon receptor agonists and antagonists revealed by site-directed mutagenesis. Mol Cell Biol. 2014;34:1707–1719. doi: 10.1128/MCB.01183-13. This paper was the first to comprehensively analyze the ability of selective amino acid substitutions within the AhR LBD to differentially affect the ability of structurally diverse ligands to bind to the AhR and stimulate AhR DNA binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science (New York, NY) 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 48.Watkins RE, Noble SM, Redinbo MR. Structural insights into the promiscuity and function of the human pregnane X receptor. Curr Opin Drug Discov Devel. 2002;5:150–158. [PubMed] [Google Scholar]
- 49.Wu B, Li S, Dong D. 3D structures and ligand specificities of nuclear xenobiotic receptors CAR, PXR and VDR. Drug Discov Today. 2013;18:574–581. doi: 10.1016/j.drudis.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Denison MS. Summary of probe development efforts to identify activators of the Aryl Hydrocarbon Receptor (AHR), PubChem BioAssay Record for AID 602173, NCBI. 2010 https://pubchem.ncbi.nlm.nih.gov/bioassay/602173#section=Same-Project-BioAssays.
- 51.Wincent E, Bengtsson J, Mohammadi Bardbori A, Alsberg T, Luecke S, Rannug U, Rannug A. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc Natl Acad Sci (USA) 2012;109:4479–4484. doi: 10.1073/pnas.1118467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohammadi-Bardori A, Bengtsson J, Rannug U, Rannug A, Wincent E. Quercetin, resveritrol and curcumin are indirect activators of the aryl hydrocarbon receptor (AHR) Chem Res Toxicol. 2012;25:1878–1884. doi: 10.1021/tx300169e. [DOI] [PubMed] [Google Scholar]
- 53••.DeGroot DE, Denison MS. Nucleotide specificity of DNA binding of the aryl hydrocarbon receptor:ARNT complex is unaffected by ligand structure. Toxicol Sci. 2014;137:102–113. doi: 10.1093/toxsci/kft234. This paper demonstrated for the first time that the binding to and activation of the AhR by structurally diverse ligands does not alter the nucleotide specificity of AhR DNA binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, Laine J, Sakai T, Korsmeyer SJ, Casper RF, Sherr DH, Tilly JL. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- 55.Gouedard C, Barouki R, Morel Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol Cell Biol. 2004;24:5209–5222. doi: 10.1128/MCB.24.12.5209-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harper TA, Jr, Joshi AD, Elferink CJ. Identification of stanniocalcin 2 as a novel aryl hydrocarbon receptor target gene. J Pharmacol Exper Therapeutics. 2013;344:579–588. doi: 10.1124/jpet.112.201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guyot E, Chevallier A, Barouki R, Coumoul X. The AhR twist: ligand-dependent AhR signaling and pharmaco-toxicological implications. Drug Discov Today. 2013;18:479–486. doi: 10.1016/j.drudis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 58.DeGroot DE, Hayashi A, Denison MS. Lack of ligand-selective binding of the aryl hydrocarbon receptor to putative DNA binding sites regulating expression of Bax and paraoxonase 1 genes. Arch Biochem Biophys. 2014;541:13–20. doi: 10.1016/j.abb.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paige LA, Christensen DJ, Gron H, Norris JD, Gottlin EB, Padilla KM, Chang CY, Ballas LM, Hamilton PT, McDonnell DP, Fowlkes DM. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc Natl Acad Sci (USA) 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar R, McEwan IJ. Allosteric modulators of steroid hormone receptors: structural dynamics and gene regulation. Endocr Rev. 2012;33:271–299. doi: 10.1210/er.2011-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aarts JM, Wang S, Houtman R, van Beuningen RM, Westerink WM, Van De Waart BJ, Rietjens IM, Bovee TF. Robust array-based coregulator binding assay predicting ERalpha-agonist potency and generating binding profiles reflecting ligand structure. Chem Res Toxicol. 2013;26:336–346. doi: 10.1021/tx300463b. [DOI] [PubMed] [Google Scholar]
- 62.Wang S, Houtman R, Melchers D, Aarts J, Peijnenburg A, van Beuningen R, Rietjens I, Bovee TF. A 155-plex high-throughput in vitro coregulator binding assay for (anti-)estrogenicity testing evaluated with 23 reference compounds. ALTEX. 2013;30:145–157. doi: 10.14573/altex.2013.2.145. [DOI] [PubMed] [Google Scholar]
- 63••.Zhang S, Rowlands C, Safe S. Ligand-dependent interactions of the Ah receptor with coactivators in a mammalian two-hybrid assay. Toxicol Appl Pharmacol. 2008;227:196–206. doi: 10.1016/j.taap.2007.10.019. This paper reported for the first time that AhR bound by different ligands could recruit different coactivator proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. J Biol Chem. 2008;283:32995–33005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.Joshi AD, Carter DE, Harper TA, Jr, Elferink CJ. Aryl hydrocarbon receptor-dependent stanniocalcin 2 induction by cinnabarinic acid provides cytoprotection against endoplasmic reticulum and oxidative stress. J Pharmacol Exper Ther. 2015;353:201–212. doi: 10.1124/jpet.114.222265. This paper demonstrated for the first time differential DNA binding and DRE-dependent gene expression by two different AhR ligands/agonists (TCDD and cinnabarinic acid), suggesting ligand-selective differences in AhR structure/function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lowe MM, Mold JE, Kanwar B, Huang Y, Louie A, Pollastri MP, Wang C, Patel G, Franks DG, Schlezinger J, Sherr DH, Silverstone AE, Hahn ME, McCune JM. Identification of cinnabarinic acid as a novel endogenous aryl hydrocarbon receptor ligand that drives IL-22 production. PloS one. 2014;9:e87877. doi: 10.1371/journal.pone.0087877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradshaw TD, Bell DR. Relevance of the aryl hydrocarbon receptor (AhR) for clinical toxicology. Clin Toxicol (Phila) 2009;47:632–642. doi: 10.1080/15563650903140423. [DOI] [PubMed] [Google Scholar]
- 68.Francis JE, Smith AG. Polycyclic aromatic hydrocarbons cause hepatic porphyria in iron-loaded C57BL/10 mice: comparison of uroporphyrinogen decarboxylase inhibition with induction of alkoxyphenoxazone dealkylations. Biochem Biophys Res Commun. 1987;146:13–20. doi: 10.1016/0006-291x(87)90683-8. [DOI] [PubMed] [Google Scholar]
- 69.Karras JG, Morris DL, Matulka RA, Kramer CM, Holsapple MP. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) elevates basal B-cell intracellular calcium concentration and suppresses surface Ig- but not CD40-induced antibody secretion. Toxicol Appl Pharmacol. 1996;137:275–284. doi: 10.1006/taap.1996.0081. [DOI] [PubMed] [Google Scholar]
- 70•.Puga A, Hoffer A, Zhou S, Bohm JM, Leikauf GD, Shertzer HG. Sustained increase in intracellular free calcium and activation of cyclooxygenase-2 expression in mouse hepatoma cells treated with dioxin. Biochem Pharmacol. 1997;54:1287–1296. doi: 10.1016/s0006-2952(97)00417-6. This paper was one of the earliest that suggested that TCDD could produce AhR-independent responses in cells. [DOI] [PubMed] [Google Scholar]
- 71.Park SJ, Yoon WK, Kim HJ, Son HY, Cho SW, Jeong KS, Kim TH, Kim SH, Kim SR, Ryu SY. 2,3,7,8-Tetrachlorodibenzo-p-dioxin activates ERK and p38 mitogen-activated protein kinases in RAW 264.7 cells. Anticancer Res. 2005;25:2831–2836. [PubMed] [Google Scholar]
- 72.Kobayashi D, Ahmed S, Ishida M, Kasai S, Kikuchi H. Calcium/calmodulin signaling elicits release of cytochrome c during 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced apoptosis in the human lymphoblastic T-cell line, L-MAT. Toxicol. 2009;258:25–32. doi: 10.1016/j.tox.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 73.Yoshioka H, Hiromori Y, Aoki A, Kimura T, Fujii-Kuriyama Y, Nagase H, Nakanishi T. Possible aryl hydrocarbon receptor-independent pathway of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced antiproliferative response in human breast cancer cells. Toxicol Lett. 2012;211:257–265. doi: 10.1016/j.toxlet.2012.04.005. [DOI] [PubMed] [Google Scholar]


