Abstract
Amplification of a DNA target by the polymerase chain reaction (PCR) often requires laborious optimization efforts. In this regard, the use of certain organic chemicals such as dimethyl sulfoxide, polyethylene glycol, betaine and formamide as cosolvents has been found to be very helpful. Unfortunately, very little is known about the precise structural features that make these additives effective and, accordingly, the number of such chemicals currently known to enhance PCR is limited. In order to address these issues, we decided to focus on formamide and undertook an extensive study of low molecular weight amides as a class to see how changing the substituents in the amide structure influences its effect on PCR. We describe here the results of this study, which involved 11 different amides, and present observations that provide a cohesive picture of structure–activity relations in this group of additives. We found several of these amides to be exceptionally effective and introduce them as novel PCR enhancers.
INTRODUCTION
Since it was first described in 1988 (1), the polymerase chain reaction (PCR) has rapidly become one of the central techniques of molecular biology. Despite its widespread application; however, the technique is often fraught with difficulties. In many cases, the standard procedure fails to produce meaningful amplification or any amplification at all. In others, the amplification of the target gene is non-specific, meaning that its amplification is accompanied by similar amplification of non-target DNA fragments. Accordingly, improvement of amplification and specificity has been the focus of a number of studies (2–4). It has been found that various organic additives can often yield significant improvements in this regard, the most successful of the additives tested being DMSO, glycerol, polyethylene glycol, betaine and formamide (5–9).
Formamide is one of the most widely used additives. It has been particularly noted for its ability to improve specificity in PCR (10). It seemed likely that other amides could improve PCR amplification and specificity. Given the potential of such chemicals to enhance the PCR process, we undertook a comprehensive structure–activity investigation with a group of low molecular weight amides to see how variations in the amide structure influence PCR amplification and specificity.
We report below the results of this investigation, which included the following amides: formamide, N-methylformamide (MMF), N,N-dimethylformamide (DMF), 2-pyrrolidone, N-methylpyrrolidone (NMP), N-hydroxyethylpyrrolidone (HEP), acetamide, N-methylacetamide (MMA), N,N-dimethylacetamide (DMA), propionamide and isobutyramide. We identify a number of amides from this group that are novel and potent enhancers of PCR.
MATERIALS AND METHODS
Amplification reactions were carried out under the following conditions: 10 mM Tris–HCl pH 8.8, 50 mM KCl, 1.5 mM MgCl2, 0.01% (w/v) gelatin, 0.2 µM primers, 0.06 ng/µl template, 0.2 mM each dNTP, 0.04 U/µl Taq polymerase. The templates used were bovine brain N-WASP cDNA (1518 bp), a 996 bp segment of human myeloid leukocyte c-jun cDNA, a 511 bp segment of human prostate-specific membrane antigen (PSM) cDNA, and bovine brain glycolipid transfer protein (GTP) cDNA (660 bp). cDNA synthesis was carried out using the First-Strand RT–PCR kit from Stratagene on the respective mRNAs purchased from Clontech. All amplification reactions for each gene were carried out using a single master batch of cDNA. Taq polymerase and dNTPs were obtained from Stratagene. Primers were obtained from Genosys. Primer melting temperatures (Tms) were calculated using the Genosys oligo calculator program. The sequences of the primers were as follows: N-WASP primer n1, d(ATGAGCTCCGGCCAGCAGC), and primer n2, d(TCAGTCTTCCCATTCATCATCATCCTC); c-jun primer j1, d(ATGACTGCAAAGATGGAAACG), and primer j2, d(TCAAAATGTTTGCAACTGCTGCG); PSM primer p1, d(AAACACTGCTGTGGTGGA), and primer p2 d(TAGCTCAACAGAATCCAGGC); GTP primer g1, d(GAATTCGAAATGGCGCTGCTGG), and primer g2, d(CTCGAGGTCCAGAGTACCCGCTGTG). Calculated Tms of the primers were as follows: n1, 72.4°C; n2, 70.7°C; j1, 63.9°C; j2, 70.8°C; p1, 60.3°C; p2, 61.9°C; g1, 73.3°C; g2, 74.4°C.
Additive chemicals were obtained from the following sources. MMF, DMF, acetamide, MMA, DMA and propionamide were purchased from Acros. 2-Pyrrolidone, HEP and NMP were obtained as free samples from BASF Corporation. Isobutyramide was purchased from Aldrich. Formamide was purchased from Gibco BRL.
Polymerase chain reactions for the N-WASP gene were conducted on a Robocycler Gradient 40 thermal cycler from Stratagene using 100 µl solutions in 600 µl thin-walled tubes. For expediency, amplification of c-jun, PSM and GTP was carried out in a Robocycler Gradient 96 thermal cycler from Stratagene using 50 µl solutions in 200 µl thin-walled tubes. Prior to Taq polymerase addition, a hotstart protocol was carried out that consisted of an initial cycle of 95°C for 5 min (11) to ensure complete first-strand separation, followed by a cycle of 54°C for 5 min. Amplifications were run for 30 cycles. Denaturation was done for 1 min at the minimum temperatures that yielded discernable amplification: 92°C for N-WASP and 95°C for c-jun, PSM and GTP. Extension was done at 72°C for periods depending on target length: 2 min for N-WASP, 1.5 min for c-jun and GTP, and 1 min for PSM. Annealings were carried out either at fixed concentrations of additives using temperature gradients of 38–56°C for N-WASP and 44–58°C for the other targets, or at varying concentrations of additives using fixed temperatures of 41°C for N-WASP, 50°C for c-jun and PSM, and 54°C for GTP.
Amplification products were analyzed by agarose gel electrophoresis on 0.8% agarose gels in which 20 µl reaction products were loaded with 4 µl loading buffer. Gels were stained with ethidium bromide, visualized on a UV transilluminator (Fisher), and documented by photography. Densitometric quantitation of amplification products was carried out using an Image Scanner and Image Master software from Amersham Pharmacia Biotech. Background correction was done using the software’s rolling disc method.
RESULTS
The bovine brain N-WASP gene (1518 bp, 49% GC) was selected as the primary subject of this study because it offered a good opportunity to differentiate structure–activity correlations of additives. Under the conditions used, it failed to amplify to any meaningful extent in the absence of additives and in the presence of DMSO, one of the most widely used additives, PCR resulted in rather low amplification and poor specificity (Fig. 1). Multiple low molecular weight bands appeared in the latter case. Various concentrations of DMSO were tested using an annealing temperature gradient of 38–56°C; 5% concentration gave the best results, which were nonetheless unsatisfactory (Fig. 1).
Figure 1.
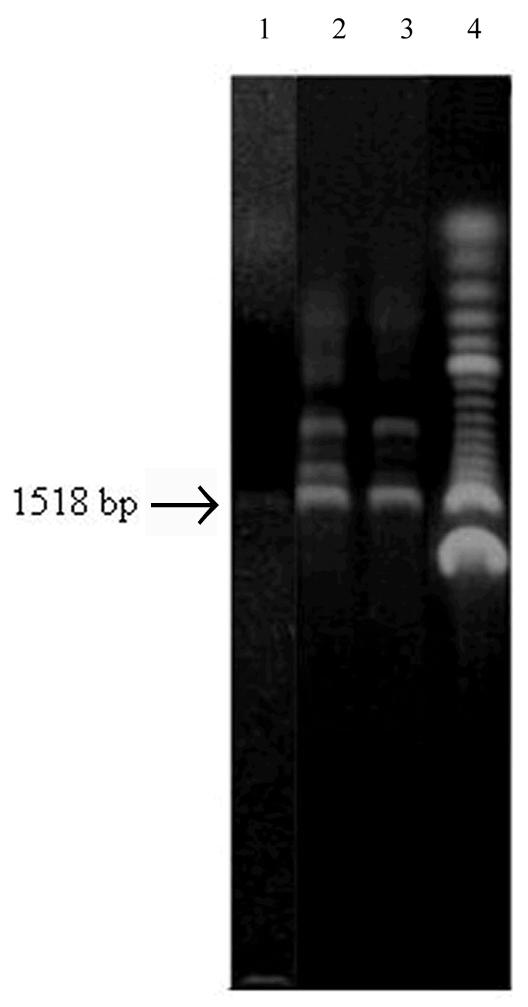
Best results obtained in the amplification of N-WASP cDNA (1518 bp) in the absence of additives and in the presence of DMSO. Lane 1, control (no additive), 56°C annealing temperature; lane 2, 5% DMSO, 54°C annealing temperature; lane 3, 5% DMSO, 58°C annealing temperature; lane 4, 100 bp DNA ladder (Gibco).
The additives tested can be divided into four subclasses. These are the pyrrolidones, the formamides, the acetamides and the higher-chain primary amides. Figure 2 lists the additives by class and provides their structures and abbreviated nomenclatures.
Figure 2.
Structures of the additives tested. Abbreviations used in this paper are indicated in parentheses.
For the N-WASP target, additives were initially tested at a few disparate concentrations using an annealing temperature gradient of 38–56°C. The result was that for all the additives tested the optimal annealing temperature (yield and specificity) was 41°C; concentration did not influence the optimal annealing temperature. The relatively low value (41°C) for the optimal annealing temperature is consistent with the findings that both formamide and NMP lower template melting temperature (12). Additives were then run at a spectrum of concentrations using an annealing temperature of 41°C, and the results in each case compared. The standard concentrations tested were: 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 and 5.0% and 1 M. In the cases of formamide, 2-pyrrolidone and HEP, additional concentrations were tested: 3.5, 4.0 and 4.5% for formamide and 6.0, 7.0 and 7.5% for 2-pyrrolidone and HEP (see below). MMA and isobutyramide were only tested at regular incremental concentrations up to 3.0%, and then at 1 M.
Before presenting the results, it is necessary to define precisely two terms that are central to our investigation. The potency of an additive is defined as the maximum densitometric volume of target band amplification observed at any concentration of that additive. Potencies of the additives were normalized to that of formamide, which was assigned a value of 1. The specificity of an additive is inversely related to the volume of secondary undesirable bands, and is quantitatively defined as the ratio of target band volume to the total volume of all bands, expressed as a percent. With the additives we tested, the highest specificity invariably occurred at a concentration where the volume of the target band was also at its maximum. The data in its entirety (results of PCR amplification for each additive at each concentration tested) is too extensive to present here. Potencies and best specificities of the additives are tabulated in Table 1.
Table 1. Potency, specificity and effective range of additives (N-WASP).
| Additive | Potencya | Specificity | Effective range (molar) | |
| |
|
(%)b |
Leveling-off |
Cutoff |
| 2-Pyrrolidone | 1.18 | 86 | 0.12 | 0.88 |
| HEP | 0.98 | 79 | 0.08 | >0.58, <1.0 |
| NMP | 1.10 | 81 | 0.15 | >0.51, <1.0 |
| Formamide | 1.00 | 87 | 0.67 | 0.89 |
| MMF | 1.07 | 79 | 0.51 | >0.84, <1.0 |
| DMF | 0.98 | 77 | 0.41 | >0.55, <1.0 |
| Acetamide | 1.07 | 98 | 0.25 | >0.84, <1.0 |
| MMA | 0.81 | 85 | 0.27 | >0.41, <1.0 |
| DMA | 0.88 | 80 | 0.29 | >0.57, <1.0 |
| Propionamide | 1.13 | 82 | 0.21 | >0.68, <1.0 |
| Isobutyramide | 1.10 | 85 | 0.23 | >0.35, <1.0 |
| Control | negligible | NA | NA | NA |
aNormalized densitometric volume of target band averaged over effective range: formamide = 1.
bRepresents best specificity (densitometric volume of target as percent of total volume) over the effective range.
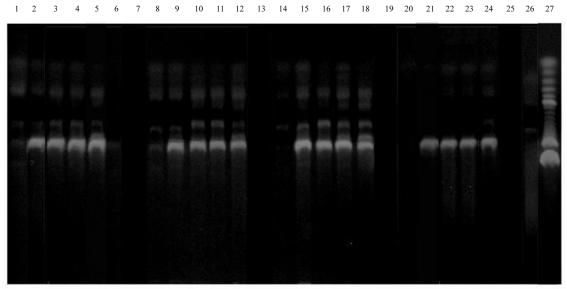
Analysis of the entire body of data generated with N-WASP reveals a number of interesting common features shared by all the additives. The first is that in the case of each additive there exists a range of concentrations over which the amplification is maximal. We term this phenomenon leveling-off; after a certain concentration (up to a point, as we will see below) the amplification levels off at its maximum value. Quantitation of the densitometric volumes of target band amplification over the leveled-off range showed that the volumes fall within ±5% of the average values (data not shown). The leveling-off concentration for each additive corresponds to the concentration of highest specificity, and hence to its optimal concentration, except for MMA which showed optimal performance at a concentration higher than its leveling-off concentration (Fig. 3). Leveling-off data for the various additives are listed in Table 1. A second characteristic shared by all of the additives is that specificity decreases with increasing concentration. The third common feature has only begun to be investigated at this point. We refer to this characteristic as cutoff. For the N-WASP gene, all the additives display complete absence of amplification at 1 M concentration. For formamide and 2-pyrrolidone, the location of cutoff was precisely determined and found to correspond to ∼0.9 M concentration in each case. Formamide exhibited drastically lower amplification at 5.0%, and was subsequently tested at 3.5, 4.0 and 4.5%, whereupon cutoff was observed at 4.0% (0.89 M). 2-Pyrrolidone was tested at 6.0, 7.0 and 7.5% concentration, and cutoff was observed at 7.5% (0.88 M). In the cases of the other additives, cutoff occurs somewhere between 5.0% and 1 M (between 3.0% and 1 M in the cases of MMA and isobutyramide, 7.5% and 1 M in the case of HEP). Cutoff data for the various additives are shown in Table 1. The concentration 0.9 M falls within the cutoff interval for each of the additives, and in the cases of acetamide and MMF the intervals are sufficiently narrow to establish the cutoff at ∼0.9 M. The data thus appear to suggest that, in the case of N-WASP, cutoff occurs universally for low molecular weight amides at ∼0.9 M. Figure 3 displays the above three phenomena in the particular cases of 2-pyrrolidone, HEP, NMP and acetamide.
Figure 3.
Variation in amplification of N-WASP cDNA (1518 bp) with concentration of 2-pyrrolidone, HEP, NMP and acetamide. Lanes 1–6, 2-pyrrolidone 0.5, 1.0, 2.5, 3.0, 5.0 and 7.5%; lane 7, spacer; lanes 8–12, HEP 0.5, 1.0, 2.5, 3.0 and 5.0%; lane 13, spacer; lanes 14–18, NMP 0.5, 1.5, 2.5, 3.0 and 5.0%; lane 19, spacer; lanes 20–24, acetamide 0.5, 1.5, 2.5, 3.0 and 5.0%; lane 25, spacer; lane 26, control (no additive); lane 27, 100 bp DNA ladder (Gibco).
In addition to N-WASP, three other targets—a 996 bp segment of the human c-jun gene, a 511 bp segment of the human PSM gene, and the bovine brain GTP gene (660 bp)—were studied to a limited extent to generalize our findings. These targets were selected because they were especially difficult to amplify in the absence of additives, due to their particularly high GC contents: c-jun, 64% GC; PSM, 52% GC with a 73% GC 158 bp region; GTP, 58% GC (13). We chose 2-pyrrolidone and acetamide, the two additives that performed best in the case of N-WASP, and the standard additive formamide for these studies. Even in the presence of the additives, these targets could not be amplified at denaturing temperatures <95°C. Accordingly, denaturing was carried out at 95 rather than 92°C for each. As in the case of N-WASP, the targets were initially tested at a few concentrations over an annealing temperature gradient (44–58°C), and then at varying concentrations at their optimal annealing temperatures (50°C for c-jun and PSM, 54°C for GTP). Densitometric data are collected in Table 2. The best results in the case of c-jun are displayed in Figure 4. The study of these targets also displayed the phenomena of leveling-off and cutoff concentrations for the additives tested (data not shown).
Table 2. Potency and specificity of select additives (GTP, PSM, c-jun)a,b.
| Additive | GTP | PSM | c-jun | |||
| |
Potency |
Specificity (%) |
Potency |
Specificity (%) |
Potency |
Specificity (%) |
| Control | 0.38 | 20 | 0.88 | 29 | 0 | NA |
| Formamide | 1.00 | 100 | 1.00 | 83 | 1.00 | 100 |
| 2-Pyrrolidone | 1.37 | 100 | 2.62 | 83 | 2.90 | 100 |
| Acetamide | 0.53 | 100 | 0.95 | 66 | 0.99 | 100 |
aDetermined at optimal concentrations of additives: formamide, 0.6 M for PSM and 0.8 M for GTP and c-jun; 2-pyrrolidone, 0.4 M for all targets; acetamide, 0.8 M for all targets.
bSpecificity = densitometric volume of target band as percent of total volume.
Figure 4.
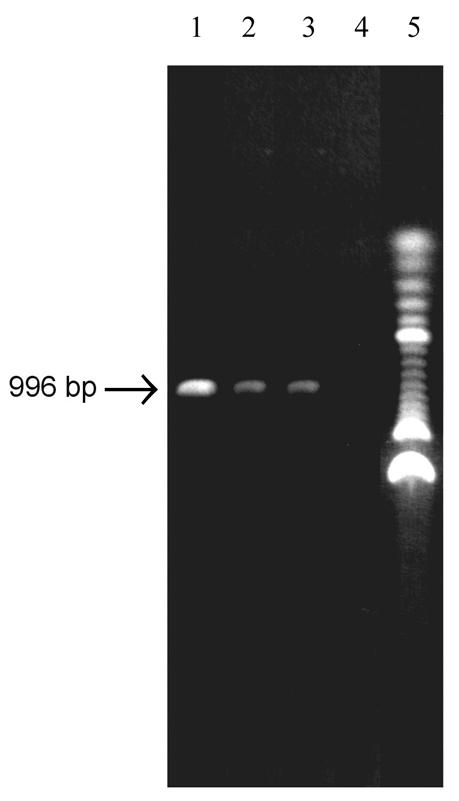
Enhancement of PCR amplification of c-jun (996 bp segment) cDNA by 2-pyrrolidone, formamide and acetamide. Lane 1, 0.4 M 2-pyrrolidone; lane 2, 0.8 M formamide; lane 3, 0.8 M acetamide; lane 4, control (no additive); lane 5, 100 bp DNA ladder (Gibco).
DISCUSSION
The most important outcome of this study is the identification of novel PCR additives that have the potential to function as well as or better than the current state of the art. There are three factors that make a particular additive desirable: high potency, high specificity and a wide effective range, or window of applicability. A wide window of applicability increases the likelihood of identifying an effective additive during initial screenings, and also increases the probability of an additive yielding positive results for a variety of amplicons. The effective ranges of the various additives for the N-WASP gene are shown in Table 1. The data, as discussed, suggest a cutoff of 0.9 M for each of the compounds.
Based on the above criteria, we found 2-pyrrolidone to be the most effective additive in the case of N-WASP. It gives the highest level of amplification of the target band and one of the highest specificities, and has the widest range of all the additives tested. After 2-pyrrolidone, acetamide is the most effective. It gives good amplification of the target band and the highest specificity, and has an effective range near the top of the list. HEP and NMP, the two other pyrrolidones, behave similarly to 2-pyrrolidone with respect to effective range, but their potencies and specificities are compromised to a certain extent by N-substitution. We therefore place them third. Propionamide also gives a high level of amplification of the target band and moderate specificity, but displays a slightly inferior range and thus we assign it to the fourth level of effectiveness. DMA gives modest amplification of the target band and moderate specificity, lower than acetamide as a result of N-substitution, and exhibits an effective range somewhat inferior to that of propionamide. We therefore assign it to the fifth level of effectiveness. The formamides are placed last on the list because of their inferior effective ranges. They begin functioning at significantly higher concentrations than the other compounds. Formamide, the current state of the art, displayed in these experiments the narrowest effective range of all the additives tested. Thus, while it exhibits a high specificity, its limited effective range renders it overall a less desirable additive. MMF and DMF also fall short in terms of effective range, and once again, as in the case of the pyrrolidone and acetamide series, N-substitution seems to reduce specificity. We withhold judgment on MMA and isobutyramide because the data regarding their effective ranges are limited.
As mentioned previously, acetamide and 2-pyrrolidone, the two additives that performed best with N-WASP, were chosen along with the standard additive formamide for further studies with three additional targets, GTP, PSM and c-jun, which are especially difficult to amplify. As can be seen from Table 2, all three compounds exhibit significant enhancement of potency and specificity. However, with these high-GC targets, 2-pyrrolidone clearly stands out with respect to potency, being 1.4 to 2.9 times as effective as formamide. It is also effective at the lowest concentration of the additives tested, as in the case of N-WASP. Another interesting feature of these high-GC targets is that acetamide, which stood a close second in rank in the N-WASP study, now trails significantly behind 2-pyrrolidone and even formamide. Based on the collective results of the N-WASP and high-GC target studies, we identify 2-pyrrolidone as the most effective low molecular weight amide in the enhancement of PCR amplification and specificity.
It has been known for some time that organic solvents that enhance PCR amplification can also unwind the DNA double-helix (14). It has accordingly been proposed by a number of investigators that organic additives such as formamide exert their effect in PCR by binding in the major and minor grooves of DNA and destabilizing the template double-helix (15,16 and references therein). The superior overall performance of 2-pyrrolidone compared to formamide and other acyclic amides may, in part, be due to a greater affinity of 2-pyrrolidone for the grooves of the double-stranded template. The cyclic structure of the pyrrolidone ring might constrain the molecule in a conformation that is optimal for complementary hydrogen bonding with donor and acceptor moieties in the major and minor grooves. It may also minimize steric repulsion with neighboring groups on the DNA backbone by restricting free rotation. Detailed understanding of the differing potencies and specificities of the various low-molecular weight amides, and of the phenomena of leveling-off and cutoff, will require further structural and mechanistic studies.
In conclusion, we have shown that low molecular weight amides, irrespective of their structure, are effective in the enhancement of PCR. Differences exist, however, from structure to structure that appear to be at least somewhat sensitive to the nature of the target. Acetamide was found to be very effective in the case of the medium-GC content gene N-WASP, but not so in the case of high-GC content targets. Further work will be necessary to determine whether it consistently yields exemplary results with medium-GC templates. 2-Pyrrolidone was identified as being an especially effective enhancer across a wide variety of amplicons, demonstrating particularly good performance with high-GC targets.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank BASF Corporation for providing free samples of 2-pyrrolidone, HEP and NMP. R.C. also wishes to thank the National Science Foundation for its support in offering a 3 year research fellowship. C.E.S. is supported by a grant from the National Institutes of Health (GM 44038).
References
- 1.Saiki R.K., Gelfand,D.H., Stoffel,S., Scharf,S.J., Higuchi,R., Horn,G.T., Mullis,K.B. and Erlich,H.A. (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–491. [DOI] [PubMed] [Google Scholar]
- 2.Roux K.H. (1995) Optimization and troubleshooting in PCR. In Dieffenbach,C.W. and Dveksler,G.S. (eds), PCR Primer – A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 55–66.
- 3.Newton C.R. and Graham,A. (1994) PCR. Bios Scientific, Oxford.
- 4.Henegariu O., Heerema,N.A., Dlouhy,S.R., Vance,G.H. and Vogt,P.H. (1997) Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques, 23, 504–511. [DOI] [PubMed] [Google Scholar]
- 5.Winship P.R. (1989) An improved method for directly sequencing PCR amplified material using dimethylsulfoxide. Nucleic Acids Res., 17, 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman B., Luke,W. and Hunsmann,G. (1990) Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res., 18, 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomp D. and Madrano,J.F. (1991) Organic solvents as facilitators of polymerase chain reaction. Biotechniques, 10, 58–59. [PubMed] [Google Scholar]
- 8.Smith T.K., Long,C.M., Bowman,B. and Manos,M.M. (1990) Using cosolvents to enhance PCR amplification. Amplifications, 5, 16–17. [Google Scholar]
- 9.Weissensteiner T. and Lanchbury,J.S. (1996) Strategy for controlling preferential amplification and avoiding false negatives in PCR typing reactions. Biotechniques, 21, 1102–1108. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar G., Kapelner,S. and Sommer,S.S. (1990) Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res., 18, 7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivinson A.J. and Taylor,G.R. (1991) PCR in genetic diagnosis. In McPherson,M.J., Quirke,P. and Taylor,G.R. (eds), PCR: A Practical Approach. Oxford University Press, New York, NY, p. 19.
- 12.Landre P.A., Gelfand,D.H. and Watson,R.M. (1995) The use of cosolvents to enhance amplification by the polymerase chain reaction. In Gelfand,D.H., Innis,M.A., Sninsky,J.J. and Sninsky,J.I. (eds), PCR Strategies. Academic Press, New York, NY, p. 3–16.
- 13.Henke W., Herdel,K., Jung,J., Schnorr,D. and Loenig,S.A. (1997) Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res., 19, 3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C.-H., Mizusawa,H. and Kakefuda,T. (1981) Unwinding of double-stranded DNA helix by dehydration. Proc. Natl Acad. Sci. USA, 78, 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varadaraj K. and Skinner,D.M. (1994) Denaturants or cosolvents improve the specificity of PCR amplification of a G+C rich DNA using genetically engineered DNA polymerases. Gene, 140, 1–5. [DOI] [PubMed] [Google Scholar]
- 16.Cheng S., Fockler,C., Barnes,W.M. and Higuchi,R. (1994) Effective amplification of long targets from cloned inserts and human genomic DNA. Proc. Natl Acad. Sci. USA, 91, 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]