Abstract
Citrullus colocynthis (L.) Schrad. (Cucurbitaceae) shows high levels of variation in fruit color, fruit stripe pattern, seed coat color, and size. Thirty-eight accessions of C. colocynthis plants from different parts of semi-arid Rajasthan were collected and genetic diversity was assessed using random-amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. Out of 65 RAPD decamer primers, 50 primers produced 549 scorable bands of which 318 were polymorphic. Polymorphic banding patterns with the number of amplified fragments varied from 5 (OPA-08 and OPF-9) to 19 (OPT-20) in the molecular size range of 150–6000 bp. Percent polymorphism ranged from 22.2% (OPA-09) to 83.3% (OPE-12) with 55.14% polymorphism. Out of the 20 ISSR primers screened, 13 primers produced 166 amplification products, of which 99 were polymorphic. The number of bands amplified per primer varied between 9 (UBC-807, 802) and 16 (UBC-803, 812) with average band size between 250 and 4000 bp. Percent polymorphism ranged from 45.4% (UBC-815) to 73.3% (UBC-814) with 65.05% polymorphism. Dendrogram constructed on the basis of RAPD + ISSR polymorphism separated the accessions into four distinct clusters at 72% variation with Jaccard’s similarity coefficient ranging from minimum 0.64 to 0.95. The matrices for RAPD and ISSR were also compared using Mantel’s test and obtained correlation value (r = 0.7947). Discriminating power of RAPD and ISSR markers was assessed by calculating polymorphic information content, multiplex ratio, marker index, and resolving power. Approx. 50% RAPD and ISSR markers showed PIC value and heterozygosity (H) ≥ 0.50, indicating marker as informative. The primers that showed higher polymorphism had higher RP, MR, and MI values.
Keywords: ISSR, Polymorphism, Citrullus colocynthis, RAPD, Genetic diversity, Similarity matrix, Heterozygosity (H), Polymorphic information content (PIC), Resolution power (RP), Multiplex ratio (MR), Marker index (MI)
Introduction
Genetic variation and diversity are a prerequisite for the short- and long-term survival of the plant species in their natural habitat. The evaluation of genetic diversity of any species is very important for their conservation and gene manipulation (Khan et al. 2012).
Cucurbits are among the largest and most diverse plant families, cultivated worldwide in diverse environmental conditions, and have paid consideration globally because the fruits, seeds, and vegetables are traditionally, economically, and medicinally consumed in various ways. C. colocynthis (L.) Schrad. species belongs to genus Citrullus, family Cucurbitaceae. It is also known as “bitter melon”, “bitter gourd”, or “tumba” related to watermelon and is a creeping herb, small scarbid, non-hardy drought resistant, and perennial with prostate or climbing stem, bearing smooth spherical green and yellow-ripe fruits (Shah and Qadry 1985). The pharmacological properties of C. colocynthis are attributed to fruits. The white flesh of the fruit is associated with bitterness caused by Cucurbitacin E, glycoside, or colocynthine (Novot et al. 1990). It is more prominently used in anti-tumorous and antidiabetic drugs as it contains a mixture of Cucurbitacins (A, B, C, D, E, I, J, K, and L). Beside this, several important bioactive primary and secondary metabolites of this plant have been reported which attributed medicinal importance to C. colocynthis (Sturm et al. 2009). It is a drought-resistant desert plant with greater survival rate under severe stress and drought conditions (Dane et al. 2006). The plant is native to dry areas of North Africa, being common throughout the Sahara, areas of Morocco, Egypt and Sudan, eastward through Iran to India, Pakistan, and Afghanistan (Bhandari 1995).
C. colocynthis is the vegetation of desert and sand dunes and distributed mainly in desert areas of Rajasthan viz. Jaisalmer, Barmer, Bikaner, and Jodhpur, but it can also be seen excessively at Jaipur, Jhunjhunu, Churu, Ganganagar, Alwar, Hanumaangarh, and Udaipur in Rajasthan. The sand dunes vegetation is totally a different plant community with remarkable ability to tolerate a hostile environment of drought, nutrient deficiency, high winds, high temperatures, salts sprays, and sand blast.
C. colocynthis show high levels of variations within the species. The variations are more prominent in fruit color, fruit stripe pattern, seed coat color, and seed size. The fundamental source of biodiversity is genetic diversity—the total number of genetic characters contributing to variation within species. Genetic diversity described by the presence of various alleles in the gene pool and different genotypes within populations. Genetic variability on the other hand describes the tendency of the genetic traits within populations to vary (Laikre et al. 2009). Molecular phylogenetic and genetic diversity analysis can explain the taxonomic identity and evolutionary relationships amongst the wild species. Variability and genetic diversity are important factors in evolution and determine the response of a given organism to environmental stress, natural selection, and susceptibility to different diseases (Jain 2002). Finding and analysis of genetic diversity can help in understanding the molecular basis of various biological phenomena. The availability of different molecular markers and increased computing power has helped in the development of sophisticated techniques that have helped in understanding the various biological phenomena (Csillery et al. 2010).
Advances in the field of molecular biology provided many tools for studying genetic diversity at the genome level to investigate phylogenetic relationships among inter or intra-species. The development and use of molecular markers for the detection and exploitation of DNA polymorphism is one of the significant achievements in the field of molecular genetics. Molecular markers are reliable indicators of genetic diversity, because they are neutral to environmental influence and reveal differences at the whole genome level. Amid the several markers, no molecular markers are available that complete all the requirements needed. Different molecular markers used in genetic fingerprinting viz. restriction fragment length polymorphism (RFLP), random-amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), inter-simple sequence repeats (ISSRs), sequence characterized amplified regions (SCARs), sequence-based amplified polymorphism (SBAPs), and sequence tag sites (STSs) (Sharma et al. 2012), have their own advantages and disadvantages.
RAPD and ISSR are PCR-based markers, require only small amounts of DNA sample without involving radioactive labels, and are simpler as well as faster. RAPD has proven to be quite efficient in detecting genetic variations and used for diversity assessment as well as identifying germplasm in several plant species, bacteria, and microorganism (Kapteyn et al. 2002; Moghaieb et al. 2017). RAPDs are very quick and easy to develop due to the arbitrary sequence of the primers. It resolves most of the technical obstacle owing to its cost-effective and easy to perform (Williams et al. 1990). Therefore, RAPDs have been extensively used in assessing genetic relationship among various accessions of different plant species. ISSR has been shown to provide a powerful, rapid, simple, reproducible, and inexpensive means to assess genetic diversity and identify differences between closely related cultivars in many plant species, where the sequence information is limited; ISSR markers are easy to handle quick and more reproducible than other markers (Gonzalez et al. 2000). The nucleotide repeats (inter-simple sequence repeats) are distributed throughout the genome and has potential discriminatory power which has been seen in the clonal plant species for the assessment of genetic diversity. Earlier, these marker systems have been used in investigating genetic relatedness, for resolving intra- and intergenomic relationships, genetic diversity of plant populations, and cultivars (Verma et al. 2013; Khurana-Kaul et al. 2012; Velasco-Ramirez et al. 2014). Several other studies have been successfully effective in population genetics and in detecting genetic diversity in many species using RAPD or ISSR markers or both.
The natural habitat of a plant species determines its genetic diversity which is important for long-term survival and evolution under abiotic and biotic stresses. Diversity is influenced by various geographical factors which leads to development of subspecies, races, or ecotypes (Sreekumar and Renuka 2006). Genetic variation is required for the short- and long-term survival of plant species in the wild conditions. Identification of genetic diversity within and among widespread, restricted, and endangered species is necessary, prior to their conservation (Gitzendanner and Soltis 2000) in situ as well as in vitro. The decline in genetic diversity has resulted from the loss of alleles which may be due to the reduction in population size as genetic drift can induce in genetic variation and thus affects the survival of natural population (Reed and Frankham 2003).
The present study is the first attempt on genetic variation among several accessions of C. colocynthis collected from different locations of Thar desert and other parts of Rajasthan using random-amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. The study was aimed to characterize the morphological and molecular diversity employing both markers.
Materials and methods
Plant material and DNA extraction
To characterize the genetic diversity, a germplasm exploration survey was undertaken to identify C. colocynthis from the different locations of the University of Rajasthan, Jaipur, Agricultural Research Centre Durgapura, Jaipur, Ramgarh Dam, Jaipur, Amber, Alwar, Sikar, Jhunjhunu, Churu, and mainly dry areas of Rajasthan (Jaisalmer, baadmer, Bikaner, and Jodhpur; Table 1). The selection was made on the basis of phenotypic assessment of various characters viz. growth habitats, fruit color, fruit stripe pattern, and seed color.
Table 1.
Details of Citrullus colocynthis germplasm collected from different locations in Rajasthan, India
| S. no. | Accession code | Place | Location | Latitude and longitude | Collection site |
|---|---|---|---|---|---|
| 1 | A1 | Rajgarh | Alwar | 27°14′09N 76°37′20E | Wild |
| 2 | A2 | Bahrod | Alwar | 27°53′12N 76°16′51E | Wild |
| 3 | J1 | University of Rajasthan | Jaipur | 26°53′06N 75°49′11E | Wild |
| 4 | J2 | Durgapura agriculture Inst. | Jaipur | 26°50′22N 75°47′19E | Farm Land |
| 5 | J3 | Sekh Ki Dhani, Ramgarh | Jaipur | 26°37′59N 76°16′00E | Farm land |
| 6 | J4 | Ragarh Dam | Jaipur | 27°02′51N 76°03′22E | Farm land |
| 7 | J5 | Amber | Jaipur | 26°58′59N 75°52′00E | Wild |
| 8 | S1 | Neem Ka thana | Sikar | 27°44′14N 75°47′11E | Farm land |
| 9 | S2 | Rengus | Sikar | 27°21′35N 75°33′59E | Farm land |
| 10 | S3 | Shri Madhopur | Sikar | 27°28′00N 75°35′59E | Wild |
| 11 | Jh1 | Chidawa | Jhunjhunu | 28°14′24N 75°38′44E | Farm Land |
| 12 | Jh2 | Bisaau | Jhunjhunu | 28°14′41N 75°04′32E | Farm land |
| 13 | Jh3 | Surajgarh | Jhunjhunu | 28°18′33N 75°43′57E | Wild |
| 14 | C1 | Ratangarh | Churu | 28°04′47N 74°37′06E | Wild |
| 15 | C2 | Sujaangarh | Churu | 27°41′59N 74°28′00E | Farm land |
| 16 | C3 | Churu outer area | Churu | 28°18′02N 74°58′39E | Wild |
| 17 | H1 | Hanumangarh city | Hanumangarh | 29°37′03N 74°17′20E | Wild |
| 18 | H2 | Bhadra | Hanumangarh | 26°06′06N 75°10′16E | Wild |
| 19 | G1 | Anupgarh | Ganganagar | 29°11′20N 73°12′34E | Farm land |
| 20 | G2 | Jaitsar | Ganganagar | 29°19′27N 73°40′21E | Farm land |
| 21 | G3 | Ganganagar Canal Area | Ganganagar | 30°07′31N 74°06′03E | Wild |
| 22 | B1 | Rasisar | Bikaner | 27°44′56N 73°22′25E | Wild |
| 23 | B2 | Kolayat | Bikaner | 27°50′08N 72°57′09E | Wild |
| 24 | B3 | Nokha | Bikaner | 27°33′52N 73°28′33E | Farm land |
| 25 | B4 | Deshnokh | Bikaner | 27°47′34N 73°20′16E | Farm land |
| 26 | Js1 | Sum | Jaisalmer | 26°54′56N 70°54′30E | Wild |
| 27 | Js2 | Tanot | Jaisalmer | 27°47′47N 70°21′14E | Wild |
| 28 | Js3 | Pokhran | Jaisalmer | 27°05′41N 71°45′10E | Wild |
| 29 | Js4 | Jaisalmer canal Area | Jaisalmer | 26°54′56N 70°54′30E | Farm land |
| 30 | B1 | Hodu | Baadmer | 25°26′37N 71°45′28E | Farm land |
| 31 | B2 | Baadmer City | Baadmer | 25°45′00N 71°22′59E | Wild |
| 32 | B3 | Balotra | Baadmer | 25°49′51N 72°14′24E | Wild |
| 33 | Jd1 | Falodi | Jodhpur | 27°07′57N 72°21′44E | Wild |
| 34 | Jd2 | kalda | Jodhpur | 26°14′20N 73°01′27E | Wild |
| 35 | Jd3 | Jodhpur outer area | Jodhpur | 26°15′17N 73°00′13E | Wild |
| 36 | U1 | Salumbar | Udaipur | 24°08′05N 74°02′40E | Wild |
| 37 | U2 | Lasadia | Udaipur | 24°34′16N 73°41′29E | Farm land |
| 38 | U3 | Kherwara | Udaipur | 23°59′07N 73°35′46E | Wild |
A representative set of 38 accessions of C. colocynthis were collected from different regions of Rajasthan in the year 2009–2013 for assessing genetic diversity among them. Total genomic DNA was extracted from young leaves following the standard CTAB method as described by Doyle and Doyle (1990) with minor modifications.
C. colocynthis leaves (5 g) were ground in liquid nitrogen, then homogenized in 25 ml of extraction buffer (2% CTAB, 20 mM EDTA, 2% PVP, 1.4 M NaCl, 100 mM Tris–HCl pH 8.0, and 1% β-mercaptoethanol) add 4 μl of RNAse incubated at 65 °C for 1 h. The supernatant was extracted twice with chloroform: isoamylalcohol (24:1 v/v). RNase A was again added and incubated at room temperature for 30 min. The DNA was pelleted with chilled isopropanol and washed twice with 70% ethanol. The washed pellet DNA was air-dried, dissolved in 500 μL TE buffer, and stored at −20 °C. DNA concentration was estimated by agarose (Himedia, India) gel electrophoresis using a known amount of λ DNA as standard and nanodrop (ND1000, Thermo Scientific, US) spectrophotometer.
RAPD amplification
A total of 65 random primers (Operon Technologies Inc., USA) were initially screened to assess genetic diversity among 38 accessions of C. colocynthis. The composition of the PCR reaction mixture in 20 µL of RAPD reaction volume contains 2.5 µl DNA (25 ng), 2 µl Taq Buffer A (1×), 0.5 µl dNTP mix (100 µM each), 2 µl Primer (0.4 µM), 0.25 µl Taq Polymerase (0.25 U µl−1), and Milli-Q water to complete the reaction. PCR reactions were performed at an initial denaturation at 94 °C for 5 min followed by 40 cycles at 94 °C for 1 min denaturation, annealing at 37 °C for 45 s. and 2 min extension at 72 °C with a final extension of 10 min at 72 °C using a DNA Engine (Bio-Rad, Germany). Amplified products were separated on 1.2% agarose (Himedia, India) gel in 1× TAE buffer by electrophoresis at 100 V, visualized with EtBr staining and photographed using Gel Documentation System (Bio-Rad, USA). The size of the amplification products was determined by comparison using 100 bp and 1 kb ladder (Bangalore Genei, India).
ISSR amplification
In the case of ISSR primers, the optimal annealing temperature was found to vary according to base composition of the primers. 20 ISSR primers (University of British Columbia, primer set no. 9, Vancouver, Canada) were initially screened to assess genetic diversity in 38 accessions of C. colocynthis out of which 13 primers gave amplification. PCR reaction was carried out in 20 μl reaction volume containing 25 ng genomic DNA (2.5 μl) as template, 2.5 μl MgCl2, 2 μl Taq buffer B 0.5 μl of 100 μM dNTP, 2.5 μl of ISSR primers (4 μM), and 0.35 μl of Taq DNA polymerase (Bangalore Genei, India). PCR amplifications were performed with the initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 92 °C for 1 min, 1 min at the annealing temperature (depending on the primer Tm), and 2 min extension at 72 °C with a final extension at 72 °C for 7 min using DNA Engine (Bio-Rad, Germany). The PCR products were separated on 1.5% agarose gel (Himedia, India) using 100 bp and 1 kb markers (Bangalore Genei, India) as the band size standard and photographed in a gel documentation system (Bio-Rad, Germany).
Data analysis
The DNA fingerprint patterns obtained were converted into binary data matrices containing arrays of 0 and 1 s. The RAPD and ISSR bands were scored visually for the presence (1) or absence (0) of bands of various molecular weight sizes. Only polymorphic and reproducible bands were considered for the analysis. Data were analyzed using SIMQUAL route to generate Jaccard’s similarity coefficient using NTSYS-pc version 2.02e22 (Numerical Taxonomy System) (Rohlf 1998). Similarity matrices were utilized to construct dendrograms independently for both the marker systems and on pooled marker data using UPGMA (Unweighted Pair Group Method with Arithmetic Average) algorithm and SAHN clustering (Sneath and Sokal 1973). Finally, a principal coordinate analysis was performed to highlight the resolving power of the ordination. A two- (2D) and three-dimensional (3D) principal component analyses were constructed to provide another means of testing the relationships among accessions using EIGEN program (NTSYS-PC). The robustness of each phenogram was evaluated by a bootstrap analysis of each data set using the computer program WINBOOT (Yap and Nelson 1996). There are two important measures of the quality or informativeness of a polymorphism as a genetic marker: heterozygosity (H) and polymorphic information content (PIC). To simplify the work of molecular studies, we used a useful online tool (http://www.genomics.liv.ac.uk/animal/pic.html) to facilitate the calculation of H and PIC values. This program, PIC Calculator Extra, can calculate these values from manually uploaded allelic frequencies or from a given file containing binary data. Primer banding characteristics such as number of scored bands (NSB), number of polymorphic band (NPB), and percentage of polymorphic bands (PPB) were obtained. Polymorphism information content (PICi) and heterozygosity of a band were calculated according to Liu (1998)
Informativeness of a band (BIi) was calculated as
where p is the proportion of the total genotypes containing a particular band. It is useful in calculating the resolving power (RP), which in turn enabled us to know the ability of a primer to distinguish various genotypes. Then, the resolving power (RP) of each primer was calculated as (Prevost and Wilkinson 1999)
Multiplex ratio was calculated as MR = L/T, where T = total number of primer combinations. The Marker Index (MI) was obtained by multiplying the average heterozygosity by Multiplex Ratio: MI = Hav × MR (Anderson et al. 1993).
Results
The characterization of different accessions selected on the basis of mainly fruit stripe pattern is presented in Fig. 1. The rind of C. colocynthis fruit can be striped or solid colored. The stripe patterns of C. colocynthis can be characterized using different stripe widths (narrow, medium, and wide), stripe colors, and background colors (dark green, medium green, and light green); besides stripe width, the stripe pattern can also have different characteristics. The stripe patterns are two sets of alternating light- and dark-colored stripes on the rind. The stripe pattern of fruit is an important factor to characterize genetic diversity. It might be possible that this pattern is controlled by some set of gene. Seed traits are important for characterizing diversity in C. colocynthis. Seed characters, such as seed size and seed coat color, may to a certain extent affect the diversity profile of C. colocynthis. In terms of seed coat color ‘grey, dark grey, brown, light brown, and creamish seeds’ often observed in the field but the observation regarding seed coat color, since light-colored or white seed coat colors are seen in immature fruit. Seed coat color is also difficult to classify due to the shades of different colors. The characterization of seed coat color pattern in different accessions is presented in Fig. 2.
Fig. 1.
a–t Different stripe patterns in fruit of Citrullus colocynthis (L.) Schard. collected from different locations in Rajasthan, India
Fig. 2.
a–i Different seed coat color patterns in seeds of Citrullus colocynthis (L.) Schard. collected from different locations in Rajasthan, India
RAPD analysis
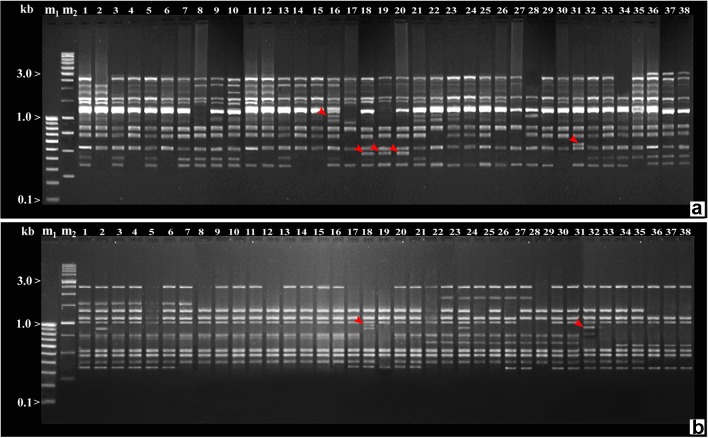
In the case of RAPD analysis, 65 RAPD primers were used for the initial screening of C. colocynthis genotypes, of which 50 primers revealed polymorphic banding patterns. The 50-decamer primers amplified DNA fragments across the 38 genotypes, with the number of amplified fragments varying from 5 (OPA-08 and OPF-9) to 19 (OPT-20) in the molecular size range of 150–6000 bp. A total of 549 bands were produced that could be scored, out of which 318 bands were polymorphic with an average of 6.36 polymorphic bands per primer. Percent polymorphism ranged from 22.2% (OPA-09) to 83.3% (OPE-12) with an average of 55.14% polymorphism (Table 2). The extent of polymorphism observed among the C. colocynthis genotypes as revealed by various RAPD primers is shown in Fig. 3a, b.
Table 2.
List of RAPD primers and amplification details used for the study of genetic diversity of 38 Citrullus colocynthis accessions
| S. no. | Primmer | Sequence (5′–3′) | NSB = L | NPB | NMB | PP | PIC | H | BIi | Rp | T | MR = (L/T) | MI = H × MR | Size range (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | OPA-1 | CAGGCCCTTC | 10 | 6 | 4 | 60.00 | 0.46 | 0.48 | 0.44 | 2.63 | 1 | 10 | 4.8 | 450–2000 |
| 2 | OPA-2 | TGCCGAGCTG | 11 | 6 | 5 | 54.54 | 0.39 | 0.40 | 0.54 | 3.21 | 1 | 11 | 4.4 | 150–1000 |
| 3 | OPA-3 | AGTCAGCCAC | 7 | 3 | 4 | 42.85 | 0.46 | 0.48 | 0.49 | 1.47 | 1 | 7 | 3.36 | 600–1500 |
| 4 | OPA-4 | AATCGGGCTG | 11 | 6 | 5 | 54.54 | 0.46 | 0.47 | 0.68 | 4.11 | 1 | 11 | 5.17 | 200–1500 |
| 5 | OPA-5 | AGGGGTCTTG | 16 | 12 | 4 | 75.00 | 0.64 | 0.65 | 0.43 | 5.11 | 1 | 16 | 10.4 | 250–2800 |
| 6 | OPA-7 | GAAACGGGTG | 8 | 5 | 3 | 62.50 | 0.32 | 0.33 | 0.83 | 4.16 | 1 | 8 | 2.64 | 200–3500 |
| 7 | OPA-8 | GTGACGTAGG | 5 | 4 | 1 | 80.00 | 0.11 | 0.22 | 0.57 | 2.26 | 1 | 5 | 1.10 | 150–1500 |
| 8 | OPA-9 | GGGTAACGCC | 9 | 2 | 7 | 22.22 | 0.87 | 0.89 | 0.92 | 1.84 | 1 | 9 | 8.01 | 150–2000 |
| 9 | OPA-10 | GTGATCGCAG | 15 | 11 | 4 | 73.33 | 0.34 | 0.35 | 0.45 | 4.95 | 1 | 15 | 5.25 | 350–3500 |
| 10 | OPA-11 | CAATCGCCGT | 13 | 7 | 6 | 53.84 | 0.55 | 0.56 | 0.69 | 4.84 | 1 | 13 | 7.28 | 450–2200 |
| 11 | OPA-14 | TCTGTGCTGG | 7 | 0 | 7 | 0.00 | 0.83 | 0.85 | 0.00 | 0.00 | 1 | 7 | 5.95 | 200–1700 |
| 12 | OPA-15 | TTCCGAACCC | 10 | 4 | 6 | 40.00 | 0.46 | 0.47 | 0.55 | 2.21 | 1 | 10 | 4.70 | 500–2000 |
| 13 | OPA-16 | AGCCAGCGAA | 9 | 5 | 4 | 55.55 | 0.23 | 0.24 | 0.60 | 3.00 | 1 | 9 | 2.16 | 300–1200 |
| 14 | OPA-18 | AGGTGACCGT | 14 | 10 | 4 | 71.42 | 0.44 | 0.45 | 0.40 | 4.00 | 1 | 14 | 6.30 | 600–5000 |
| 15 | OPA-19 | CAAACGTCGG | 14 | 9 | 5 | 64.28 | 0.55 | 0.56 | 0.27 | 2.47 | 1 | 14 | 7.84 | 230–1800 |
| 16 | OPB-04 | GGACTGGAGT | 18 | 13 | 5 | 72.22 | 0.52 | 0.53 | 0.33 | 4.26 | 1 | 18 | 9.54 | 320–3500 |
| 17 | OPB-08 | GTCCACACGG | 6 | 0 | 6 | 0.00 | 0.81 | 0.83 | 0.00 | 0.00 | 1 | 6 | 4.98 | 300–2500 |
| 18 | OPE-12 | TTATCGCCCC | 12 | 10 | 2 | 83.33 | 0.46 | 0.47 | 0.38 | 3.84 | 1 | 12 | 5.64 | 450–2200 |
| 19 | OPF-1 | ACGGATCCTG | 14 | 10 | 4 | 71.42 | 0.12 | 0.15 | 0.50 | 5.00 | 1 | 14 | 2.10 | 320–3200 |
| 20 | OPF-4 | GGTGATCAGG | 11 | 7 | 4 | 63.63 | 0.16 | 0.18 | 0.52 | 3.63 | 1 | 11 | 1.98 | 200–3000 |
| 21 | OPF-5 | CCGAATTCCC | 8 | 3 | 5 | 37.50 | 0.51 | 0.61 | 0.23 | 0.68 | 1 | 8 | 4.88 | 600–2000 |
| 22 | OPF-6 | GGGAATTCGG | 6 | 4 | 2 | 66.66 | 0.42 | 0.49 | 0.50 | 5.00 | 1 | 6 | 2.94 | 200–1500 |
| 23 | OPF-7 | CCGATATCCC | 15 | 10 | 5 | 66.66 | 0.36 | 0.37 | 0.33 | 3.26 | 1 | 15 | 5.55 | 250–4000 |
| 24 | OPF-9 | CCAAGCTTCC | 5 | 3 | 2 | 60.00 | 0.11 | 0.22 | 0.58 | 1.74 | 1 | 5 | 1.10 | 200–1000 |
| 25 | OPF- 10 | GGAAGCTTGG | 16 | 11 | 5 | 68.75 | 0.18 | 0.18 | 0.44 | 4.79 | 1 | 16 | 2.88 | 220–4200 |
| 26 | OPF-13 | GGCTGCAGAA | 17 | 9 | 8 | 52.94 | 0.81 | 0.82 | 0.46 | 4.16 | 1 | 17 | 13.94 | 250–2000 |
| 27 | OPF-14 | TGCTGCAGGT | 11 | 4 | 7 | 36.36 | 0.30 | 0.31 | 0.45 | 1.79 | 1 | 11 | 3.41 | 200–1700 |
| 28 | OPF-17 | AACCCGGGAA | 11 | 5 | 6 | 45.45 | 0.68 | 0.69 | 0.53 | 2.63 | 1 | 11 | 7.59 | 450–2000 |
| 29 | OPF-19 | CCTCTAGACC | 13 | 10 | 3 | 76.92 | 0.23 | 0.24 | 0.43 | 4.32 | 1 | 13 | 3.12 | 300–2250 |
| 30 | OPF-20 | GGTCTAGAGG | 9 | 7 | 2 | 77.77 | 0.23 | 0.24 | 0.56 | 3.95 | 1 | 9 | 2.16 | 300–3000 |
| 31 | OPJ-11 | ACTCCTGCGA | 13 | 5 | 8 | 38.46 | 0.59 | 0.60 | 0.85 | 4.26 | 1 | 13 | 7.8 | 350–2200 |
| 32 | OPJ-12 | GTCCCGTGGT | 12 | 8 | 4 | 66.66 | 0.18 | 0.18 | 0.59 | 4.74 | 1 | 12 | 2.16 | 200–1500 |
| 33 | OPK-19 | CACAGGCGGA | 14 | 5 | 9 | 35.71 | 0.29 | 0.30 | 0.38 | 1.89 | 1 | 14 | 4.2 | 200–1700 |
| 34 | OPP-12 | AAGGGCGAGT | 12 | 7 | 5 | 58.33 | 0.23 | 0.24 | 0.46 | 3.21 | 1 | 12 | 2.88 | 400–3000 |
| 35 | OPR-02 | CACAGCTGCC | 6 | 0 | 6 | 0.00 | 0.81 | 0.83 | 0.00 | 0.00 | 1 | 6 | 4.98 | 500–1200 |
| 36 | OPS-08 | TTCAGGGTGG | 8 | 6 | 2 | 75.00 | 0.55 | 0.56 | 0.72 | 4.42 | 1 | 8 | 4.88 | 50–3500 |
| 37 | OPS-13 | GTCGTTCCTG | 9 | 6 | 3 | 66.66 | 0.64 | 0.65 | 0.35 | 2.11 | 1 | 9 | 5.85 | 200–2100 |
| 38 | OPT-1 | GGGCCACTCA | 11 | 7 | 4 | 63.63 | 0.71 | 0.72 | 0.62 | 4.32 | 1 | 11 | 7.92 | 300–2500 |
| 39 | OPT-4 | CACAGAGGGA | 14 | 11 | 3 | 78.57 | 0.25 | 0.29 | 0.19 | 2.11 | 1 | 14 | 4.06 | 250–3500 |
| 140 | OPT-6 | CAAGGGCAGA | 10 | 6 | 4 | 60.00 | 0.86 | 0.87 | 0.61 | 3.68 | 1 | 10 | 8.70 | 450–3000 |
| 411 | OPT-7 | GGCAGGCTGT | 6 | 4 | 2 | 66.66 | 0.11 | 0.22 | 0.41 | 1.63 | 1 | 6 | 1.32 | 300–1800 |
| 412 | OPT-8 | AACGGCGACA | 9 | 3 | 6 | 33.33 | 0.51 | 0.61 | 0.21 | 0.63 | 1 | 9 | 5.49 | 100–2000 |
| 413 | OPT-9 | CACCCCTGAG | 11 | 6 | 5 | 54.54 | 0.39 | 0.40 | 0.39 | 2.37 | 1 | 11 | 4.40 | 350–3500 |
| 414 | OPT-11 | TTCCCCGCGA | 8 | 2 | 6 | 25.00 | 0.59 | 0.61 | 0.56 | 1.16 | 1 | 8 | 4.88 | 450–2000 |
| 415 | OPT-12 | GGGTGTGTAG | 9 | 3 | 6 | 33.33 | 0.87 | 0.88 | 0.58 | 1.74 | 1 | 9 | 7.92 | 150–2000 |
| 416 | OPT-14 | AATGCCGCAG | 17 | 14 | 3 | 82.35 | 0.62 | 0.62 | 0.30 | 4.26 | 1 | 17 | 10.54 | 450–2700 |
| 417 | OPT-15 | GGATGCCACT | 7 | 4 | 3 | 57.14 | 0.54 | 0.55 | 0.28 | 1.11 | 1 | 7 | 3.85 | 650–1800 |
| 48 | OPT-16 | GGTGAACGCT | 12 | 7 | 5 | 58.33 | 0.71 | 0.72 | 0.53 | 3.63 | 1 | 12 | 8.64 | 400–2500 |
| 49 | OPT-19 | GTCCGTATGG | 11 | 5 | 6 | 45.45 | 0.71 | 0.72 | 0.44 | 2.21 | 1 | 11 | 7.92 | 100–1200 |
| 50 | OPT-20 | GACCAATGCC | 19 | 13 | 6 | 68.42 | 0.38 | 0.38 | 0.32 | 4.21 | 1 | 19 | 7.22 | 350–6000 |
| – | Total | 549 | 318 | 231 | 2757.25 | 23.55 | 24.68 | 22.8 | 149 | 50 | 549 | 266.38 | – | |
| – | Average | 10.98 | 6.36 | 4.62 | 55.14 | 0.47 | 0.49 | 0.45 | 2.98 | 1 | 10.98 | 5.32 | – |
NSB number of scorable bands, NPB number of polymorphic bands, NMB number of monomorphic bands, PP % polymorphism, PIC polymorphic information content, H heterozygosity,BIi average informativeness of a band, RP resolving power, T total no. of assays/primers combination, MR multiplex ratio, MI marker index
Fig. 3.
a–b RAPD profile of Citrullus colocynthis genotypes produced with different primers (a OPA-05, b OPA-10) m1-100 bp ladder (Lane 1), m2-1 kb ladder (Lane 2), 1–38 Citrullus colocynthis accessions
Similarity matrix values using Jaccard’s coefficient ranged from 0.64 between U1 and B3 to 0.96 between Jh1 and Jh2 (Table 6). At 72% similarity, the accessions separated into three clusters (Fig. 5a). Cluster I comprised of most of the genotypes and is divided into two sub-clusters (Ia and Ib). Sub-cluster Ia comprised of the accessions A1, A2, Jh1, Jh2, Jh3, C1, C2, C3, Jd1, Jd2, and Jd3. Sub-cluster Ib comprised of 13 accessions J1, J2, J3, J4, J5, S1, S2, S3, H1, H2, H3, G1, G2, and G3. Cluster II divided into two sub-clusters (IIa and IIb). Sub-cluster IIa comprised of the accessions U1 and U2 and sub-cluster IIb contains only one accession U3. Cluster III divided into two sub-clusters (IIIa and IIIb). Sub-cluster IIIa comprised of four accessions B1, B2, B3, and B4 and sub-cluster IIIb comprised of the accessions Js1, Js2, Js3, Js4, Bd1, Bd2, and Bd3. The result of principal coordinate analysis was comparable to the cluster analysis with minor deviations (Fig. 6a). Approx. 50% RAPD markers showed PIC value and heterozygosity ≥0.50 suggesting the informativeness of a marker. The highest PIC value (0.86) and heterozygosity (0.88) revealed by OPT 6, OPT 12, and lowest PIC value (0.11) and heterozygosity (0.22) shown by OPF 9 and OPT 7. The estimates of marker attributes for RAPD primers varied widely. The multiplex ratio (MR) was the highest for OPT-20 (19) and lowest for OPF-9 (5) and OPA-8 (5). Similarly, marker index (MI) value was highest for OPF-13 (13.94) and lowest for OPF-9 (1.10) and OPF-8 (1.10). The average value of MR and MI was 10.98 and 5.32 for RAPD markers. The values for average informativeness of a band (BIi) were observed maximal for OPA-9 (0.92) and minimal for OPT-4 (0.19). The resolving power (RP) is a parameter that indicates the discriminatory potential of the selected primers. The highest RP value was observed with the primer OPA-5 (5.11) and the lowest with the primer OPT-8 (0.63). The average informativeness of a band (BIi) and resolving power (RP) was 0.45 and 2.98 for RAPD markers (Table 2).
Table 6.
Jaccard’s similarity coefficient of 38 Citrullus colocynthis accessions based on RAPD data analysis
| A1 | A2 | J1 | J2 | J3 | J4 | J5 | S1 | S2 | S3 | Jh1 | Jh2 | Jh3 | C1 | C2 | C3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 1.000 | |||||||||||||||
| A2 | 0.885 | 1.000 | ||||||||||||||
| J1 | 0.737 | 0.749 | 1.000 | |||||||||||||
| J2 | 0.742 | 0.742 | 0.870 | 1.000 | ||||||||||||
| J3 | 0.746 | 0.734 | 0.887 | 0.926 | 1.000 | |||||||||||
| J4 | 0.741 | 0.736 | 0.858 | 0.890 | 0.926 | 1.000 | ||||||||||
| J5 | 0.761 | 0.740 | 0.876 | 0.894 | 0.921 | 0.931 | 1.000 | |||||||||
| S1 | 0.762 | 0.741 | 0.836 | 0.853 | 0.884 | 0.873 | 0.908 | 1.000 | ||||||||
| S2 | 0.757 | 0.727 | 0.808 | 0.824 | 0.855 | 0.840 | 0.873 | 0.901 | 1.000 | |||||||
| S3 | 0.767 | 0.737 | 0.804 | 0.824 | 0.846 | 0.845 | 0.868 | 0.885 | 0.927 | 1.000 | ||||||
| Jh1 | 0.772 | 0.787 | 0.733 | 0.730 | 0.746 | 0.737 | 0.765 | 0.792 | 0.791 | 0.823 | 1.000 | |||||
| Jh2 | 0.770 | 0.773 | 0.715 | 0.720 | 0.744 | 0.734 | 0.762 | 0.786 | 0.780 | 0.812 | 0.961 | 1.000 | ||||
| Jh3 | 0.778 | 0.781 | 0.735 | 0.728 | 0.752 | 0.755 | 0.771 | 0.803 | 0.785 | 0.799 | 0.932 | 0.939 | 1.000 | |||
| C1 | 0.771 | 0.778 | 0.745 | 0.742 | 0.762 | 0.752 | 0.773 | 0.796 | 0.764 | 0.796 | 0.922 | 0.925 | 0.939 | 1.000 | ||
| C2 | 0.761 | 0.761 | 0.739 | 0.752 | 0.752 | 0.746 | 0.762 | 0.794 | 0.767 | 0.799 | 0.907 | 0.895 | 0.910 | 0.949 | 1.000 | |
| C3 | 0.760 | 0.760 | 0.734 | 0.747 | 0.751 | 0.733 | 0.761 | 0.793 | 0.770 | 0.793 | 0.910 | 0.898 | 0.903 | 0.932 | 0.951 | 1.000 |
| H1 | 0.728 | 0.739 | 0.749 | 0.746 | 0.762 | 0.761 | 0.760 | 0.758 | 0.727 | 0.737 | 0.784 | 0.786 | 0.786 | 0.800 | 0.781 | 0.797 |
| H2 | 0.729 | 0.729 | 0.803 | 0.819 | 0.812 | 0.816 | 0.825 | 0.804 | 0.772 | 0.782 | 0.725 | 0.735 | 0.731 | 0.733 | 0.719 | 0.730 |
| G1 | 0.696 | 0.711 | 0.800 | 0.794 | 0.810 | 0.804 | 0.827 | 0.788 | 0.769 | 0.775 | 0.707 | 0.717 | 0.713 | 0.722 | 0.709 | 0.708 |
| G2 | 0.706 | 0.698 | 0.796 | 0.797 | 0.800 | 0.827 | 0.831 | 0.806 | 0.773 | 0.783 | 0.713 | 0.715 | 0.732 | 0.717 | 0.711 | 0.714 |
| G3 | 0.741 | 0.720 | 0.790 | 0.814 | 0.826 | 0.839 | 0.863 | 0.822 | 0.812 | 0.827 | 0.757 | 0.755 | 0.751 | 0.736 | 0.734 | 0.733 |
| B1 | 0.706 | 0.722 | 0.683 | 0.695 | 0.703 | 0.710 | 0.730 | 0.748 | 0.729 | 0.727 | 0.762 | 0.752 | 0.757 | 0.737 | 0.723 | 0.718 |
| B2 | 0.718 | 0.738 | 0.676 | 0.684 | 0.689 | 0.687 | 0.706 | 0.720 | 0.701 | 0.704 | 0.762 | 0.748 | 0.748 | 0.741 | 0.732 | 0.735 |
| B3 | 0.718 | 0.722 | 0.671 | 0.691 | 0.680 | 0.686 | 0.693 | 0.703 | 0.696 | 0.703 | 0.734 | 0.728 | 0.724 | 0.730 | 0.720 | 0.715 |
| B4 | 0.722 | 0.726 | 0.683 | 0.711 | 0.703 | 0.706 | 0.726 | 0.731 | 0.729 | 0.727 | 0.758 | 0.744 | 0.752 | 0.741 | 0.748 | 0.738 |
| Js1 | 0.692 | 0.688 | 0.658 | 0.677 | 0.677 | 0.695 | 0.702 | 0.692 | 0.693 | 0.704 | 0.714 | 0.716 | 0.717 | 0.702 | 0.689 | 0.684 |
| Js2 | 0.674 | 0.671 | 0.681 | 0.692 | 0.708 | 0.707 | 0.721 | 0.707 | 0.682 | 0.688 | 0.718 | 0.712 | 0.724 | 0.721 | 0.697 | 0.688 |
| Js3 | 0.695 | 0.688 | 0.707 | 0.732 | 0.731 | 0.734 | 0.750 | 0.727 | 0.709 | 0.711 | 0.733 | 0.727 | 0.736 | 0.725 | 0.711 | 0.707 |
| Js4 | 0.682 | 0.682 | 0.730 | 0.744 | 0.748 | 0.742 | 0.767 | 0.765 | 0.728 | 0.726 | 0.754 | 0.747 | 0.743 | 0.749 | 0.747 | 0.746 |
| Bd1 | 0.698 | 0.709 | 0.726 | 0.756 | 0.751 | 0.776 | 0.766 | 0.764 | 0.741 | 0.747 | 0.753 | 0.742 | 0.759 | 0.752 | 0.751 | 0.749 |
| Bd2 | 0.699 | 0.691 | 0.687 | 0.732 | 0.719 | 0.722 | 0.746 | 0.735 | 0.725 | 0.723 | 0.741 | 0.739 | 0.736 | 0.741 | 0.731 | 0.734 |
| Bd3 | 0.700 | 0.693 | 0.697 | 0.733 | 0.729 | 0.728 | 0.748 | 0.733 | 0.710 | 0.717 | 0.723 | 0.725 | 0.729 | 0.735 | 0.717 | 0.724 |
| Jd1 | 0.766 | 0.778 | 0.720 | 0.733 | 0.737 | 0.732 | 0.751 | 0.757 | 0.731 | 0.749 | 0.787 | 0.785 | 0.785 | 0.790 | 0.785 | 0.792 |
| Jd2 | 0.759 | 0.742 | 0.696 | 0.733 | 0.721 | 0.715 | 0.735 | 0.741 | 0.710 | 0.720 | 0.759 | 0.760 | 0.765 | 0.762 | 0.773 | 0.763 |
| Jd3 | 0.764 | 0.787 | 0.729 | 0.726 | 0.742 | 0.721 | 0.740 | 0.738 | 0.744 | 0.750 | 0.797 | 0.794 | 0.782 | 0.783 | 0.774 | 0.776 |
| U1 | 0.682 | 0.690 | 0.694 | 0.711 | 0.723 | 0.721 | 0.733 | 0.743 | 0.737 | 0.752 | 0.741 | 0.731 | 0.739 | 0.717 | 0.703 | 0.718 |
| U2 | 0.683 | 0.684 | 0.723 | 0.712 | 0.728 | 0.718 | 0.751 | 0.740 | 0.725 | 0.745 | 0.722 | 0.708 | 0.712 | 0.706 | 0.684 | 0.699 |
| U3 | 0.685 | 0.704 | 0.704 | 0.726 | 0.730 | 0.733 | 0.749 | 0.742 | 0.727 | 0.738 | 0.720 | 0.709 | 0.722 | 0.703 | 0.694 | 0.697 |
| H1 | H2 | G1 | G2 | G3 | B1 | B2 | B3 | B4 | Js1 | Js2 | Js3 | Js4 | Bd1 | Bd2 | Bd3 | Jd1 | Jd2 | Jd3 | U1 | U2 | U3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | ||||||||||||||||||||||
| A2 | ||||||||||||||||||||||
| J1 | ||||||||||||||||||||||
| J2 | ||||||||||||||||||||||
| J3 | ||||||||||||||||||||||
| J4 | ||||||||||||||||||||||
| J5 | ||||||||||||||||||||||
| S1 | ||||||||||||||||||||||
| S2 | ||||||||||||||||||||||
| S3 | ||||||||||||||||||||||
| Jh1 | ||||||||||||||||||||||
| Jh2 | ||||||||||||||||||||||
| Jh3 | ||||||||||||||||||||||
| C1 | ||||||||||||||||||||||
| C2 | ||||||||||||||||||||||
| C3 | ||||||||||||||||||||||
| H1 | 1.000 | |||||||||||||||||||||
| H2 | 0.852 | 1.000 | ||||||||||||||||||||
| G1 | 0.801 | 0.899 | 1.000 | |||||||||||||||||||
| G2 | 0.796 | 0.890 | 0.912 | 1.000 | ||||||||||||||||||
| G3 | 0.790 | 0.888 | 0.870 | 0.896 | 1.000 | |||||||||||||||||
| B1 | 0.686 | 0.699 | 0.692 | 0.711 | 0.756 | 1.000 | ||||||||||||||||
| B2 | 0.691 | 0.665 | 0.663 | 0.688 | 0.710 | 0.912 | 1.000 | |||||||||||||||
| B3 | 0.675 | 0.664 | 0.657 | 0.663 | 0.690 | 0.853 | 0.899 | 1.000 | ||||||||||||||
| B4 | 0.694 | 0.687 | 0.688 | 0.706 | 0.738 | 0.866 | 0.868 | 0.877 | 1.000 | |||||||||||||
| Js1 | 0.654 | 0.661 | 0.659 | 0.676 | 0.706 | 0.752 | 0.728 | 0.720 | 0.795 | 1.000 | ||||||||||||
| Js2 | 0.673 | 0.666 | 0.675 | 0.684 | 0.695 | 0.743 | 0.704 | 0.685 | 0.751 | 0.889 | 1.000 | |||||||||||
| Js3 | 0.694 | 0.695 | 0.700 | 0.719 | 0.726 | 0.748 | 0.720 | 0.699 | 0.764 | 0.833 | 0.913 | 1.000 | ||||||||||
| Js4 | 0.720 | 0.714 | 0.707 | 0.726 | 0.725 | 0.778 | 0.756 | 0.744 | 0.783 | 0.760 | 0.806 | 0.864 | 1.000 | |||||||||
| Bd1 | 0.693 | 0.726 | 0.719 | 0.738 | 0.746 | 0.795 | 0.764 | 0.739 | 0.795 | 0.789 | 0.796 | 0.815 | 0.860 | 1.000 | ||||||||
| Bd2 | 0.709 | 0.703 | 0.697 | 0.703 | 0.730 | 0.756 | 0.756 | 0.752 | 0.803 | 0.760 | 0.779 | 0.789 | 0.813 | 0.885 | 1.000 | |||||||
| Bd3 | 0.703 | 0.712 | 0.706 | 0.704 | 0.723 | 0.737 | 0.729 | 0.742 | 0.771 | 0.758 | 0.777 | 0.770 | 0.801 | 0.873 | 0.925 | 1.000 | ||||||
| Jd1 | 0.782 | 0.740 | 0.742 | 0.737 | 0.735 | 0.690 | 0.695 | 0.686 | 0.694 | 0.676 | 0.698 | 0.698 | 0.696 | 0.700 | 0.705 | 0.726 | 1.000 | |||||
| Jd2 | 0.746 | 0.724 | 0.714 | 0.712 | 0.719 | 0.662 | 0.679 | 0.677 | 0.666 | 0.646 | 0.665 | 0.674 | 0.668 | 0.669 | 0.682 | 0.698 | 0.906 | 1.000 | ||||
| Jd3 | 0.767 | 0.741 | 0.743 | 0.718 | 0.737 | 0.687 | 0.692 | 0.676 | 0.695 | 0.684 | 0.681 | 0.680 | 0.670 | 0.701 | 0.695 | 0.689 | 0.873 | 0.852 | 1.000 | |||
| U1 | 0.682 | 0.726 | 0.724 | 0.739 | 0.751 | 0.690 | 0.664 | 0.640 | 0.682 | 0.695 | 0.688 | 0.687 | 0.697 | 0.746 | 0.718 | 0.712 | 0.697 | 0.677 | 0.717 | 1.000 | ||
| U2 | 0.690 | 0.719 | 0.725 | 0.719 | 0.743 | 0.679 | 0.654 | 0.652 | 0.679 | 0.654 | 0.677 | 0.680 | 0.702 | 0.702 | 0.707 | 0.705 | 0.698 | 0.677 | 0.676 | 0.882 | 1.000 | |
| U3 | 0.707 | 0.721 | 0.718 | 0.733 | 0.745 | 0.685 | 0.659 | 0.646 | 0.681 | 0.670 | 0.682 | 0.693 | 0.691 | 0.692 | 0.693 | 0.687 | 0.727 | 0.706 | 0.712 | 0.864 | 0.861 | 1.000 |
Fig. 5.
Dendogram (UPGMA) representing genetic relationship among 38 accessions of Citrullus colocynthis using Jaccard’s similarity coefficients. Numbers on the nodes of the cluster indicate the bootstrap values generated by 1000 replications using the program WINBOOT. Figure shows a RAPD data-based dendrogram, b ISSR data-based dendrogram, and c combined (RAPD + ISSR) data-based dendrogram
Fig. 6.
Two-dimensional plot of 38 accessions of Citrullus colocynthis by principle coordinate analysis (PCA) using Jaccard’s similarity coefficients
ISSR analysis
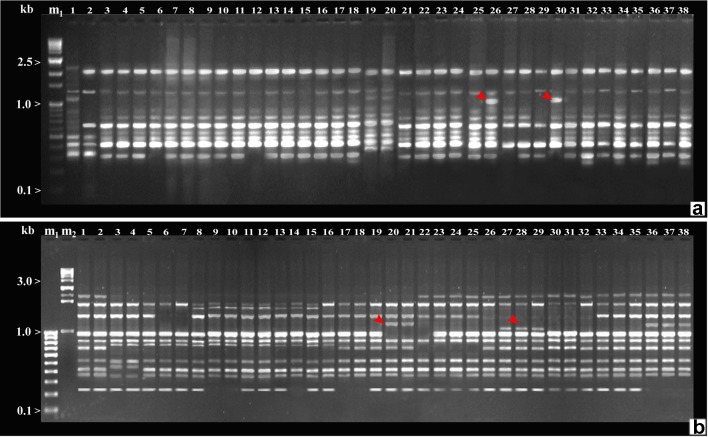
Among the 20 ISSR primers screened, 13 primers amplifying polymorphic banding patterns were selected for further study (Table 3). ISSR primers produced different numbers of DNA fragments, depending upon their simple sequence repeat motifs. Thirteen primers produced 166 amplification products, and out of which 99 were polymorphic, with an average of 7.6 polymorphic bands per primer. The number of bands amplified per primer varied between 9 (UBC-807 and 802) and 16 (UBC-803 and 812) with average band size between 250 and 4000 bp. Percent polymorphism ranged from 45.4% (UBC-815) to 73.3% (UBC-814) with an average of 65.05% polymorphism (Table 3). The extent of polymorphism observed among the C. colocynthis genotypes as revealed by various ISSR primers is shown in Fig. 4a, b. Jaccard’s similarity coefficient ranged from 0.58 between A1 and Js2 to 0.96 between B2 and B3 (Table 7). A dendrogram based on UPGMA analysis with ISSR data is shown in Fig. 5b. Dendrogram analysis separated the accessions into four clusters at 72% similarity. Cluster I comprised of five OTUs and is divided into two sub-clusters (Ia and Ib). Sub-cluster Ia comprised of A1 and A2. Sub-cluster Ib comprised of Jd1, Jd2, and Jd3. Within sub-cluster 1a, A1 and A2 appeared to be closer to each other with similarity coefficient equal to 0.90 and accessions Jd1, Jd2, and Jd3 appeared to be closer to each other with similarity coefficient equal to 0.93 0.96, and 0.69 in sub-cluster Ib, respectively. Accessions Jd1, Jd2, and Jd3 had distinct OTU as in case of RAPD analysis. Cluster II divided into two sub-clusters (IIa and IIb). Sub-cluster IIa comprised of the accessions Jh1–Jh3 and C1–C3 and sub-cluster IIb contains accessions G1–G3, H1, and H2. Cluster III divided into two sub-clusters (IIIa and IIIb). Sub-cluster IIIa comprised of eight accessions J1–J5 and S1–S3, sub-cluster IIIb comprised of the accessions U1, U2, and U3, and last IV cluster divided into two sub-clusters (IVa and IVb) which comprised of the accessions B1–B4, Js1–Js4, and Bd1–Bd3. The result of PCA was comparable to cluster analysis with minor deviations (Fig. 6b). Approx. 50% ISSR markers showed PIC value and heterozygosity ≥0.50 suggesting the informativeness of a marker. The highest PIC value (0.91) and heterozygosity (0.92) revealed by UBC 808 and lowest PIC value (0.18) and heterozygosity (0.20) shown by UBC 814. The estimates of marker attributes for ISSR primers varied widely. The multiplex ratio (MR) was the highest for UBC-803 (16) and lowest for UBC 803 and 807 (5). Similarly, marker index (MI) value was highest for UBC-808 (12.88) and lowest for UBC-809 (2.16). The average value of MR and MI was 12.76 and 5.86 for ISSR markers. The values for average informativeness of a band (BIi) were observed maximal for UBC-803 (0.20) and minimal for UBC-803 (0.82). The highest RP value was observed with the primer UBC-803 (8.18) and the lowest with UBC-811 (1.56). The average informativeness of a band (BIi) and resolving power (RP) was 0.46 and 4.10 for RAPD markers (Table 3).
Table 3.
List of ISSR primers and amplification details used for the study of genetic diversity of 38 Citrullus colocynthis accessions
| S. no. | Primmer | Sequence (5′–3′) | Tm (°C) | NSB = L | NPB | NMB | PP | PIC | H | BIi | Rp | T | MR = (L/T) | MI = H × MR | Size range (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | UBC 802 | (AT)8G | 30 | 9 | 0 | 9 | 100 | 0.87 | 0.88 | 0.00 | 0.00 | 1 | 9 | 7.92 | 250–2500 |
| 2 | UBC 803 | (AT)8C | 30 | 16 | 10 | 6 | 62.5 | 0.50 | 0.59 | 0.82 | 8.18 | 1 | 16 | 9.44 | 300–2500 |
| 3 | UBC 805 | (TA)8C | 30 | 14 | 10 | 4 | 71.42 | 0.18 | 0.18 | 0.65 | 6.48 | 1 | 14 | 2.52 | 250–2000 |
| 4 | UBC 807 | (AG)8T | 51 | 9 | 5 | 4 | 55.5 | 0.23 | 0.24 | 0.41 | 2.86 | 1 | 9 | 2.16 | 600–4000 |
| 5 | UBC 808 | (AG)8C | 51.4 | 14 | 10 | 4 | 71.4 | 0.91 | 0.92 | 0.64 | 6.42 | 1 | 14 | 12.88 | 150–2500 |
| 6 | UBC 809 | (AG)8G | 51 | 13 | 9 | 4 | 69.2 | 0.55 | 0.56 | 0.32 | 2.84 | 1 | 13 | 7.28 | 250–1500 |
| 7 | UBC 810 | (GA)8T | 49 | 10 | 5 | 5 | 50 | 0.29 | 0.30 | 0.44 | 4.88 | 1 | 10 | 3.0 | 400–2000 |
| 8 | UBC 811 | (GA)8C | 50 | 13 | 7 | 6 | 53.8 | 0.39 | 0.40 | 0.20 | 1.56 | 1 | 13 | 5.2 | 450–3500 |
| 9 | UBC 812 | (GA)8A | 48.5 | 16 | 10 | 6 | 62.5 | 0.45 | 0.46 | 0.47 | 4.70 | 1 | 16 | 7.36 | 450–4000 |
| 10 | UBC 813 | (CT)8T | 48 | 13 | 9 | 4 | 69.2 | 0.27 | 0.28 | 0.40 | 3.58 | 1 | 13 | 3.64 | 450–2500 |
| 11 | UBC 814 | (CT)8A | 49 | 15 | 11 | 4 | 73.3 | 0.18 | 0.20 | 0.58 | 5.78 | 1 | 15 | 3.0 | 100–1000 |
| 12 | UBC 815 | (CT)8G | 50 | 11 | 5 | 5 | 45.4 | 0.51 | 0.53 | 0.61 | 3.06 | 1 | 11 | 5.83 | 150–2000 |
| 13 | UBC 819 | (GT)8A | 53 | 13 | 8 | 5 | 61.5 | 0.51 | 0.52 | 0.50 | 3.02 | 1 | 13 | 6.76 | 700–4000 |
| – | Total | – | 166 | 99 | 66 | 845.72 | 5.84 | 6.06 | 6.04 | 53.36 | 13 | 166 | 76.99 | – | |
| – | Average | – | 12.76 | 7.6 | 5.07 | 65.05 | 0.44 | 0.46 | 0.46 | 4.10 | 1 | 12.76 | 5.86 | – |
NSB number of scorable bands, NPB number of polymorphic bands, NMB number of monomorphic bands, PP % polymorphism, PIC polymorphic information content, H heterozygosity,BIi average informativeness of a band, RP resolving power, T total no. of assays/primers combination, MR multiplex ratio, MI marker index
Fig. 4.
a–b ISSR profile of Citrullus colocynthis genotypes produced with different primers (a UBC-803, b UBS-805) m1-100 bp ladder (Lane 1), m2-1 kb ladder (Lane 2), and 1–38 Citrullus colocynthis accessions
Table 7.
Jaccard’s similarity coefficient of 38 Citrullus colocynthis accessions based on ISSR data analysis
| A1 | A2 | J1 | J2 | J3 | J4 | J5 | S1 | S2 | S3 | Jh1 | Jh2 | Jh3 | C1 | C2 | C3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 1.000 | |||||||||||||||
| A2 | 0.949 | 1.000 | ||||||||||||||
| J1 | 0.760 | 0.770 | 1.000 | |||||||||||||
| J2 | 0.781 | 0.778 | 0.922 | 1.000 | ||||||||||||
| J3 | 0.738 | 0.734 | 0.889 | 0.897 | 1.000 | |||||||||||
| J4 | 0.754 | 0.736 | 0.874 | 0.915 | 0.930 | 1.000 | ||||||||||
| J5 | 0.769 | 0.738 | 0.875 | 0.899 | 0.915 | 0.965 | 1.000 | |||||||||
| S1 | 0.769 | 0.752 | 0.844 | 0.852 | 0.851 | 0.883 | 0.916 | 1.000 | ||||||||
| S2 | 0.735 | 0.718 | 0.821 | 0.844 | 0.843 | 0.875 | 0.908 | 0.940 | 1.000 | |||||||
| S3 | 0.733 | 0.715 | 0.835 | 0.843 | 0.842 | 0.874 | 0.907 | 0.923 | 0.931 | 1.000 | ||||||
| Jh1 | 0.704 | 0.699 | 0.692 | 0.699 | 0.736 | 0.725 | 0.740 | 0.754 | 0.746 | 0.800 | 1.000 | |||||
| Jh2 | 0.699 | 0.682 | 0.649 | 0.682 | 0.705 | 0.708 | 0.710 | 0.710 | 0.702 | 0.754 | 0.948 | 1.000 | ||||
| Jh3 | 0.687 | 0.682 | 0.674 | 0.682 | 0.705 | 0.695 | 0.697 | 0.697 | 0.689 | 0.740 | 0.931 | 0.897 | 1.000 | |||
| C1 | 0.672 | 0.654 | 0.659 | 0.642 | 0.690 | 0.667 | 0.669 | 0.682 | 0.674 | 0.724 | 0.897 | 0.879 | 0.946 | 1.000 | ||
| C2 | 0.699 | 0.681 | 0.687 | 0.694 | 0.744 | 0.733 | 0.748 | 0.735 | 0.727 | 0.780 | 0.974 | 0.957 | 0.923 | 0.906 | 1.000 | |
| C3 | 0.689 | 0.697 | 0.702 | 0.697 | 0.748 | 0.723 | 0.738 | 0.712 | 0.718 | 0.756 | 0.948 | 0.914 | 0.914 | 0.864 | 0.957 | 1.000 |
| H1 | 0.721 | 0.717 | 0.674 | 0.681 | 0.715 | 0.705 | 0.719 | 0.707 | 0.712 | 0.723 | 0.838 | 0.808 | 0.808 | 0.779 | 0.846 | 0.852 |
| H2 | 0.681 | 0.676 | 0.681 | 0.676 | 0.711 | 0.688 | 0.691 | 0.679 | 0.683 | 0.706 | 0.780 | 0.750 | 0.819 | 0.803 | 0.788 | 0.792 |
| G1 | 0.664 | 0.660 | 0.676 | 0.671 | 0.706 | 0.696 | 0.686 | 0.674 | 0.667 | 0.688 | 0.761 | 0.731 | 0.785 | 0.756 | 0.756 | 0.759 |
| G2 | 0.685 | 0.669 | 0.674 | 0.693 | 0.715 | 0.717 | 0.719 | 0.695 | 0.688 | 0.710 | 0.784 | 0.767 | 0.754 | 0.713 | 0.791 | 0.795 |
| G3 | 0.690 | 0.686 | 0.703 | 0.710 | 0.746 | 0.735 | 0.750 | 0.737 | 0.717 | 0.741 | 0.831 | 0.786 | 0.786 | 0.744 | 0.824 | 0.829 |
| B1 | 0.632 | 0.638 | 0.691 | 0.674 | 0.709 | 0.686 | 0.701 | 0.701 | 0.681 | 0.704 | 0.664 | 0.636 | 0.636 | 0.599 | 0.660 | 0.674 |
| B2 | 0.634 | 0.641 | 0.693 | 0.676 | 0.711 | 0.688 | 0.703 | 0.715 | 0.696 | 0.719 | 0.679 | 0.650 | 0.638 | 0.613 | 0.674 | 0.676 |
| B3 | 0.626 | 0.621 | 0.671 | 0.667 | 0.701 | 0.691 | 0.705 | 0.717 | 0.698 | 0.721 | 0.681 | 0.652 | 0.641 | 0.615 | 0.676 | 0.667 |
| B4 | 0.620 | 0.626 | 0.667 | 0.650 | 0.697 | 0.674 | 0.676 | 0.689 | 0.669 | 0.692 | 0.652 | 0.635 | 0.672 | 0.644 | 0.647 | 0.650 |
| Js1 | 0.625 | 0.619 | 0.674 | 0.656 | 0.680 | 0.669 | 0.685 | 0.685 | 0.664 | 0.714 | 0.685 | 0.654 | 0.667 | 0.651 | 0.679 | 0.682 |
| Js2 | 0.583 | 0.588 | 0.629 | 0.612 | 0.659 | 0.662 | 0.664 | 0.664 | 0.644 | 0.693 | 0.677 | 0.672 | 0.634 | 0.606 | 0.672 | 0.674 |
| Js3 | 0.620 | 0.627 | 0.669 | 0.652 | 0.701 | 0.690 | 0.705 | 0.692 | 0.672 | 0.709 | 0.692 | 0.662 | 0.636 | 0.609 | 0.674 | 0.677 |
| Js4 | 0.615 | 0.646 | 0.677 | 0.646 | 0.696 | 0.672 | 0.674 | 0.662 | 0.641 | 0.677 | 0.662 | 0.618 | 0.618 | 0.603 | 0.644 | 0.646 |
| Bd1 | 0.639 | 0.634 | 0.690 | 0.672 | 0.696 | 0.698 | 0.701 | 0.688 | 0.667 | 0.704 | 0.674 | 0.656 | 0.643 | 0.615 | 0.656 | 0.659 |
| Bd2 | 0.637 | 0.632 | 0.688 | 0.695 | 0.706 | 0.709 | 0.711 | 0.711 | 0.677 | 0.714 | 0.698 | 0.654 | 0.654 | 0.638 | 0.679 | 0.669 |
| Bd3 | 0.609 | 0.603 | 0.656 | 0.652 | 0.662 | 0.664 | 0.667 | 0.679 | 0.647 | 0.695 | 0.679 | 0.649 | 0.649 | 0.646 | 0.674 | 0.664 |
| Jd1 | 0.733 | 0.742 | 0.672 | 0.679 | 0.652 | 0.679 | 0.681 | 0.693 | 0.674 | 0.722 | 0.744 | 0.714 | 0.714 | 0.712 | 0.739 | 0.729 |
| Jd2 | 0.761 | 0.758 | 0.699 | 0.693 | 0.667 | 0.693 | 0.708 | 0.721 | 0.701 | 0.750 | 0.773 | 0.729 | 0.729 | 0.727 | 0.754 | 0.744 |
| Jd3 | 0.771 | 0.767 | 0.720 | 0.701 | 0.687 | 0.701 | 0.704 | 0.716 | 0.696 | 0.746 | 0.769 | 0.725 | 0.738 | 0.736 | 0.750 | 0.740 |
| U1 | 0.692 | 0.700 | 0.746 | 0.727 | 0.752 | 0.727 | 0.742 | 0.756 | 0.748 | 0.760 | 0.770 | 0.724 | 0.724 | 0.722 | 0.750 | 0.754 |
| U2 | 0.692 | 0.700 | 0.732 | 0.727 | 0.738 | 0.727 | 0.742 | 0.756 | 0.748 | 0.746 | 0.756 | 0.711 | 0.711 | 0.709 | 0.750 | 0.740 |
| U3 | 0.650 | 0.657 | 0.713 | 0.695 | 0.719 | 0.695 | 0.710 | 0.710 | 0.702 | 0.713 | 0.723 | 0.679 | 0.732 | 0.744 | 0.718 | 0.721 |
| H1 | H2 | G1 | G2 | G3 | B1 | B2 | B3 | B4 | Js1 | Js2 | Js3 | Js4 | Bd1 | Bd2 | Bd3 | Jd1 | Jd2 | Jd3 | U1 | U2 | U3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | ||||||||||||||||||||||
| A2 | ||||||||||||||||||||||
| J1 | ||||||||||||||||||||||
| J2 | ||||||||||||||||||||||
| J3 | ||||||||||||||||||||||
| J4 | ||||||||||||||||||||||
| J5 | ||||||||||||||||||||||
| S1 | ||||||||||||||||||||||
| S2 | ||||||||||||||||||||||
| S3 | ||||||||||||||||||||||
| Jh1 | ||||||||||||||||||||||
| Jh2 | ||||||||||||||||||||||
| Jh3 | ||||||||||||||||||||||
| C1 | ||||||||||||||||||||||
| C2 | ||||||||||||||||||||||
| C3 | ||||||||||||||||||||||
| H1 | 1.000 | |||||||||||||||||||||
| H2 | 0.878 | 1.000 | ||||||||||||||||||||
| G1 | 0.830 | 0.913 | 1.000 | |||||||||||||||||||
| G2 | 0.880 | 0.850 | 0.885 | 1.000 | ||||||||||||||||||
| G3 | 0.901 | 0.870 | 0.850 | 0.930 | 1.000 | |||||||||||||||||
| B1 | 0.718 | 0.702 | 0.697 | 0.743 | 0.774 | 1.000 | ||||||||||||||||
| B2 | 0.720 | 0.716 | 0.711 | 0.732 | 0.763 | 0.967 | 1.000 | |||||||||||||||
| B3 | 0.722 | 0.706 | 0.701 | 0.722 | 0.764 | 0.936 | 0.968 | 1.000 | ||||||||||||||
| B4 | 0.707 | 0.767 | 0.761 | 0.695 | 0.725 | 0.864 | 0.895 | 0.896 | 1.000 | |||||||||||||
| Js1 | 0.631 | 0.638 | 0.645 | 0.631 | 0.636 | 0.792 | 0.823 | 0.810 | 0.810 | 1.000 | ||||||||||||
| Js2 | 0.624 | 0.607 | 0.614 | 0.613 | 0.629 | 0.770 | 0.800 | 0.802 | 0.802 | 0.900 | 1.000 | |||||||||||
| Js3 | 0.638 | 0.621 | 0.629 | 0.615 | 0.655 | 0.786 | 0.816 | 0.817 | 0.818 | 0.884 | 0.944 | 1.000 | ||||||||||
| Js4 | 0.610 | 0.604 | 0.612 | 0.599 | 0.626 | 0.754 | 0.784 | 0.786 | 0.785 | 0.865 | 0.873 | 0.926 | 1.000 | |||||||||
| Bd1 | 0.621 | 0.628 | 0.659 | 0.633 | 0.638 | 0.768 | 0.798 | 0.786 | 0.815 | 0.917 | 0.873 | 0.908 | 0.889 | 1.000 | ||||||||
| Bd2 | 0.631 | 0.614 | 0.633 | 0.631 | 0.647 | 0.764 | 0.794 | 0.795 | 0.780 | 0.892 | 0.850 | 0.884 | 0.865 | 0.917 | 1.000 | |||||||
| Bd3 | 0.627 | 0.621 | 0.629 | 0.638 | 0.631 | 0.758 | 0.787 | 0.789 | 0.774 | 0.901 | 0.842 | 0.843 | 0.825 | 0.874 | 0.936 | 1.000 | ||||||
| Jd1 | 0.761 | 0.719 | 0.702 | 0.736 | 0.729 | 0.693 | 0.719 | 0.721 | 0.706 | 0.729 | 0.682 | 0.684 | 0.679 | 0.705 | 0.756 | 0.764 | 1.000 | |||||
| Jd2 | 0.775 | 0.746 | 0.716 | 0.750 | 0.768 | 0.707 | 0.734 | 0.723 | 0.696 | 0.718 | 0.672 | 0.687 | 0.669 | 0.695 | 0.731 | 0.738 | 0.935 | 1.000 | ||||
| Jd3 | 0.772 | 0.743 | 0.725 | 0.746 | 0.765 | 0.703 | 0.730 | 0.719 | 0.704 | 0.713 | 0.667 | 0.682 | 0.690 | 0.703 | 0.727 | 0.708 | 0.918 | 0.967 | 1.000 | |||
| U1 | 0.721 | 0.742 | 0.699 | 0.733 | 0.752 | 0.714 | 0.742 | 0.719 | 0.702 | 0.726 | 0.690 | 0.720 | 0.702 | 0.730 | 0.740 | 0.734 | 0.657 | 0.709 | 0.692 | 1.000 | ||
| U2 | 0.733 | 0.716 | 0.686 | 0.733 | 0.752 | 0.689 | 0.716 | 0.706 | 0.677 | 0.698 | 0.664 | 0.693 | 0.688 | 0.702 | 0.726 | 0.706 | 0.657 | 0.684 | 0.679 | 0.946 | 1.000 | |
| U3 | 0.691 | 0.763 | 0.719 | 0.703 | 0.721 | 0.672 | 0.699 | 0.676 | 0.710 | 0.693 | 0.646 | 0.674 | 0.656 | 0.683 | 0.693 | 0.701 | 0.629 | 0.679 | 0.650 | 0.938 | 0.921 | 1.000 |
Molecular markers specific to 17 accessions were identified (Table 4). These markers could be converted to SCAR primers for enhancing the reliability of the markers.
Table 4.
Accession-specific molecular markers for Citrullus colocynthis
| S. no. | Genotype | Marker type | Primer identity | Amplicons size (bp) |
|---|---|---|---|---|
| 1. | Js1, Bd1 | ISSR | UBC-803, UBC-809 | 1100, 550 |
| 2. | G2, Js3 | ISSR | UBC-805 | 1100, 1250 |
| 3. | C3 | ISSR | UBC-807 | 450 |
| 4. | Js2 | ISSR | UBC-809 | 550 |
| 5. | S2 | ISSR | UBC-813 | 1250 |
| 6. | Bd3 | ISSR | UBC-14 | 500 |
| 7. | H1, Bd2 | RAPD | OPA-05 | 1000, 400 |
| 8. | H2, Bd3 | RAPD | OPA-10 | 1000 |
| 9. | B2 | RAPD | OPT-04 | 1500 |
| 10. | G2 | RAPD | OPA-18, OPA-19, OPA-05 | 1750, 600, 550 |
| 11. | Js4 | RAPD | OPB-04 | 2200 |
| 12 | Js2, Js3 | RAPD | OPE-12 | 450 |
| 13 | Jh3 | RAPD | OPF-1 | 600 |
| 14 | J1 | RAPD | OPF-19 | 600 |
| 15 | G1 | RAPD | OPF-19, OPT-14 | 600, 250 |
| 16 | B3, B4 | RAPD | OPT-04, OPR-02 | 2000, 800 |
| 17 | H2 | RAPD | OPT-14 | 250 |
Combined RAPD and ISSR analyses
The combined RAPD and ISSR data showed that total 63 primers were used which produce 417 polymorphic bands out of 715 total bands with 60.09% polymorphism (Table 5). The RAPD and ISSR data were also combined for UPGMA cluster analysis. The UPGMA dendrogram thus obtained from the cluster analysis of RAPD and ISSR data is shown in Fig. 5c. Dendrogram constructed on the basis of RAPD + ISSR polymorphism separated the accessions into four distinct clusters at 72% variation (Fig. 5c) with Jaccard’s similarity coefficient ranging from 0.64 (Js1 and H1) to 0.95 (Jh2 and Jh2) (Table 8). A cluster consisting of 11 accessions and is divided into two sub-clusters (Ia and Ib). Sub-cluster Ia comprised of A1 and A2. Sub-cluster Ib comprised of Jh1–Jh3, C1–C3, and Jd1–Jd3. Cluster II was the largest cluster consisting 13 accessions divided into two sub-clusters IIa and IIb comprised of J1–J5, S1–S3, H1–H2, and G1–G3. Groups III and IV also divided into two sub-clusters (IIIa, IIIb and IVa, IVb) which consisting 14 accessions including U1–U3, B1–B4, Js1–Js4, and Bd1–Bd3. The clustering pattern of the genotypes in the combined analysis remained akin to the ISSR dendrogram, while the RAPD-based dendrogram showed some variation in the clustering of genotypes.
Table 5.
Relative efficiency of molecular markers for determining polymorphism in Citrullus colocynthis
| S. no. | Parameters for marker efficiency | Molecular marker system | ||
|---|---|---|---|---|
| RAPD | ISSR | RAPD + ISSR | ||
| 1. | Number of individuals | 38 | 38 | 38 |
| 2. | Total number of scorable bands (NSB = L) | 549 | 166 | 715 |
| 3. | Total no. of assays/primers combination (T) | 50 | 13 | 63 |
| 4. | Total number of Polymorphic bands (p) | 318 | 99 | 417 |
| 5. | Multiplex ratio (MR) = (L/T) | 10.98 | 12.76 | 11.34 |
| 6. | % Polymorphism | 55.14 | 65.04 | 60.09 |
| 7. | Average polymorphism (bands primer−1) | 6.36 | 7.60 | 6.98 |
| 8. | PIC average (PICavg) | 0.47 | 0.44 | 0.46 |
| 9. | Average heterozygosity (H avg) | 0.49 | 0.46 | 0.48 |
| 10. | Marker index (MI) = H avg × MR | 5.32 | 5.86 | 5.45 |
| 11. | Average informativeness of a band (BIi) | 0.45 | 0.46 | 0.45 |
| 12. | Resolving power (RP) | 2.98 | 4.10 | 3.21 |
Table 8.
Jaccard’s similarity coefficient of 38 Citrullus colocynthis accessions based on RAPD + ISSR data analysis
| A1 | A2 | J1 | J2 | J3 | J4 | J5 | S1 | S2 | S3 | Jh1 | Jh2 | Jh3 | C1 | C2 | C3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 1.000 | |||||||||||||||
| A2 | 0.899 | 1.000 | ||||||||||||||
| J1 | 0.742 | 0.754 | 1.000 | |||||||||||||
| J2 | 0.751 | 0.750 | 0.882 | 1.000 | ||||||||||||
| J3 | 0.744 | 0.734 | 0.887 | 0.919 | 1.000 | |||||||||||
| J4 | 0.744 | 0.736 | 0.861 | 0.896 | 0.927 | 1.000 | ||||||||||
| J5 | 0.763 | 0.739 | 0.876 | 0.896 | 0.920 | 0.939 | 1.000 | |||||||||
| S1 | 0.764 | 0.744 | 0.838 | 0.853 | 0.876 | 0.876 | 0.910 | 1.000 | ||||||||
| S2 | 0.751 | 0.725 | 0.811 | 0.829 | 0.852 | 0.848 | 0.882 | 0.910 | 1.000 | |||||||
| S3 | 0.759 | 0.732 | 0.811 | 0.829 | 0.845 | 0.852 | 0.877 | 0.894 | 0.928 | 1.000 | ||||||
| Jh1 | 0.756 | 0.767 | 0.723 | 0.723 | 0.744 | 0.734 | 0.759 | 0.783 | 0.780 | 0.818 | 1.000 | |||||
| Jh2 | 0.753 | 0.752 | 0.699 | 0.711 | 0.735 | 0.728 | 0.750 | 0.767 | 0.761 | 0.798 | 0.958 | 1.000 | ||||
| Jh3 | 0.757 | 0.758 | 0.721 | 0.717 | 0.741 | 0.741 | 0.754 | 0.778 | 0.762 | 0.785 | 0.932 | 0.929 | 1.000 | |||
| C1 | 0.748 | 0.750 | 0.725 | 0.718 | 0.745 | 0.732 | 0.748 | 0.768 | 0.743 | 0.779 | 0.917 | 0.914 | 0.941 | 1.000 | ||
| C2 | 0.747 | 0.742 | 0.726 | 0.738 | 0.750 | 0.743 | 0.759 | 0.780 | 0.757 | 0.794 | 0.922 | 0.909 | 0.913 | 0.939 | 1.000 | |
| C3 | 0.743 | 0.745 | 0.726 | 0.735 | 0.750 | 0.731 | 0.756 | 0.773 | 0.757 | 0.784 | 0.919 | 0.902 | 0.905 | 0.917 | 0.953 | 1.000 |
| H1 | 0.726 | 0.734 | 0.730 | 0.730 | 0.750 | 0.747 | 0.750 | 0.745 | 0.723 | 0.734 | 0.796 | 0.791 | 0.791 | 0.795 | 0.796 | 0.809 |
| H2 | 0.718 | 0.716 | 0.772 | 0.782 | 0.787 | 0.783 | 0.790 | 0.772 | 0.749 | 0.763 | 0.738 | 0.738 | 0.751 | 0.749 | 0.735 | 0.744 |
| G1 | 0.688 | 0.699 | 0.769 | 0.762 | 0.784 | 0.777 | 0.790 | 0.759 | 0.743 | 0.753 | 0.720 | 0.720 | 0.730 | 0.730 | 0.720 | 0.720 |
| G2 | 0.701 | 0.691 | 0.764 | 0.770 | 0.779 | 0.798 | 0.802 | 0.777 | 0.750 | 0.764 | 0.730 | 0.727 | 0.737 | 0.716 | 0.730 | 0.733 |
| G3 | 0.728 | 0.712 | 0.768 | 0.787 | 0.806 | 0.813 | 0.833 | 0.800 | 0.787 | 0.805 | 0.775 | 0.762 | 0.759 | 0.738 | 0.755 | 0.755 |
| B1 | 0.688 | 0.702 | 0.685 | 0.690 | 0.705 | 0.704 | 0.723 | 0.736 | 0.717 | 0.722 | 0.738 | 0.723 | 0.726 | 0.703 | 0.708 | 0.708 |
| B2 | 0.698 | 0.714 | 0.680 | 0.682 | 0.694 | 0.688 | 0.705 | 0.719 | 0.700 | 0.707 | 0.742 | 0.724 | 0.721 | 0.710 | 0.718 | 0.720 |
| B3 | 0.695 | 0.697 | 0.671 | 0.685 | 0.685 | 0.687 | 0.696 | 0.707 | 0.697 | 0.707 | 0.721 | 0.709 | 0.704 | 0.701 | 0.709 | 0.703 |
| B4 | 0.697 | 0.702 | 0.679 | 0.696 | 0.702 | 0.698 | 0.714 | 0.721 | 0.714 | 0.719 | 0.732 | 0.717 | 0.733 | 0.718 | 0.723 | 0.717 |
| Js1 | 0.676 | 0.673 | 0.661 | 0.672 | 0.678 | 0.689 | 0.698 | 0.690 | 0.686 | 0.706 | 0.708 | 0.702 | 0.705 | 0.691 | 0.687 | 0.684 |
| Js2 | 0.653 | 0.652 | 0.669 | 0.674 | 0.697 | 0.696 | 0.708 | 0.698 | 0.673 | 0.689 | 0.709 | 0.703 | 0.703 | 0.695 | 0.691 | 0.685 |
| Js3 | 0.677 | 0.674 | 0.698 | 0.712 | 0.724 | 0.724 | 0.740 | 0.719 | 0.700 | 0.710 | 0.724 | 0.712 | 0.713 | 0.698 | 0.703 | 0.700 |
| Js4 | 0.666 | 0.674 | 0.718 | 0.720 | 0.736 | 0.725 | 0.745 | 0.740 | 0.707 | 0.715 | 0.732 | 0.716 | 0.714 | 0.714 | 0.722 | 0.722 |
| Bd1 | 0.684 | 0.692 | 0.718 | 0.736 | 0.738 | 0.757 | 0.751 | 0.746 | 0.723 | 0.737 | 0.735 | 0.723 | 0.732 | 0.720 | 0.728 | 0.728 |
| Bd2 | 0.684 | 0.678 | 0.687 | 0.723 | 0.716 | 0.719 | 0.738 | 0.730 | 0.714 | 0.721 | 0.731 | 0.719 | 0.717 | 0.717 | 0.719 | 0.719 |
| Bd3 | 0.679 | 0.672 | 0.687 | 0.714 | 0.713 | 0.713 | 0.728 | 0.720 | 0.695 | 0.712 | 0.713 | 0.707 | 0.711 | 0.714 | 0.707 | 0.710 |
| Jd1 | 0.759 | 0.769 | 0.709 | 0.720 | 0.717 | 0.719 | 0.734 | 0.742 | 0.717 | 0.742 | 0.777 | 0.768 | 0.769 | 0.772 | 0.774 | 0.777 |
| Jd2 | 0.759 | 0.746 | 0.697 | 0.723 | 0.708 | 0.710 | 0.728 | 0.736 | 0.708 | 0.727 | 0.762 | 0.753 | 0.757 | 0.754 | 0.768 | 0.759 |
| Jd3 | 0.765 | 0.782 | 0.727 | 0.720 | 0.729 | 0.716 | 0.731 | 0.733 | 0.732 | 0.749 | 0.790 | 0.778 | 0.772 | 0.773 | 0.768 | 0.768 |
| U1 | 0.685 | 0.693 | 0.706 | 0.714 | 0.729 | 0.723 | 0.735 | 0.746 | 0.740 | 0.754 | 0.748 | 0.729 | 0.736 | 0.718 | 0.714 | 0.726 |
| U2 | 0.685 | 0.687 | 0.725 | 0.715 | 0.730 | 0.720 | 0.749 | 0.744 | 0.731 | 0.745 | 0.730 | 0.708 | 0.712 | 0.706 | 0.699 | 0.708 |
| U3 | 0.676 | 0.693 | 0.707 | 0.718 | 0.727 | 0.723 | 0.739 | 0.734 | 0.721 | 0.732 | 0.720 | 0.702 | 0.724 | 0.713 | 0.699 | 0.702 |
| H1 | H2 | G1 | G2 | G3 | B1 | B2 | B3 | B4 | Js1 | Js2 | Js3 | Js4 | Bd1 | Bd2 | Bd3 | Jd1 | Jd2 | Jd3 | U1 | U2 | U3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | ||||||||||||||||||||||
| A2 | ||||||||||||||||||||||
| J1 | ||||||||||||||||||||||
| J2 | ||||||||||||||||||||||
| J3 | ||||||||||||||||||||||
| J4 | ||||||||||||||||||||||
| J5 | ||||||||||||||||||||||
| S1 | ||||||||||||||||||||||
| S2 | ||||||||||||||||||||||
| S3 | ||||||||||||||||||||||
| Jh1 | ||||||||||||||||||||||
| Jh2 | ||||||||||||||||||||||
| Jh3 | ||||||||||||||||||||||
| C1 | ||||||||||||||||||||||
| C2 | ||||||||||||||||||||||
| C3 | ||||||||||||||||||||||
| H1 | 1.000 | |||||||||||||||||||||
| H2 | 0.858 | 1.000 | ||||||||||||||||||||
| G1 | 0.808 | 0.902 | 1.000 | |||||||||||||||||||
| G2 | 0.817 | 0.879 | 0.905 | 1.000 | ||||||||||||||||||
| G3 | 0.817 | 0.883 | 0.865 | 0.905 | 1.000 | |||||||||||||||||
| B1 | 0.694 | 0.699 | 0.694 | 0.719 | 0.760 | 1.000 | ||||||||||||||||
| B2 | 0.698 | 0.677 | 0.675 | 0.699 | 0.723 | 0.925 | 1.000 | |||||||||||||||
| B3 | 0.687 | 0.674 | 0.668 | 0.678 | 0.708 | 0.873 | 0.916 | 1.000 | ||||||||||||||
| B4 | 0.697 | 0.706 | 0.706 | 0.704 | 0.735 | 0.865 | 0.875 | 0.882 | 1.000 | |||||||||||||
| Js1 | 0.649 | 0.656 | 0.656 | 0.665 | 0.689 | 0.761 | 0.749 | 0.740 | 0.798 | 1.000 | ||||||||||||
| Js2 | 0.662 | 0.652 | 0.660 | 0.667 | 0.679 | 0.749 | 0.725 | 0.711 | 0.762 | 0.891 | 1.000 | |||||||||||
| Js3 | 0.681 | 0.677 | 0.683 | 0.693 | 0.708 | 0.756 | 0.741 | 0.726 | 0.777 | 0.844 | 0.919 | 1.000 | ||||||||||
| Js4 | 0.693 | 0.686 | 0.683 | 0.693 | 0.700 | 0.772 | 0.763 | 0.754 | 0.783 | 0.782 | 0.820 | 0.877 | 1.000 | |||||||||
| Bd1 | 0.676 | 0.702 | 0.705 | 0.712 | 0.719 | 0.788 | 0.772 | 0.750 | 0.799 | 0.816 | 0.812 | 0.835 | 0.866 | 1.000 | ||||||||
| Bd2 | 0.691 | 0.681 | 0.681 | 0.685 | 0.709 | 0.758 | 0.764 | 0.762 | 0.798 | 0.787 | 0.794 | 0.809 | 0.824 | 0.892 | 1.000 | |||||||
| Bd3 | 0.685 | 0.690 | 0.687 | 0.688 | 0.700 | 0.742 | 0.742 | 0.753 | 0.771 | 0.787 | 0.790 | 0.785 | 0.806 | 0.873 | 0.927 | 1.000 | ||||||
| Jd1 | 0.777 | 0.735 | 0.732 | 0.736 | 0.734 | 0.691 | 0.701 | 0.695 | 0.697 | 0.688 | 0.695 | 0.695 | 0.692 | 0.702 | 0.717 | 0.735 | 1.000 | |||||
| Jd2 | 0.753 | 0.729 | 0.714 | 0.721 | 0.731 | 0.673 | 0.692 | 0.689 | 0.673 | 0.662 | 0.666 | 0.677 | 0.668 | 0.675 | 0.693 | 0.708 | 0.913 | 1.000 | ||||
| Jd3 | 0.768 | 0.742 | 0.739 | 0.725 | 0.743 | 0.691 | 0.701 | 0.686 | 0.697 | 0.691 | 0.678 | 0.680 | 0.675 | 0.702 | 0.702 | 0.693 | 0.883 | 0.878 | 1.000 | |||
| U1 | 0.691 | 0.730 | 0.718 | 0.738 | 0.751 | 0.696 | 0.682 | 0.658 | 0.687 | 0.702 | 0.688 | 0.694 | 0.698 | 0.743 | 0.723 | 0.717 | 0.687 | 0.684 | 0.711 | 1.000 | ||
| U2 | 0.700 | 0.718 | 0.715 | 0.723 | 0.745 | 0.681 | 0.668 | 0.665 | 0.679 | 0.664 | 0.674 | 0.683 | 0.699 | 0.702 | 0.711 | 0.705 | 0.688 | 0.679 | 0.676 | 0.896 | 1.000 | |
| U3 | 0.703 | 0.731 | 0.718 | 0.726 | 0.739 | 0.681 | 0.668 | 0.653 | 0.688 | 0.676 | 0.674 | 0.689 | 0.683 | 0.690 | 0.693 | 0.690 | 0.703 | 0.700 | 0.697 | 0.881 | 0.875 | 1.000 |
The principal component analysis based on RAPD + ISSR polymorphism grouped the accessions into four major clusters and eight minor clusters (Fig. 6c). A few differences in clustering were observed with UPGMA clustering and principal component analysis. Principal component analysis showed three major groups consisting U1–U3, A1–A2, Jd1–Jd3, G1–G2, H1–H2, and J1 in the first group, accessions J2–J5, S1–S3, C1–C3, Jh1–Jh3, and G3 in the second group, and third group consisting accessions Bd2–Bd3, B1–B4, Js1–Js4, and U1–U3 in PCA. The matrices for two markers, RAPD and ISSR, were also compared using Mantel’s test. The correlation value between the matrices is high (r = 0.7947) indicating a good correlation between the two molecular marker systems (Mantel, 1967).
The results of the relative efficiency of combined RAPD + ISSR markers for determining polymorphism in C. colocynthis showed PIC value and heterozygosity ≥0.50. The estimates of marker attributes for RAPD + ISSR showed slightly different results. The average value of MR and MI was 11.34 and 5.45 for combined RAPD + ISSR markers. The average informativeness of a band (BIi) and resolving power (RP) was 0.45 and 3.21, respectively, for RAPD markers (Table 5).
Discussion
In the present study, the molecular diversity analysis was carried out using RAPD and ISSR markers to assess the phylogenetic relation at inter-specific level amongst 38 accessions of C. colocynthis from the different geographical zones of Rajasthan district, India. The 50 RAPD primers revealed polymorphic banding patterns amongst 38 accessions of C. colocynthis with 318 polymorphic bands out of 549 bands that covered the molecular size between 150 and 6000 bp. The amplified fragments ranged from five (OPA-08 and OPF-9) to nineteen (OPT-20) with an average of 6.36 polymorphic bands (PB) per primer (P). This average polymorphic band per primer is closely related with earlier studies as 6.83 polymorphic fragments per primer in Ricinus communis as (Gajera et al. 2010), 7.6 PB/P in Pistacia khinjuk (Karimi et al. 2012) and 7.6 PB/P in Iranian Pistachios (Aliakbarkhani et al. 2015). However, earlier observations also reflect less number of polymorphic band per primer in various plant species such as, 2.9 PB/P in bread wheat (Khaled et al. 2015), 3.58 PB/P in Cucumis sativus (manohar et al. 2013), 4.17 PB/P in Citrullus lanatus (Solmaz et al. 2010) and in barley 5.67 PB/P (Guasmi et al. 2012). Contrary to this, deviations in polymorphic bands by RAPD markers were also reported extremely high values such as 13.87 PB/P in Hordeum vulgare (Eshghi et al. 2012) and 12.2 PB/P in Citrullus lanatus (Mujaju et al. 2010).
Similarly, 13 ISSRs markers out of 20 markers showed amplification profile in 38 accessions of C. colocynthis with 99 polymorphic bands out of 166 amplification bands with a molecular size between 250 and 4000 bp. The number of bands amplified per primer varied between 9 (UBC-807, 802) and 16 (UBC-803, 812) with an average of 7.6 polymorphic bands per primer (PB/P). This average result is similar to previous reports of 7.4 PB/P in sandalwood (Patel et al. 2016) and 8.0 PB/P in Jatropha curcas (Khurana-Kaul et al. 2012). However, earlier observations of less average polymorphic band per primer from the present study were also reported in different plant species using ISSR markers viz. 2.8 PB/P in Benincasa hispida (Verma et al. 2007), 4.4 PB/P in sugarcane Germplasm (Khaled et al. 2015) and 5.12 PB/P in chickpea (Rao et al. 2007). Contrary to this, high average value of the present work has been reported in other plant species such as, 17.78 PB/P in Primula obconica (Nan et al. 2003), 16.87 PB/P in Vigna radiata (Singh et al. 2012), 15 PB/P in Artemisia herba-alba (Mohsen and Ali 2008), 14.4 PB/P in sugar beet (Izzatullayeva et al. 2014), 13 PB/P in Miscanthus (Hodkinson et al. 2002), 12.6 PB/P in Citrullus lanatus (Dje et al. 2010 ) and 11.6 PB/P in Cucumis melo (Stepansky et al. 1999).
Currently, the average value of percent polymorphism for ISSR markers was 65.05% which was higher than the average value of percent polymorphism for RAPD markers that was 55.14% amongst 38 accessions of C. colocynthis. This pattern of observation complies with findings by Levi et al. (2005) in which ISSR marker was more polymorphic compared to RAPD markers. This can be ascribed as ISSR markers are the class of microsatellite region, distributed throughout the genome, and having high mutation rate along with its may link with functionally important loci (Penner 1996), while RAPD markers are located in non-coding regions and are selectively neutral. Moreover, the practicing of ISSR technologies found to be more reproducible (Goulao and Oliveira 2001), more effective for uncovering polymorphism (Mahar et al. 2011; Gorji et al. 2011) and obtained more composite marker patterns than RAPD markers (Chowdhury et al. 2002). Previous observations have been mentioned that ISSR markers found to be more effective than RAPD markers and differentiated closely related cultivars in various plant species (Singh et al. 2012). Contrary to these reports, others enough observations that RAPD markers revealed more polymorphism than ISSR markers in different plant species (Farajpour et al. 2011; Patel et al. 2016). This may be possible that these divergences are due to the proportion of coding and non-coding sequences within the genome and differences in genome composition of species.
The genetic diversity was also established amongst 38 accessions of C. colocynthis belonging to the different geographical zones of Rajasthan state (India) using RAPD, ISSR, and RAPD + ISSR markers. For the same, DNA fingerprinting employed using both markers to study the extent of genetic diversity across a set of germplasm or cultivars and group them into specific categories. Both the markers revealed above 50% percent polymorphism and almost comparable polymorphic information content (PIC). Therefore, the use of PIC analysis is required to identify most appropriate marker selection for genetic mapping and phylogenetic analysis (Powell et al. 1996). Enormous studies on the assessment of genetic diversity of the diverse group of plant species using molecular markers have been established a correlation between geographical distance and genetic similarity between individuals (Islam and Alam 2004). Beside PIC and heterozygosity, our study also evaluated the discriminatory power of RAPD and ISSR primers for genetic relationship studies through the marker index (MI), multiplex ratio (MR), average informativeness of bands (BIi), and resolving power (RP); features that to the best of our knowledge have not yet been reported in other molecular studies in C. Colocynthis. The average PIC and heterozygosity values are higher for RAPD (0.47, 0.49) markers instead of ISSR (0.44, 0.46) showed that RAPD markers have higher polymorphism than ISSR. The marker index varied from 0.302 to 3.017 (average 1.575) and has been used to assess the informativeness of various markers in several crop species, soybean (Powell et al. 1996), wheat (Bohn et al. 1999), corn salad (Muminovic et al. 2004), and jatropha (Grativol et al. 2011), but in C. colocynthis, we observed that average MI value was 5.86 higher to RAPD 5.32. Resolving powers in our study were in the range of 0.63–5.11 (average 2.98) for RAPD and ISSR 1.56–8.18 (average 4.10) per primer. According to Fang and Roose (1997) and Naik et al. (2017), ISSR markers were found to be more effective in diversity study than RAPD markers. The present study clearly supports the view. Average MI value of both the markers is more or less similar (5.32–5.86), but average RP value (4.10) of ISSR primers was more than RAPD primers (2.98). Such variation shown by ISSR markers is due to selective amplification. They amplify conserved regions present between the microsatellite repeat sequences, but RAPD markers are not selective; rather, they amplify any regions within the entire genome (Zietkiewicz et al. 1994). The values of multiplex ratio (MR), marker index (MI), average informativeness of a band (BIi), and resolving power (RP) for ISSR marker proved that these markers are more informative to detect polymorphism. Furthermore, our results represent that both RAPD and ISSR markers have the similar potential for phylogenetic relationships in 38 accessions of C. colocynthis with a moderate amount of similarity exists among the both marker systems. Therefore, arbitrary primers have enormous potential for resolving intra- and intergenomic relationships and different marker technologies target the different portions of the genome. While UPGMA-based dendrograms revealed geographic similarity and some differences in the area magnitude were also reported among the different accessions with a particular type of molecular markers technology. Moreover, high reproducibility in dendrogram topologies was obtained for both RAPD and ISSR markers with a few differences between the species and different accessions of C. colocytnhis. These differences may be attributed to marker sampling error and/or the level of polymorphism detected, reinforcing again the importance of the number of loci and their coverage of the overall genome in obtaining reliable estimates of genetic relationships among cultivars (Loarce et al. 1996). Therefore, the evolution of varieties in distinct agro-climatic zones also demonstrates significant levels of variation in response to the selection pressure in the zones (Singh et al. 1998). Likewise, a huge number of studies also evaluated to genetic relationship establishments in a variety of cultivars and plant species, such as in Curcuma longa (Singh et al. 2012), Jatropha curcas (Khurana-Kaul et al. 2012), Cajanus cajan (Yadav et al. 2014), Dioscorea spp. (Velasco-Ramirez et al. 2014), Cymbopogon flexuosus (Debajit et al. 2015), and sandalwood (Patel et al. 2016).
In conclusion, the present study was intended to assess molecular characterization, marker polymorphisms, and genetic diversity analysis amongst 38 different accessions of C. colocytnhis from the distinct geographical zone. Likewise, RAPD and ISSR fingerprinting technologies were employed for genetic analysis which provides a powerful tool for the generation of potential diagnostic of markers for cultivar analysis. Fifty RAPD markers and thirteen ISSR markers revealed clear amplification in 38 different C. colocytnhis with percent polymorphism 55.14 and 65.05%, respectively. Furthermore, genetic diversity was established through dendrogram construction amongst all the different accessions of C. colocytnhis using RAPD, ISSR, and RAPD + ISSR marker technologies. Both the markers indicate an important level of genetic differences amongst 38 distinct accessions of C. colocytnhis in relation with their different geographical distributions and some genetic differences were also obtained from different types of marker technologies. Thus, the study provides an evidence for selecting of marker technologies further for breeding, integration into the marker-assisted selection and comparative genetic studies.
Acknowledgements
Authors are thankful to CSIR for financial support and for the award of SRF during Ph.D., Interdisciplinary Program in life Science (IPLS), University with Potential for Excellence Scheme by UGC (UPE Program), DRS-II, Dept. of Botany, Bioinformatics Infrastructure Facility (BIF) by DBT and University of Rajasthan are gratefully acknowledged.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Aliakbarkhani ST, Akbari M, Hassankhah A, et al. Phenotypic and genotypic variation in Iranian Pistachios. J Genet Eng Biotechnol. 2015;13(2):235–241. doi: 10.1016/j.jgeb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JA, Churchill JE, Autrique SD, et al. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–188. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Bhandari MM. Flora of the Indian desert. Jodhpur: MPS Repros; 1995. [Google Scholar]
- Bohn MH, Utz F, Melchinger AE. Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs and SSRs and their use for predicting progeny variance. Crop Sci. 1999;39:228–237. doi: 10.2135/cropsci1999.0011183X003900010035x. [DOI] [Google Scholar]
- Chowdhury MA, Vandenberg B, Warkentin T. Cultivar identification and genetic relationship among selected breeding lines and cultivars in chickpea (Cicer arietinum L.) Euphytica. 2002;127:317–325. doi: 10.1023/A:1020366819075. [DOI] [Google Scholar]
- Csillery K, Blum MGB, Gaggiotti OE, Francois O. Approximate bayesian computation (ABC) in practice. Trends Ecol Evol. 2010;25:410–418. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Dane F, Liu J, Zhang C. Phylogeography of the bitter apple, Citrullus colocynthis. Genet Resour Crop Evol. 2006;54:327–336. doi: 10.1007/s10722-005-4897-2. [DOI] [Google Scholar]
- Debajit S, Sukriti D, Sneha G, Mohan L, et al. RAPD and ISSR based Intra-specific molecular genetic diversity analysis of Cymbopogon flexuosus L. Stapf with a distinct correlation of morpho-chemical observations. Res J Biotechnol. 2015;10(7):105–113. [Google Scholar]
- Dje Y, Tahi CG, Bi AZ, et al. Use of ISSR markers to assess genetic diversity of African edible seeded Citrullus lanatus landraces. Sci Hortic. 2010;124(2):159–164. doi: 10.1016/j.scienta.2009.12.020. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Eshghi R, Ebrahimpour F, Ojaghi J, et al. Evaluation of genetic variability in naked barley (Hordeum vulgare L.) Intl J Agric Crop Sci. 2012;4:1166–1179. [Google Scholar]
- Fang DQ, Roose ML. Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor Appl Genet. 1997;95:408–417. doi: 10.1007/s001220050577. [DOI] [Google Scholar]
- Farajpour M, Ebrahimi M, Amiri R, et al. Study of genetic variation in yarrow using inter-simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol. 2011;10(54):11137–11141. [Google Scholar]
- Gajera BB, Kumar N, Singh AS, et al. Assessment of genetic diversity in castor (Ricinus communis L.) using RAPD and ISSR markers. Ind Crops Prod. 2010;32(3):491–498. doi: 10.1016/j.indcrop.2010.06.021. [DOI] [Google Scholar]
- Gitzendanner MA, Soltis PS. Patterns of genetic variation in rare and widespread plant congeners. Am J Bot. 2000;87:783–792. doi: 10.2307/2656886. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Coulson M, Brettell R. Development of DNA markers (ISSRs) in mango. Acta Hortic. 2000;575:139–143. [Google Scholar]
- Gorji AM, Poczai P, Polgar Z, et al. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res. 2011;88(3):226–237. doi: 10.1007/s12230-011-9187-2. [DOI] [Google Scholar]
- Goulao L, Oliveira CM. Molecular characterization of cultivars of apple (Malus × domestica Borkh.) using microsatellite (SSR and ISSR) markers. Euphytica. 2001;122:81–89. doi: 10.1023/A:1012691814643. [DOI] [Google Scholar]
- Grativol C, da Fonseca Lira-Medeiros C, Hemerly AS, Ferreira PCG. High efficiency and reliability of inter-simple sequence repeats (ISSR) markers for evaluation of genetic diversity in Brazilian cultivated Jatropha curcas L. accessions. Mol Biol Rep. 2011;38:4245–4256. doi: 10.1007/s11033-010-0547-7. [DOI] [PubMed] [Google Scholar]
- Guasmi F, Elfalleh W, Hannachi H et al (2012) The use of ISSR and RAPD markers for genetic diversity among South Tunisian barley. ISrN Agronomy
- Hodkinson TR, Chase MW, Renvoize SA. Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropogoneae, Poaceae) using AFLP and ISSR PCR. Ann Bot. 2002;89(5):627–636. doi: 10.1093/aob/mcf091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Alam MS. Randomly amplified polymorphic DNA analysis of four different populations of the Indian major carp, Labeo rohita (Hamilton) J Appl Ichthyol. 2004;20(5):407–412. doi: 10.1111/j.1439-0426.2004.00588.x. [DOI] [Google Scholar]
- Izzatullayeva V, Akparov Z, Babayeva S, et al. Efficiency of using RAPD and ISSR markers in evaluation of genetic diversity in sugar beet. Turk J Biol. 2014;38(4):429–438. doi: 10.3906/biy-1312-35. [DOI] [Google Scholar]
- Jain KK. Personalized medicine. Curr Opin Mol Ther. 2002;4:548–558. [PubMed] [Google Scholar]
- Kapteyn J, Goldsbrough P, Simon J. Genetic relationships and diversity of commercially relevant Echinacea species. Theor Appl Genet. 2002;105:369–376. doi: 10.1007/s00122-002-0960-y. [DOI] [PubMed] [Google Scholar]
- Karimi HR, Abadi MHH, Kohbanani AM. Genetic diversity of Pistacia khinjuk Stocks. using RAPD markers and leaf morphological characters. Plant Syst Evol. 2012;298(5):963–968. doi: 10.1007/s00606-012-0605-9. [DOI] [Google Scholar]
- Khaled AGA, Motawea MH, Said AA. Identification of ISSR and RAPD markers linked to yield traits in bread wheat under normal and drought conditions. J Genet Eng Biotechnol. 2015;13(2):243–252. doi: 10.1016/j.jgeb.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Al-Qurainy F, Nadeem M. Biotechnological approaches for conservation and improvement of rare and endangered plants of Saudi Arabia. Saudi J Biol Sci. 2012;19(1):1–11. doi: 10.1016/j.sjbs.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana-Kaul V, Kachhwaha S, Kothari SL. Characterization of genetic diversity in Jatropha curcas L. germplasm using RAPD and ISSR markers. Indian J Biotechnol. 2012;11(1):54–61. [Google Scholar]
- Laikre L, Allendorf FW, Aroner LC, et al. Neglect of genetic diversity in implementation of the conservation on biological diversity. Conserv Biol. 2009;24:86–88. doi: 10.1111/j.1523-1739.2009.01425.x. [DOI] [PubMed] [Google Scholar]
- Levi A, Thomas CE, Simmons AM, Thies JA. Analysis based on RAPD and ISSR markers reveals closer similarities among Citrullus and Cucumis species than with Praecitrullus fistulosus (Stocks) Pangalo. Genet Resour Crop Evol. 2005;52:465–472. doi: 10.1007/s10722-005-2260-2. [DOI] [Google Scholar]
- Liu BH. Statistical genomics: linkage, mapping and QTL analysis. Boca Raton: CRC Press; 1998. [Google Scholar]
- Loarce Y, Gallego R, Ferrer E. A comparative analysis of the genetic relationships between rye cultivars using RFLP and RAPD markers. Euphytica. 1996;88:107–115. doi: 10.1007/BF00032441. [DOI] [Google Scholar]
- Mahar KS, Rana TS, Ranade SA, Meena B. Genetic variability and population structure in Sapindus emarginatus Vahl from India. Gene. 2011;485(1):32–39. doi: 10.1016/j.gene.2011.05.036. [DOI] [PubMed] [Google Scholar]
- Manohar SH, Murthy HN, Ravishankar KV. Genetic diversity in a collection of Cucumis sativus L. assessed by RAPD and ISSR markers. J Plant Biochem Biotechnol. 2013;22(2):241–244. doi: 10.1007/s13562-012-0123-4. [DOI] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Moghaieb REA, Abdelhadi AA, El-Sadaw HA, et al. Molecular identification and genetic diversity among Photorhabdus and Xenorhabdus isolates. 3 Biotech. 2017;7(1):6. doi: 10.1007/s13205-016-0594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen H, Ali F. Study of genetic polymorphism of Artemisia herba-alba from Tunisia using ISSR markers. Afr J Biotechnol. 2008;7(1):44–50. [Google Scholar]
- Mujaju C, Sehic J, Werlemark G, et al. Genetic diversity in watermelon (Citrullus lanatus) landraces from Zimbabwe revealed by RAPD and SSR markers. Hereditas. 2010;147(4):142–153. doi: 10.1111/j.1601-5223.2010.02165.x. [DOI] [PubMed] [Google Scholar]
- Muminovic J, Melchinger AE, Lubberstedt T. Genetic diversity in corn salad (Valerianella locusta) and related species as determined by AFLP markers. Plant Breed. 2004;123:460–466. doi: 10.1111/j.1439-0523.2004.00998.x. [DOI] [Google Scholar]
- Naik A, Prajapat P, Krishnamurthy R, et al. Assessment of genetic diversity in Costus pictus accessions based on RAPD and ISSR markers. 3 Biotech. 2017;7:70. doi: 10.1007/s13205-017-0667-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan P, Shi S, Peng S, et al. Genetic diversity in Primula obconica (Primulaceae) from Central and South-west China as revealed by ISSR markers. Ann Bot. 2003;91(3):329–333. doi: 10.1093/aob/mcg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novot N, Sarfatti M, Zamir D. Linkage relationship of genes affecting bitterness and flesh color in watermelon. J Hered. 1990;81:162–165. [Google Scholar]
- Patel DM, Fougat RS, Sakure AA, et al. Detection of genetic variation in sandalwood using various DNA markers. 3 Biotech. 2016;6(1):1–11. doi: 10.1007/s13205-016-0391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner GA. RAPD analysis of plant genomes. In: Jauhar PP, editor. Methods of genome analysis in plants. Boca Raton: CRC; 1996. pp. 251–268. [Google Scholar]
- Powell W, Morgante M, Andre C, et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;2:225–238. doi: 10.1007/BF00564200. [DOI] [Google Scholar]
- Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato accessions. Theor Appl Genet. 1999;98:107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- Rao LS, Rani PU, Deshmukh PS, et al. RAPD and ISSR fingerprinting in cultivated chickpea (Cicer arietinum L.) and its wild progenitor Cicer reticulatum Ladizinsky. Genet Resour Crop Evol. 2007;54(6):1235–1244. doi: 10.1007/s10722-006-9104-6. [DOI] [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv Biol. 2003;17(1):230–237. doi: 10.1046/j.1523-1739.2003.01236.x. [DOI] [Google Scholar]
- Rohlf FJ. NTSYSpc: numerical taxonomy and multivariate analysis system, version 2.02e. Setauket: Exeter Software; 1998. [Google Scholar]
- Shah CS, Qadry JS. A text book of pharmacognosy. 5. Ahmedabad: B. S. Shah Prakashan; 1985. p. 284. [Google Scholar]
- Sharma R, Joshi A, Maloo SR, et al. Assessment of genetic finger printing using molecular marker in plants: a review. Sci Res Impact. 2012;1:29–36. [Google Scholar]
- Singh AK, Smart J, Simpson CE, et al. Genetic variation vis-a-vis molecular polymorphism in groundnut, Arachis hypogaea L. Genet Resour Crop Evol. 1998;45:119–126. doi: 10.1023/A:1008646422730. [DOI] [Google Scholar]
- Singh S, Reddy SK, Jawali N. Genetic diversity analyses of Mungbean (Vigna radiata [L]. Wilczek) by ISSR. Int J Plant Breed. 2012;6(2):73–83. [Google Scholar]
- Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco: W H Freeman and Company; 1973. [Google Scholar]
- Solmaz I, Sari N, Aka-Kacar Y, et al. The genetic characterization of Turkish watermelon (Citrullus lanatus) accessions using RAPD markers. Genet Resour Crop Evol. 2010;57(5):763–771. doi: 10.1007/s10722-009-9515-2. [DOI] [Google Scholar]
- Sreekumar VB, Renuka C. Assessment of genetic diversity in Calamus thwaitesii BECC (Arecaceae) using RAPD markers. Biochem Syst Ecol. 2006;34:397–405. doi: 10.1016/j.bse.2005.12.002. [DOI] [Google Scholar]
- Stepansky A, Kovalski I, Perl-Treves R. Intraspecific classification of melons (Cucumis melo L.) in view of their phenotypic and molecular variation. Plant Syst Evol. 1999;217(3–4):313–332. doi: 10.1007/BF00984373. [DOI] [Google Scholar]
- Sturm SP, Schveider C, Seger SH. Analysis of Citrullus colocynthis cucurbitacin derivatives with HPLC-SPE-NMR. Sci Pharm. 2009;77:254–257. doi: 10.3797/scipharm.oephg.21.PO-55. [DOI] [Google Scholar]
- Velasco-Ramirez AP, Torres-Moran MI, Molina-Moret S, et al. Efficiency of RAPD, ISSR, AFLP and ISTR markers for the detection of polymorphisms and genetic relationships in camote de cerro (Dioscorea spp.) Electron J Biotechnol. 2014;17(2):65–71. doi: 10.1016/j.ejbt.2014.01.002. [DOI] [Google Scholar]
- Verma VK, Behera TK, Munshi AD, et al. Genetic diversity of ash gourd [Benincasa hispida (Thunb.) Cogn.] inbred lines based on RAPD and ISSR markers and their hybrid performance. Sci Hortic. 2007;113(3):231–237. doi: 10.1016/j.scienta.2007.03.019. [DOI] [Google Scholar]
- Verma KS, Kachhwaha S, Kothari SL. In vitro plant regeneration of Citrullus colocynthis (L.) Schard. and assessment of genetic fidelity using ISSR and RAPD markers. Indian J Biotechnol. 2013;12:409–414. [Google Scholar]
- Williams JG, Kubelik AR, Livak KJ, et al. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav K, Yadav SK, Yadav A, et al. Comparative analysis of genetic diversity among cultivated Pigeon pea (Cajanus cajan (L) Millsp.) and its wild relatives (C. albicans and C. lineatus) using randomly amplified polymorphic DNA (RAPD) and inter simple sequence repeat (ISSR) fingerprinting. Am J Plant Sci. 2014;5(11):1665. doi: 10.4236/ajps.2014.511181. [DOI] [Google Scholar]
- Yap IV, Nelson RJ (1996) Winboot: a program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-Based dendrograms, IRRI Discussion Paper Series No. 14. International Rice Research Institute, Manila, Philippines
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–783. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]