Abstract
This study aimed to assess genetic variability at molecular and phytochemical levels among the four most commonly grown olive cultivars and the wild-type olive of Saudi Arabia. Sixty-six and 80 amplicons were generated from 9 random amplified polymorphic DNA (RAPD) and inter simple sequence repeats (ISSR) primers, each, producing an average of 95.9 and 86.44% polymorphism for the two markers, respectively. The PIC values were 82.2% for the RAPD and 85.4% for the ISSR markers and the discrimination power for both the markers was 11.1%. The UPGMA cluster analysis based on the RAPD and ISSR data resulted in the aggregation of cultivars and wild accession with a good bootstrapping value according to their origin. Furthermore, a total of 199 compounds were identified in the cultivars based on peak area, retention time, and molecular formula using GC–MS analyses of methanolic and ethanolic extracts. These compounds were classified according to their chemical class; most of them were fatty acids, alcoholic compounds, carboxylic acids, aldehydes, heterocyclic compounds, ketones, alkanes, and phenols. Genetic and phytochemical distances were significantly correlated, based on the Mantel test. The Saudi wild accession also had high numbers of fatty acids and their esters, and can be used in breeding programs for generating new genotypes with interesting characters.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0920-5) contains supplementary material, which is available to authorized users.
Keywords: Genetic variability, Olea europaea, Olea europaea subsp. europaea var. sylvestris, Molecular markers, Phytochemicals
Introduction
Olive (Olea europaea L.), which belongs to the family Oleaceae, is a subtropical, evergreen, oil-producing tree. The genus Olea comprises more than 40 species, including the cultivated, wild, and feral forms (Heywood 1978). The number of chromosomes (2n = 46) in this genus is very high, which is an indicator of its polyploid (tetraploid) origin (Brousse 1987). Microsatellite patterns and flow cytometry analyses have confirmed the hexaploid, tetraploid, and triploid nature of O. europaea (Besnard et al. 2008; Brito et al. 2008). Olive products have been valued since ancient times. The oil extracted from mesocarp of the fruit is a valuable and healthy food. It is also used as lamp fuel and in wool treatment, medicine, and cosmetic and soap production. As a food, it is used in salads, for cooking, and in the preservation of other foods. Table olives are also a typical component of the Mediterranean diet and are consumed after processing and pickling in different ways.
The origin of olive is very ancient and its cultivation goes back to the prehistoric period. Zohary and Hopf (1994) suggested that domestication of olive took place between 5500 and 5700 years ago and the most accepted opinion among researchers is that the olive tree originated in Eastern Mediterranean, more precisely, in the north of the Dead Sea (Loukas and Krimbas 1983; Zohary and Spiegel-Roy 1975). Cultivated olive harbors enormous genetic variability. To date, more than 2600 different olive cultivars have been described (Rugini and Lavee 1992) and large numbers of mislabeling, homonyms, and synonyms have been reported (Barranco and Rallo 2000). The preservation of this valuable genetic patrimony in olive is important to prevent its erosion, which would lead to an irreversible narrowing of the genetic background, as it is occurring in many other crops.
Before the availability of molecular markers, identification of olive tree cultivars was performed using morphological, agronomical, or biochemical traits (Barranco et al. 2000). In recent times, molecular markers have been widely applied to characterize and identify the olive cultivars. In olive, RAPDs have been extensively used for cultivar identification and was the first class of molecular markers to be considered in olive and have been broadly employed for cultivar identification started by early study of Bogani et al. (1994) and then many studies conducted to characterized olive cultivars utilizing RAPD (Gomes et al. 2008; Martins-Lopes et al. 2009; Muzzalupo and Perri 2009; Brake et al. 2014) and ISSRs markers have been also firstly used by Pasqualone et al. (2001) and followed by many studies of genetic diversity and cultivars fingerprint of olive (Asadiar et al. 2013; Ben-Ali et al. 2015; Brake et al. 2014; Kaya 2015; Linos et al. 2014; Noormohammadi et al. 2012). In addition to these markers AFLPs (Angiolillo et al. 1999; Ipek et al. 2015), microsatellite (Sefc et al. 2000; Cipriani et al. 2002; Erre et al. 2010; Noormohammadi et al. 2014 and Abdessemed et al. 2015), SNP markers (Reale et al. 2006; Salimia et al. 2009; Hakim et al. 2010 and Biton et al. 2015) have been employed in olive inter- and intra-cultivar variability, clarifying varietal synonymy and homonymy cases.
Wild olive grows abundantly in thick forests, and is believed to be indigenous to the Mediterranean Basin (Green 2002). The importance of studying the wild olive germplasm as a valuable source of variability could become an attractive objective in olive breeding programs. In this respect, Erre et al. (2010) attempted to elucidate the genetic relationships within and between the wild and cultivated olives using microsatellites. Genetic diversity and gene flow between wild and cultivated olive has been studied. Lumaret and Ouazzani (2001) reported that the genetic diversity values of cultivars, feral olives, and wild olives in ten forest areas around the Mediterranean basin were 0.286, 0.414, and 0.506, respectively, which is consistent with the interpretation that the domesticated olive represents a sample of the genetic variation in genuine wild olive populations that persist today. Owing to their very long lifespan, these wild trees might be closely related to the Neolithic olives, which are recognized as the crop progenitor.
Olive oil is distinguished from other edible vegetable oils by its flavor. Its nutritional value is attributed to the presence of high levels of oleic acid and other minor components. It contains more than 180 different aromas and the majority of the volatile compounds consist of aldehydes, esters, hydrocarbons, ketones, and furans (Kalua et al. 2007; Ridolfi et al. 2002). The cultivar, origin, growing season, maturity stage of fruit, storage conditions of fruit, and fruit processing of olives influence the flavor components of olive oil and, therefore, its taste, aroma, and phenolic profile (Dabbou et al. 2010; Gomez-Rico et al. 2006). Phenolic compounds, phenyl ethyl alcohols, flavones, secoiridoids, including oleuropein, lignans, and ligstroside derivatives are other important parameters for determining the quality of olive oil because phenolic structures largely contribute to the olive oil flavor and prevent oxidation of the free fatty acid fraction in the oil (Gallina-Toschi et al. 2005; Servili and Montedoro 2002).
Saudi Arabia is one of the largest consumers of olives and olive oils, but its contribution to the world olive oil production is limited. The climate in the northern part of the country resembles the Mediterranean climate, which favors the growth of olive tree and, thus, the production of olive oils with the same high international quality standards (Al-Ruqaie et al. 2016). In these areas, extensive plantations of exotic and indigenous cultivars of olives have been established (Al-Khalifah et al. 2012).
Studies characterizing the wild-type and other cultivars of olive grown in Saudi Arabia using molecular markers and phytochemical analysis are limited. This study aimed at molecular and phytochemical assessment of the most commonly grown olive cultivars in Saudi Arabia with respect to the wild type.
Materials and methods
Plant materials
Plant samples from four olive cultivars were obtained from the Al-Jouf Olive Research Center. The wild sample was collected from Al-Sodah National Park at Al- BAHA Governorate, in the south west of Saudi Arabia. Fully matured fruits from the cultivars ‘Arbosana’, ‘Koroneiki’, ‘Picual’, ‘Arbequina’ and the wild accession were harvested at the same time in mid-December from the same olive orchards kept on ice and shipped directly to the lab using ice box. These cultivars were selected because they are some of the most widely planted cultivars in the new orchards of Saudi Arabia, and are highly productive and well-adapted to the modern olive-growing techniques. Only healthy fruits, without any kind of disease or physical damage, were processed. Fruits were crushed to fine paste using grain chopper (Moulinex grain grinder, France) extracted with ethanol and methanol solvents.
100 g of ground sample with 200 ml of 95% ethanol was mixed for 30 min using a magnetic vibrator. Sodium sulfate anhydrous was used to dehydrate samples then filtered and the ethanol was evaporated using rotary evaporator. Moreover, SPE (C-18: 300 mm and 22 mm L/W) columns were used for sample purification prior GC–MS analysis. Samples were dissolved in 60% ethanol and the supernatant mixed with ether (2:1v/v) and mixed and the supernatant collected three times. After that chloroform was added (2:1v/v) the mixture was shaken and the supernatant contained chemical constituents was collected.
The GC–MS analyses of methanolic and ethanolic extracts were performed using a TSQ™ 8000 Evo Triple Quadrupole GC–MS/MS (Thermo Fisher Scientific, Waltham MA, USA) equipped with an Elite-5 capillary column (30 nm × 0.25 mm ID × 0.25 µm df); the mass detector was operated in the electron impact (EI) mode with full scan (50–550 amu). Helium was the carrier gas, which was used at a flow rate of 1 mL/min. The injector was operated at 290 °C and the oven temperature was programmed to increase from 50 to 200 °C at 8 °C/min (held for 5 min) and further to 290 at 7 °C/min (held for 10 min). The peaks in the chromatogram were identified on the basis of their mass spectra. Mass spectrum obtained from GC–MS was interpreted using the database of National Institute Standard and Technology (NIST). The mass spectra of the phytochemicals were compared with the spectra of known compounds stored in the NIST library.
DNA extraction and PCR amplification
DNA extraction was performed using a modified SDS protocol (Alghamdi et al. 2012). Fresh leaf samples were ground in liquid N2 and 200 mg of the powder was mixed with 800 μL of the extraction buffer (100 mM Tris–HCl pH 8, 50 mM EDTA pH 8, 1.4 M NaCl, 2% SDS v/v, 2% v/v PVP, and 0.1% mercaptoethanol), and incubated at 65 °C for 30 min. Thereafter, 3 μL RNase1 (10 mg/mL) was added to the extract and it was incubated at 37 °C for 15 min. An equal volume of chloroform–isoamyl alcohol (24:1) was added, mixed, and centrifuged at 13,680×g for 20 min. One-third volume of 5 M potassium acetate was added to the supernatant, mixed vigorously, and centrifuged at 13,680×g for 20 min. The supernatant was removed and ½ volume of cold isopropanol was added, mixed well, and incubated at 4 °C for 1 h. The samples were then centrifuged at 13,680×g for 15 min at 4 °C. The supernatant was decanted and the pellets were allowed to air dry for 10 min. The pellets were re-suspended in 300 μL of TE (10 mMTris, 1 mM EDTA, pH 8.0) and 1/10th volume of 3 M sodium acetate, and 2/3 volume of ice-cold isopropanol were added. The samples were mixed well, incubated at 4 °C for 1 h, and centrifuged at 13,680×g for 10 min at 4 °C to pellet the DNA. The supernatant was discarded and the pellets were washed with 80% EtOH and centrifuged at 13,680×g for 5 min at 4 °C; the supernatant was discarded again and the tubes were inverted to dry for 30 min. The DNA samples were resuspended in 100 μL of TE and incubated at 4 °C overnight. The quality and concentration of the extracted DNA were determined by electrophoresis on a 0.8% agarose gel and spectrophotometry using Thermo Scientific NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA was diluted with TE to a final concentration of 100 ng/µL.
Polymerase chain reactions were performed in 20-μL volumes containing 1X GoTaq Green Master Mix (Promega Corporation, Madison, WI, USA), 0.1 μM of each primer, 50 ng DNA templates, and nuclease-free water. The PCR amplification was performed in a TC-5000 thermal cycler (Bibby Scientific, Staffordshire, UK) using the following thermal profile: denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 1 min, annealing at 35 °C for 1 min for RAPD reactions and 50 °C for ISSR reactions, and elongation at 72 °C for 1 min, followed by a final elongation step at 72 °C for 7 min. The amplified DNA fragments were separated by electrophoresis at 80 V for 100 min on a 1.5% agarose gel in TBE buffer (0.1 M Tris base, 0.1 M boric acid, 2 mM EDTA) and visualized by staining with acridine orange (10 mg/mL). The amplification reactions were repeated at least twice and only reproducible and intense bands were scored. The gel was viewed under ultraviolet light and photographed using Bio-Rad Gel Doc EZ System. The molecular sizes of the amplified products were estimated by using a 100-bp DNA ladder (Sigma Chemical Company, Darmstadt, Germany).
Data analysis
The DNA profiles were scored visually from gel photographs. The clear and reproducible amplified bands were chosen for the analyses. Polymorphism information content (PIC) value was estimated using the following equation described by Anderson et al. (1993): , where Pij is the frequency of the ith allele for marker j and the summation extends over n amplicon calculated for each locus. The discrimination power was calculated by dividing the number of polymorphic markers amplified for each primer by the total number of polymorphic bands obtained (Khierallah et al. 2011). The presence of a band was designated as “1” and the absence of a band was recorded as “0”. The data obtained by scoring the RAPD and ISSR profiles, both individually as well as collectively, were subjected to the calculation of a similarity matrix using Jaccard’s coefficients. Cluster analysis was performed to construct dendrograms with the unweighted pair-group method by arithmetic averages (UPGMA) from the similarity data matrices using PAST3.11 software (Hammer et al. 2001). The co-phenetic correlation coefficient was used to check the goodness of fit a cluster analysis to the associated similarity matrix. A bootstrap analysis of 1000 replicates was performed using PAST3.11 software to estimate structural stability of clusters. Mantel test (Mantel 1967) was also performed using the PAST3.11 software in order to investigate the relationship between the genetic and phytochemical distances of the cultivars.
Results
Molecular analysis
Out of the 25 primers used, 9 primers that produced unambiguous fragments with repeatable patterns when tested two times with the same cultivar were considered reproducible amplicons and were used in the analysis (Table 1). A total of 66 amplicons (loci) were obtained, out of which 64 amplicons were polymorphic; the average number of polymorphic amplicons obtained per primer was 7.11 and the range was from 3 (for primer P9) to 13 (for primer P6). The polymorphism percentage ranged from 75 to 100% with an average of 95.89%. Across cultivars, the primers produced 159 bands with an average of 17.67 bands per primer, ranging from 9 for primer P1 to 33 bands for primer P6. The polymorphic information content (PIC) ranged from 66 for primer P9 to 91 for P6, with an average of 82.22%. Primer P6 had the highest discrimination power (DP) with a value of 20% and P9 had the lowest (5%) value. All the primers showed an average DP of 11.11%. A total of 80 reproducible ISSR amplicons were generated from 9 primers out of the 16 ISSRs screened, of which 70 were polymorphic, and ranged from 2 for primer P9 to 11 amplicons for primer P6; this accounted for a high percentage of polymorphism (86.44%), ranging from 50% for primer P9 to 100% for primers P4, P5, and P8. The primers produced 236 bands across cultivars with an average of 26.22 bands per primer ranging from 13 bands for primer P8 to 39 bands for primer P1. The PIC ranged from 74 for primer P9 to 91 for primer P1, with an average of 85.44%. Primer P6 had the highest discrimination power with a value of 16% and P9 had the lowest (3%) value. All the primers showed an average DP of 11.11% (Table 2).
Table 1.
Primers used for RAPD analyses: total number and polymorphic amplicons, % of polymorphism obtained, total number of bands across cultivars, polymorphism information content (PIC) and discrimination power (DP)
| RAPD primers | 5′-3′ Sequence | Total number of amplicons | Polymorphic amplicons | Polymorphism (%) | Total bands | PIC | DP |
|---|---|---|---|---|---|---|---|
| P1 | CACACTCCAG | 7 | 7 | 100 | 9 | 84 | 11 |
| P2 | ACGACCGACA | 7 | 7 | 100 | 15 | 82 | 11 |
| P3 | AGGTGACCGT | 8 | 8 | 100 | 22 | 86 | 13 |
| P4 | GGCTCATGTG | 6 | 6 | 100 | 14 | 82 | 9 |
| P5 | GGACTGCAGA | 8 | 7 | 88 | 21 | 85 | 11 |
| P6 | AGTCAGCCAC | 13 | 13 | 100 | 33 | 91 | 20 |
| P7 | AAAGCTGCGG | 7 | 7 | 100 | 19 | 84 | 11 |
| P8 | GTCAGGGCAA | 6 | 6 | 100 | 16 | 80 | 9 |
| P9 | CCTTGACGCA | 4 | 3 | 75 | 10 | 66 | 5 |
| Total | 66 | 64 | – | 159 | – | – | |
| Mean | 7.34 | 7.11 | 95.89 | 17.67 | 82.22 | 11.11 | |
| Min | 4 | 3 | 75 | 9 | 66 | 5 | |
| Max | 13 | 13 | 100 | 33 | 91 | 20 |
Table 2.
Primers used for ISSR analyses: total number and polymorphic amplicons, % of polymorphism obtained, total number of bands across cultivars, polymorphism information content (PIC) and discrimination power (DP)
| ISSR primers | 5′-3′ Sequence | Total number of amplicons | Polymorphic amplicons | Polymorphism % | Total bands | PIC | DP |
|---|---|---|---|---|---|---|---|
| P1 | (AAC)7G | 13 | 10 | 77 | 39 | 91 | 14 |
| P2 | (AAC)7A | 8 | 7 | 88 | 20 | 84 | 10 |
| P3 | (GTT)7C | 10 | 9 | 90 | 30 | 88 | 13 |
| P4 | (GTT)7T | 10 | 10 | 100 | 26 | 89 | 14 |
| P5 | (CA)10G | 9 | 9 | 100 | 27 | 88 | 13 |
| P6 | (CA)10A | 13 | 11 | 85 | 36 | 91 | 16 |
| P7 | (CA)10T | 8 | 7 | 88 | 28 | 87 | 10 |
| P8 | (TG)10C | 5 | 5 | 100 | 13 | 77 | 7 |
| P9 | (TG)10A | 4 | 2 | 50 | 17 | 74 | 3 |
| Total | 80 | 70 | – | 236 | – | – | |
| Mean | 8.89 | 7.78 | 86.44 | 26.22 | 85.44 | 11.11 | |
| Min | 4 | 2 | 50 | 13 | 74 | 3 | |
| Max | 13 | 11 | 100 | 39 | 91 | 16 |
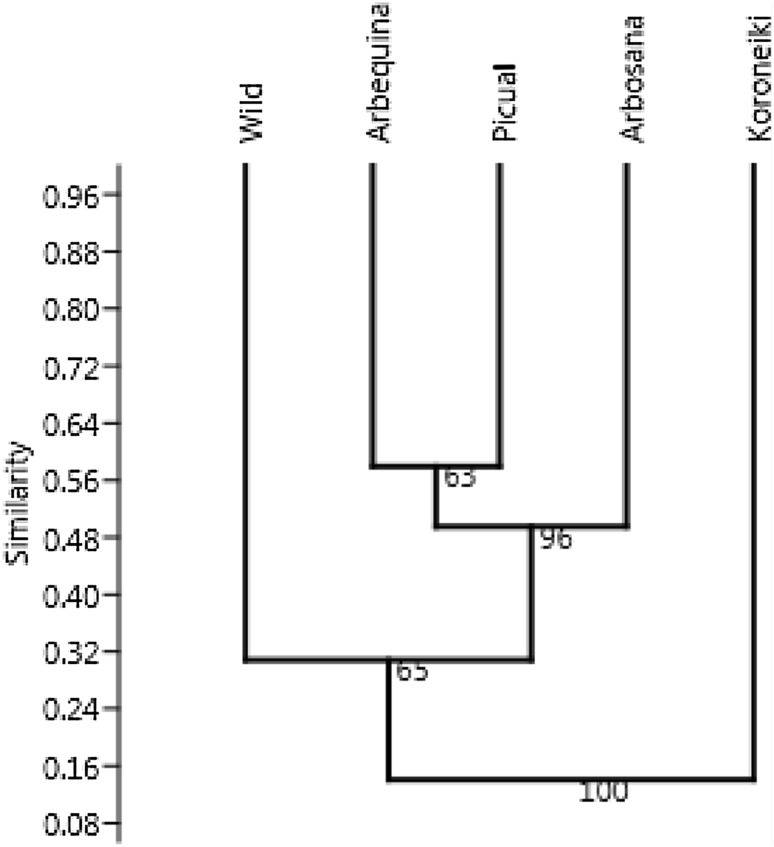
Based on the Jaccard’s similarity coefficient, a genetic similarity matrix was constructed using the RAPD and ISSR data to assess the genetic relatedness among the four olive cultivars and one Saudi wild olive accession. The RAPD data showed a similarity coefficient ranging from 0.05 (between Arbosana and Koroneiki cultivars) to 0.58 (between Arbequina and Picual cultivars). All the cultivars and wild accession showed an overall genetic similarity value of 0.31. The UPGMA cluster analysis of the cultivars and wild accession based on the RAPD data was cut at a similarity of 0.45 (which represented 50% of the distance from the maximum similarity of 0.58 to the minimum of 0.12). Cutting the dendrogram at this similarity value resulted in aggregating three cultivars, namely Arbequina, Picual, and Arbosana, with a good bootstrapping value (96), whereas Koroniki and wild accession failed to form a cluster and were individually separated (Fig. 1). However, at 60% of similarity index, all the cultivars and wild accession were identified and individually separated.
Fig. 1.
UPGMA dendrogram based on Jaccard’s coefficient illustrating the genetic similarities among olive cultivars and wild accession based on RAPD data
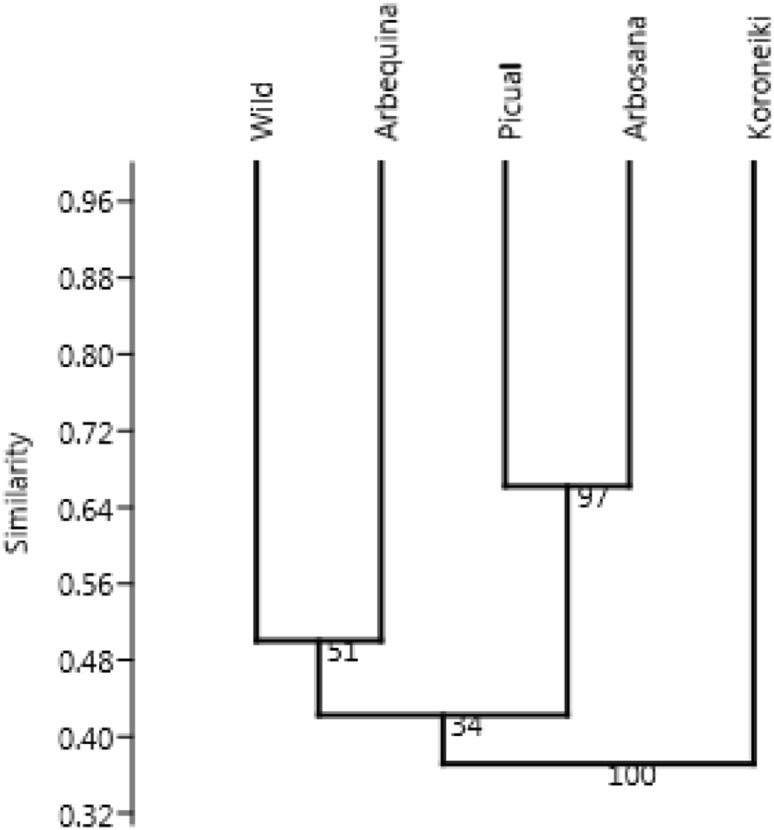
For the ISSR data, the similarity coefficient ranged from 0.36 (between Saudi wild accession and both Picual and Arbozana cultivars) to 0.66 (between Picual and Arbosana cultivars). All the cultivars and wild accession showed an overall genetic similarity value of 0.43. The UPGMA cluster analysis of the cultivars and wild accession based on the ISSR data was cut at a similarity of 0.58 (which represented 50% of the distance from the maximum similarity of 0.66 to the minimum of 0.38). Cutting the dendrogram at this similarity value resulted in aggregating Picual and Arbosana with a good bootstrapping value (97), whereas the other cultivars and the wild accession failed to form a cluster and were individually separated (Fig. 2). However, at 68% similarity index, all the cultivars and the wild accession were identified and individually separated.
Fig. 2.
UPGMA dendrogram based on Jaccard’s coefficient illustrating the genetic similarities among olive cultivars and wild accession based on ISSR data
Phytochemical analysis
A large number of phytochemical compounds were identified in the methanolic and ethanolic extracts of the five olive cultivars, including one wild-type olive accession, using GC–MS analysis. A total of 199 compounds were identified based on peak area, retention time, and molecular formula (Supplementary Tables 1 and 2). The compound with the lowest retention time (4.82 min) was propanal, 2-methyl, whereas dihydrotorulosol and silane, (3.beta.)-gorgost-5 compounds had the highest retention time (57.62 min) (Supplementary Table 1). There were significant differences in the compositions of the phytochemicals in the olive genotypes based on the extraction solvent. The main compounds in the Arbequina cultivar included 2,6,10,14,18,22-tetracosahexaene (56.39%) and 2,6,10,15,19,23-hexamethyl-2,6 (56.81%) in the ethanolic and methanolic extraction solvents, respectively. In the wild accession, 72.26% of the constituents were (10.beta.h)-des-a-lupane and octane nitrile arneel (18.02%) in ethanol and methanol, respectively. The main constituents of the Picual genotype included 2,6,10,14,18,22-tetracosahexaene (27.66%) in ethanol and n-hexadecanoic acid (19.82%) in methanol. The compounds 2,6,10,14,18,22-tetracosahexaene in ethanol and 1,1-heptanediol, diacetate in methanol were the main compounds in the Koroneiki genotype, present at 53.92 and 19.73%, respectively.
The phytocompounds identified were classified into different chemical classes (Table 3). A total of 32 fatty acids, 7 fatty acid esters, 17 alcoholic compounds, 22 carboxylic acids, 16 aldehydes, 17 heterocyclic compounds, 15 ketones, 7 esters, 6 ethers, 11 alkanes, 3 phytosterols, 3 sugars and 5 steroid compounds, two compounds each of alkenes, phenols, urea, one compound from amine, organosilicone, and nitriles were determined in the fruits and correspondent oils. Thirty-two residues were classified as unknown constituents. The most abundant compounds from each class with values more than 3% are presented in Table (3). The main constituents of the extracts were the heterocyclic compound 10.beta.h-des-a-lupane (72.26), the steroid 2,6,10,15,19,23-hexamethyl-2,6 (60.11%), the aldehyde propanal, 2-methyl-1 (37.8%) and the carboxylic acid quinic acid 1,3,4,5-tetrahy (26.57%). DOCOSANOIC ACID was the most abundant fatty acid (19.07%) and the BETA-SITOSTEROL (17.04%) was the most abundant phytosterols. A typical chromatogram of one genotype is shown in Fig. (3). GC–MS analysis revealed that the ethanolic and methanolic extracts were predominantly composed of fatty acids, carboxylic acids, heterocyclic compounds, and alcohol and aldehyde compounds. The aldehydes (14-heptadecenal and octadecanal), ester (eicosyl acetate), fatty acid (9-octadecenoic acid), and heterocyclic compound (2,6,10,14,18,22-tetracosahexaene were detected in all the genotypes. However, hexadecanal and propanal, 2-methyl aldehyde compounds, hexadecanoic acid, nonadecanoic acid, and nonadecanoic acid-ethyl ester fatty acids, 1,2-benzenedicarboxylic acid, and oxalic acid-isohexyl pentyl two carboxylic acids, octadecane alkane and methyl 3′-methyllecanorate ester were predominant in four out of five genotypes. The Arbosana genotype recorded the highest total ion chromatogram (TIC) in the ethanolic and methanolic extracts (82 compounds) followed by that in the wild genotype (74 compounds) and Arbequina (62 compounds), whereas Koroneiki and Picual recorded 40 and 50 compounds, respectively.
Table 3.
Major classes of the secondary chemical constituents, number of compounds and percentage of the most abundant compounds with retention time and molecular weight determined in olive cultivars and wild accession
| Chemical class/compound | RT | Area | Area (%) | MW |
|---|---|---|---|---|
| Alcohol (17 compounds) | ||||
| 1-Octadecanol | 22.12 | 42,461 | 3.27 | 270.5 |
| 1-Hexadecanol | 32.42 | 39,380 | 3.8 | 242.45 |
| DL-3,4-dimethyl-3,4-hexanediol | 27.10 | 32,680 | 4.88 | 146.23 |
| Vitamin E | 53.70 | 75,700 | 5.87 | 430.72 |
| 1,2,3-Propanetriol | 17.76 | 208,090 | 7.84 | 92.09 |
| Aldehyde (16 compounds) | ||||
| 4-Hydroxytetradec-2-ynal | 47.88 | 72,051 | 3.77 | 224.34 |
| 7-Octadecenal | 23.66 | 84,465 | 5.42 | 266.47 |
| Octadecanal | 52.62 | 191,624 | 11.55 | 268.49 |
| Propanal, 2-methyl-I | 4.82 | 636,539 | 37.8 | 72.11 |
| Alkane (10 compounds) | ||||
| 1,3,4-Trimethyl-1,3-cyclohexan | 26.94 | 51,431 | 3.3 | 124.23 |
| Dodecane, 1,1′-oxybis- | 54.7 | 111,361 | 4.78 | 354.66 |
| 2L,4L-dihydroxyeicosane | 22.16 | 21,110 | 5.13 | 314 |
| d-Manno-(E)-tetradec-6-en-1,2, | 26.64 | 87,287 | 6.72 | 356.5 |
| Carboxylic acid (22 compounds) | ||||
| Oxalic acid, isohexyl pentyl E | 10.28 | 48,551 | 3.11 | 244.33 |
| Benzenepropanoic acid | 22.76 | 54,531 | 5.27 | 428.75 |
| Butanoic acid | 26.68 | 33,054 | 5.65 | 88.11 |
| Benzoic acid | 30.36 | 25,960 | 8.24 | 122.12 |
| Silane, [[(3.beta.)-gorgost-5-]] | 57.62 | 188,197 | 9.84 | 472.86 |
| Cantharic acid | 22.94 | 167,915 | 12.92 | 196.2 |
| Quinic acid 1,3,4,5-tetrahy | 28.84 | 414,229 | 26.57 | 192.17 |
| Cyclopropanepentanoic acid | 37.72 | 2199 | 7.09 | 310 |
| Ester (7 compounds) | ||||
| Vitamin E acetate | 53.62 | 84,397 | 3.95 | 472.74 |
| 2-Octenyl acetate | 21.76 | 80,782 | 6.22 | 170.25 |
| Ether (6 compounds) | ||||
| Severine | 45.38 | 64,047 | 3.52 | 473.7 |
| 19-Norambrox | 55.94 | 82,277 | 4.53 | – |
| Isocineole | 16.88 | 48,319 | 11.81 | 154.25 |
| Fatty acid/fatty acid ester (39 compounds) | ||||
| 4,8-Decadienoic acid, 2-acetyl | 31.22 | 48,640 | 3.12 | 280.41 |
| N-Hexadecanoic acid | 35.94 | 64,462 | 3.55 | 256.43 |
| (E)-9-Octadecenoic acid ethyl | 39.68 | 74,257 | 3.94 | 310.52 |
| Tetradecanoic acid | 23 | 21,259 | 4.24 | 228.38 |
| 3,5,7-Trioxononanoic acid N | 53.8 | 108,672 | 4.66 | 200.19 |
| (E)-9-Octadecenoic acid | 38.52 | 52,148 | 5.66 | 282.47 |
| 15-Tetracosenoic acid, methyl | 38.5 | 35,769 | 5.88 | 380.66 |
| Docosanoic acid 1-methyl-butyl | 27 | 158,721 | 5.98 | 410.73 |
| 13-Docosenoic acid | 37.04 | 28,670 | 9.1 | 338.58 |
| 9-Hexadecenoic acid | 39.22 | 69,508 | 10.5 | 254.41 |
| Docosanoic acid | 26.98 | 78,002 | 19.07 | 340.59 |
| Hexadecanoic acid, ethyl ester | 36.54 | 84,679 | 4.49 | 284.48 |
| Heptadecanoic acid, methyl EST | 35.24 | 33,151 | 5.01 | 284.48 |
| Heterocyclic compound (17 compounds) | ||||
| Stigmast-5-en-3-ol, (3.beta.,2) | 55.98 | 107,928 | 4.63 | 414.72 |
| 2H-Pyran-2-one, 5-ethylidenete | 29.66 | 74,200 | 4.76 | 170.21 |
| Stigmasterol, 22,23-dihydro | 55.92 | 96,820 | 5.13 | 414.72 |
| Adenosine 9-beta-d | 18.62 | 96,495 | 5.31 | 267.24 |
| Fluorenone oxime 9-fluoreno | 41.18 | 131,237 | 5.63 | 195.22 |
| Cytidine cyd cytos | 28.82 | 194,693 | 10.71 | 243.22 |
| 2,6,10,14,18,22-tetracosahexae | 49.62 | 357,013 | 27.66 | 410.73 |
| (10.beta.h)-des-a-lupane | 57.58 | 1,363,225 | 72.26 | 412.75 |
| Ketone (16 compounds) | ||||
| Cyclohexan-1,4,5-triol-3-one-1 | 17.8 | 57,507 | 3.16 | 190.15 |
| 2-Cyclopenten-1-one, 2-hydroxy | 8.06 | 88,703 | 3.33 | 98.1 |
| 2-Cyclopenten-1-one | 8.08 | 55,876 | 3.53 | 82.1 |
| Spiro[4.5]dec-6-ene-1,4-dione | 30.36 | 79,723 | 4.81 | 164.2 |
| Nitriles (2 compounds) | ||||
| Octanenitrile arneel | 23.22 | 327,558 | 18.02 | 125.22 |
| Phenol (2 compounds) | ||||
| 6-Amino-1-[2-(3,4-diethoxy-phe | 30.36 | 116,713 | 3.39 | 307.37 |
| 1,3-Benzenediol | 19.16 | 85,295 | 5.39 | 110.11 |
| Phytosterols (2 compounds) | ||||
| Stigmasterol | 55.88 | 102,892 | 6.2 | 412.7 |
| Gamma-sitosterol | 55.92 | 150,662 | 8.95 | 414.72 |
| Beta-sitosterol | 55.94 | 219,994 | 17.04 | 414.72 |
| Steroid (5 compounds) | ||||
| l-Gala-l-ido-octose | 29 | 25,786 | 3.9 | 240.21 |
| Ursodeoxycholic acid | 53.46 | 126,365 | 12.2 | 392.58 |
| 2,6,10,15,19,23-Hexamethyl-2,6 | 49.5 | 351,622 | 60.11 | 410.72 |
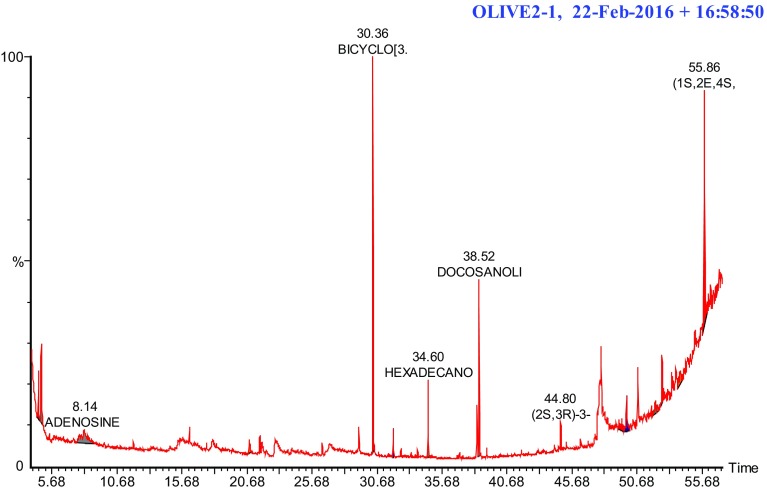
Fig. 3.
Chromatogram obtained from the GC–MS with the ethanol extract of Saudi wild accession olive oil
The GC–MS analysis also showed considerable variation in alcoholic compounds in the different genotypes investigated; the wild accession had 8 alcoholic compounds and Arbequina recorded only two alcoholic compounds. 11-tridecenol and vitamin E were recorded in three genotypes. The most dominant aldehydes were octadecanal and 14-heptadecenal in all the genotypes, followed by propanal and hexadecanal in four out of five genotypes. Out of 10 alkane compounds, octadecane was dominant in four genotypes. Two carboxylic acid compounds, benzenedicarboxylic acid and oxalic acid, and isohexyl pentyl were recorded in four genotypes. Fatty acid constituents showed high variability among the genotypes, ranging from single fatty acid specific for a genotype to one fatty acid (octadecenoic acid) dominant in all the genotypes. Nonadecanoic acid, hexadecanoic acid, and ethyl ester were common in four genotypes. Arbequina (14 fatty acid compounds) and the wild accession contained 12 fatty acids, whereas Picual and Koroneiki had 8 fatty acids. The wild accession and Arbosana recorded 8 and 7 heterocyclic compounds, respectively, whereas the other three genotypes produced five compounds each. 2,6,10,14,18,22-tetracosahexaene (squalene 30-carbon organic compound) was present in all the genotypes. Three ketone compounds were present in Koroneiki and 6 compounds were present in both Arbosana and the wild genotype. Octenone was recorded in two genotypes (Koroneiki and Wild) and bicyclooctenone was recorded in wild and Arbosana, whereas 13 other ketones were recorded in the different genotypes individually.
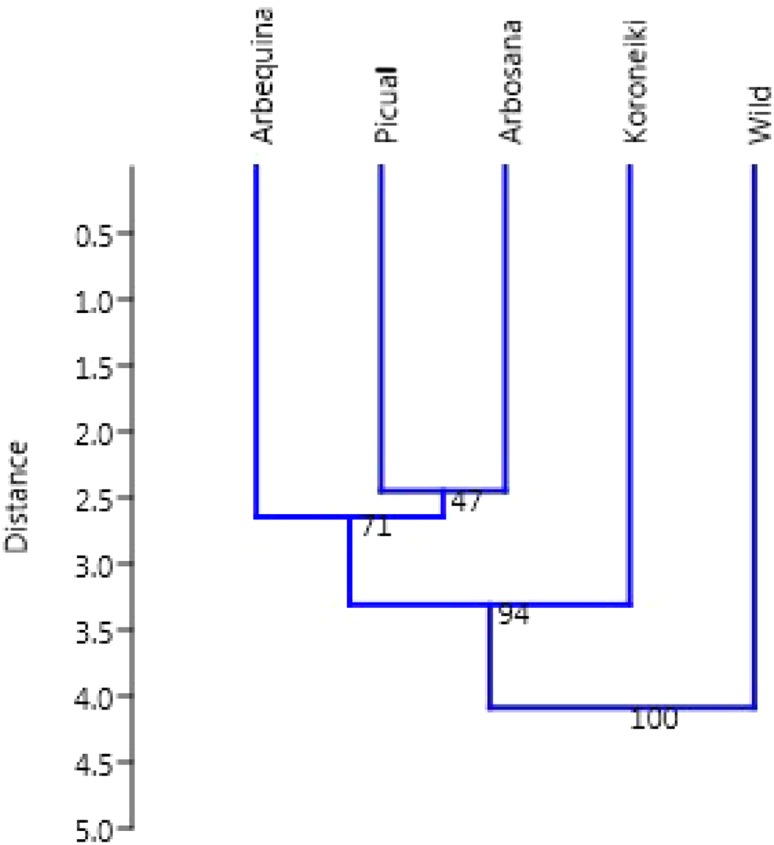
Cluster analysis showed that Spanish cultivars namely Arbequina, Picual, and Arbosana were aggregating and form one cluster and the Saudi wild accession and Koroniki cultivar failed to form a cluster and were individually separated (Fig. 4). The principal component analysis (PCA) results revealed that a considerable amount of variation (90.24%) was explained by the first three axes. Axes 1, 2 and 3 explained 50.36, 24.15 and 15.73% of the total variation, respectively (Table 4). The first principal component was positively correlated with ketone, alcohol, carboxylic acid and aldehyde. Fatty acid, carboxylic acid and steroid were the components positively correlated with the second axis and the third axis was positively correlated with alkane and carboxylic acid.
Fig. 4.
UPGMA dendrogram based on Euclidian distance coefficient illustrating the genetic similarities among olive cultivars and wild accession based on phytochemicals constitutes data
Table 4.
Eigen values, percentage variations and loadings of the phytochemical classes on the plane of the first three principal components
| PC 1 | PC 2 | PC 3 | |
|---|---|---|---|
| Eigen value | 25.28 | 12.12 | 7.90 |
| Percent of variance | 50.36 | 24.15 | 15.73 |
| Alcohol | 0.43 | −0.17 | −0.15 |
| Aldehyde | 0.35 | −0.17 | −0.21 |
| Alkane | 0.20 | −0.03 | 0.48 |
| Alkene | 0.11 | −0.07 | 0.23 |
| Carboxylic acid | 0.39 | 0.27 | 0.56 |
| Ester | 0.05 | 0.05 | 0.17 |
| Ether | 0.24 | 0.00 | −0.43 |
| Fatty acid | 0.02 | 0.87 | −0.18 |
| Heterocyclic compound | 0.25 | 0.04 | −0.24 |
| Ketone | 0.60 | −0.03 | −0.05 |
| Phenol | 0.00 | −0.10 | −0.01 |
| Phytosterols | −0.05 | −0.03 | 0.12 |
| Steroid | 0.11 | 0.28 | −0.06 |
| Sugar | 0.04 | −0.11 | 0.08 |
The detected compounds are reported to be potential therapeutic agents and of use in medical and drug formulations (Supplementary Table 3). Genetic and phytochemical distance differences among cultivars were concordant based on the Mantel test (r = 0.75, P = 0.006 for phytochemical and RAPD distances, r = 0.82, P = 0.01 for phytochemical and ISSR distances and r = 0.94, P = 0.01 for RAPD and ISSR distances) indicating that these two analysis (genetic and phytochemicals) grouped the genotypes (wild accession and cultivars) in a similar manner.
Discussion
Olea europaea L. is one of the most important trees cultivated for thousands of years in the Mediterranean area, and is known to have large genetic variability. It exists in two forms, wild (O. europaea subsp. europaea var. sylvestris) and cultivated (O. europaea subsp. europaea var. europaea). The variety ‘europaea’ is propagated by cutting or grafting, whereas ‘sylvestris’ is reproduced from seeds (Green 2002).
Although phenotypic characters and biochemical profiles are valuable in identifying the genotypes, they are controlled by minor polygenic traits and can be affected by environmental factors and cultivation conditions. RAPD and ISSR are multi-locus profiling techniques extensively used in genome studies and marker assisted selection. They are able to distinguish genotypes below the species level, such as cultivars and clones, and have been used in numerous diversity studies (Karp et al. 1997; Pasqualone et al. 2016). They have been used alone or in combination to analyze clonal variation and genetic variability in olive cultivars. The combined use of RAPD and ISSR markers facilitates a high level of genomic coverage as RAPD markers are potentially associated with functionally important loci (Penner 1996) and ISSR markers amplify the hypervariable non-coding regions (Esselman et al. 1999).
In the present study, 9 RAPD primers generated 66 amplicons (loci) with an average of 95.9% polymorphism, 82.2% PIC, and 11.1% DP values. In the ISSR analysis, 9 primers generated a total of 80 amplicons with an 86.44% polymorphism, 85.4% PIC, and 11.1% DP values. Previous studies have also indicated high polymorphism among olive cultivars grown in different parts of the world and have shown that olive germplasm encompassed high genetic variability, in Jordan (Brake et al. 2014), Syria and other Mediterranean regions (Belaj et al. 2003a, c), Cyprus (Banilas et al. 2003), Greece (Hagidimitriou et al. 2005), Morocco and western countries of the Mediterranean Basin (Essadki et al. 2006), Egypt (Hegazi et al. 2012), Iran (Noormohammadi et al. 2012), Portugal (Cordeiro et al. 2008; Martins-Lopes et al. 2007), Italy (Ganino et al. 2007), Spain (Belaj et al. 2004; Caraffa et al. 2002; Gomes et al. 2009; Sanz-Cortes et al. 2001), Turkey (Kaya 2015), and Pakistan (Irshad et al. 2014). This high genetic diversity could be due to a diverse germplasmic origin that resulted in a predominant allogamous species with a high degree of outcrossing (Bartolini et al. 1998) and the complexity of the olive genome (23 pairs of chromosomes, which is believed to have been originated by allopolyploidy (Zohary and Spiegel-Roy 1975), resulted in new cultivars found throughout the Mediterranean amid low breeding pressures (Besnard et al. 2001; Contento et al. 2002). Variations reported in the olive cultivars by different researchers may be related to the variations in the loci studied as well as to the number of genotypes and their localities (Lopes et al. 2004).
The UPGMA cluster analysis of the cultivars and wild accession based on the RAPD and ISSR data resulted in aggregation with a good bootstrapping value according to the origin. Picual, Arbosana, and Arbequina formed a single cluster using the RAPD data, whereas Picual and Arbosana formed one cluster using the ISSR data; these three cultivars were introduced from Spain. Koroneiki, the cultivar from Greece, and the wild Saudi accession failed to aggregate and were individually separated in both the DNA-based marker techniques (RAPD and ISSR). Although, there were differences in the number of amplicons (66 and 80) and polymorphism percentage (96 and 86%) generated by RAPD and ISSR, respectively, and considering the fact that the ISSR primers target specific genomic regions, whereas the RAPD primers amplify arbitrary regions (Martins-Lopes et al. 2007), there is a clear structure with the origin of the cultivars that has been observed in this study. These results are in agreement with those of several other studies conducted in both Jordan (Hassawi and Hadeib 2004) and the Mediterranean (Belaj et al. 2001, 2003a, b, 2004; Sanz-Cortes et al. 2001), where a good correlation between the banding patterns of olive cultivars and their geographical origin was obtained using RAPD markers. Moreover, the results also supported the hypotheses of autochthonal origin as well as the limited diffusion of olive cultivars from their zones of cultivation (Belaj et al. 2001; Besnard et al. 2001). However, Besnard et al. (2001), Caraffa et al. (2002), Khadari et al. (2003), Martins-Lopes et al. (2007), Poljuha et al. (2008), and Brake et al. (2014) observed no clear correlation between the olive genotypes and their geographical origin.
The dendrogram topology showed a clear separation of the cultivars from the Saudi wild olive, which agreed with the results of Erre et al. (2010) who examined the genetic relationships among and within the wild and cultivated olives and found that the differences on the allelic composition and heterozygosity levels were clear between the wild and cultivated trees. Noormohammadi et al. (2012) have reported high allelic variation and intra- and inter-population genetic diversity in wild olive trees of Iran using RAPD and ISSR markers, which is also supported by the results of Baldoni et al. (2006), who reported that the observed patterns of genetic variation were able to distinguish the wild olives from the cultivated populations and the continental olives from those found in the insular regions. The ISSR analysis also revealed that the cultivated olives from different Mediterranean countries are nested within the wild populations, indicating that either the wild and domesticated olives exchanged genetic material through hybridization or the olive tree domestication occurred more than once (Vargas and Kadereit 2001). In this study, phytochemical cluster analysis confirmed overlapping variability in the cultivars and wild accession corresponds to the geographical origin of cultivars. Similarity among cultivars within the same cluster (Picual, Arbosana, and Arbequina) is high, comparing to Saudi wild accession and Koroneiki (which showed the lowest similarity) indicated that these cultivars may have similar adaptive characteristics or were of the same origin.
In the present study, a large number of phytochemical compounds were identified in olive cultivars and the Saudi wild-type accession, which agreed with the detection of a high number of phytochemical compounds in olive oil, leaves, and other parts (Morales et al. 1994; Perez et al. 2014, 2016; Reboredo-Rodríguez et al. 2013). The identified phytocompounds were classified according to their nature into different groups, including fatty acids, alcoholic, aldehydes, phenols, phytosterols, ketones, esters, alkanes, phytosterols, sugars, and steroid compounds. Aroma, taste, and color of olive oil are considered as the main sensory properties reflective of the oil quality, which may change over time and with location (Kalua et al. 2007). Carbonyl compounds, alcohols, esters, and hydrocarbons are the main compounds found in the volatile fraction of virgin olive oil (Flath et al. 1973). The volatile compounds can be highly useful as biomarkers of the quality of virgin olive oil and show correlations with the sensory characteristics (Issaoui et al.2015). Leon et al. (2011) suggested the existence of a strong genetic influence on the fatty acid composition and several minor components and related characteristics.
The volatile composition of olive oil depends on several factors, such as on the levels and activity of the enzymes involved in various pathways (Angerosa 2002), which are genetically determined (Campeol et al. 2001), ripening cycle of the fruit (Lazzez et al. 2008), processing equipment (Di Giovacchino et al. 2001), extraction method and storage conditions (Vekiari et al. 2007), and climate and soil type (Ranalli et al. 1999). Phenolic compounds are also used as quality markers for olive oil and as traits in new cross-breeding programs because of their health promoting and organoleptic properties (Leon et al. 2011). Many studies have used the profiles of fatty acids and other minor components to characterize oils in several areas of the world, including Portugal (Matos et al. 2006), Tunisia (Haddada et al. 2008; Zarrouk et al. 2008), Spain (Pardo et al. 2010), Jordan (Al-Ismail et al. 2011), and Greece (Longobardi et al. 2012). Moreover, stigmasterol and the main fatty acids (palmitic, oleic, and linoleic acids) can be used to validate the different varieties. In addition, α-tocopherol can be used as a differentiator in bitter spicy oils (Lopez-Cortes et al. 2013).
It has been shown that olive oil consumption may reduce the risk of many diseases associated with oxidative damage, such as coronary heart diseases and cancers (Newmark 1997). The compounds detected in the present study are reported to be potential therapeutic agents, with analgesic, anti-inflammatory, and antimicrobial effects. The results obtained were consistent with many reports that have indicated that polyphenols possess potent antioxidant and anti-cancer, anti-carcinogenic, anti-bacterial, anti-viral, and anti-inflammatory activities and play a vital role in the metabolism of plants (Tapiero et al. 2002). The high demand for olive oil is based on dietary habits correlated with health benefits (Allalout et al. 2011). Monounsaturated fatty acids, tocopherols, and phenolic compounds have great importance in biological systems and act as natural antioxidants (Bendini et al. 2007). Epidemiological studies of cancer (breast and pancreatic) have demonstrated that decreased risk or no enhancement in the risk of cancer is associated with increased dietary intake of olive oil despite the higher proportion of overall lipid intake (Newmark 1997). Furthermore, phytosterols play a vital role in decreasing the blood cholesterol levels because of their antioxidant activities and their impact on health (Ostlund 2004).
Among the olive oil varieties included in the present study, were Spanish sweet oil producing variety, Arbequina, which is used as the standard in fatty acid composition (Lopez-Cortes et al. 2013) and bitter spicy oil producing variety, Picual, which is considered as the standard for the bitter spicy olive oils having the highest content of phenolic compounds (Nieto et al. 2010). In the present study, the Saudi wild accession was also recorded to have a high number of fatty acids and their esters, which can be used for generating new genotypes with interesting characters.
Conclusion
Both DNA-based markers (RAPD and ISSR) showed high allelic variation and have the power to discriminate the olive cultivars and the Saudi wild accession. A large number of phytochemical compounds were identified and the wild accession should be given more attention to understand why it was so different from the olive cultivars. Concordance of genetic and phytochemical analysis indicated that these two analyses grouped the genotypes (wild accession and cultivars) in a similar manner. Such information may prove useful in the selection of optimal varieties and help promote continued progress in olive breeding strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge with thanks for financial support to the King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, and are very grateful to Researchers Support Services Unit at King Saud University (KSU), Riyadh, Saudi Arabia, for language editing.
Compliance with ethical standards
Conflict of interest
Authors declare no conflict of interest of any type.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0920-5) contains supplementary material, which is available to authorized users.
References
- Abdessemed S, Muzzalupo I, Benbouza H. Assessment of genetic diversity among Algerian olive (Olea europaea L.) cultivars using SSR marker. SciHort. 2015;192:10–20. [Google Scholar]
- Alghamdi SS, Al-Faifi S, Migdadi HM, Khan MA, EL-Harty E, Ammar MH. Molecular diversity assessment using Sequence Related Amplified Polymorphism (SRAP) markers in Vicia faba. Int J Mol Sci. 2012;13:16457–16471. doi: 10.3390/ijms131216457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ismail KM, Ahmad R, Al-Dabbas M. Some physiochemical properties of olive and olive oil of three Jordanian olive varieties. Rivista Italiana Delle Sostanze Grasse. 2011;88:191–198. [Google Scholar]
- Al-Khalifah NS, Askari E, El-Kholy M (2012) Following olive footprints in Saudi Arabia. In: El-Kholy, M. (ed) Following olive footprints (Olea europaeaL.) cultivation and culture, Folklore and History, Traditions and Uses. Aarinena, IOC, and ISHS (Scripta Horticulturae N. 13)
- Allalout A, Krichène D, Methenni K, Taamalli A, Daoud D, Zarrouk M. Behavior of super-intensive Spanish and Greek olive cultivars grown in northern Tunisia. J Food Biochem. 2011;35:27–43. doi: 10.1111/j.1745-4514.2010.00364.x. [DOI] [Google Scholar]
- Al-Ruqaie NS, Al-Khalifah AE, Shanavaskhan Morphological cladistic analysis of eight popular Olive (Olea europaeaL.) cultivars grown in Saudi Arabia using numerical taxonomic system for personal computer to detect phyletic relationship and their proximate fruit composition. Saudi J Biol Sci. 2016;23:115–121. doi: 10.1016/j.sjbs.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Churchill G, Autrique J, Tanksley S, Sorrells M. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Angerosa F. Influence of volatile compounds on virgin olive oil quality evaluated by analytical approaches and sensor panels. Eur J Lipid Sci Technol. 2002;104:639–660. doi: 10.1002/1438-9312(200210)104:9/10<639::AID-EJLT639>3.0.CO;2-U. [DOI] [Google Scholar]
- Angiolillo A, Mencuccini M, Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor Appl Genet. 1999;98:411–421. doi: 10.1007/s001220051087. [DOI] [Google Scholar]
- Asadiar L, Rahmani F, Siami A. Assessment of genetic diversity in the Russian olive (Elaeagnus angustifolia) based on ISSR genetic markers. Revista Ciência Agronômica. 2013;44:310–316. doi: 10.1590/S1806-66902013000200013. [DOI] [Google Scholar]
- Baldoni L, Tosti N, Ricciolini C, Belaj A, Arcioni S, Pannelli G, Germana MA, Mulas M, Porceddu A. Genetic structure of wild and cultivated olives in the central Mediterranean basin. Ann Bot. 2006;98:935–942. doi: 10.1093/aob/mcl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banilas G, Minas J, Gregoriou C, Demoliou C, Kourti A, Hatzopoulos P. Genetic diversity among accessions of an ancient olive variety of Cyprus. Genome. 2003;46:370–376. doi: 10.1139/g03-011. [DOI] [PubMed] [Google Scholar]
- Barranco D, Rallo L. Olive cultivars in Spain. Hortechnology. 2000;10:107–110. [Google Scholar]
- Barranco D, Cimato A, Fiorino P, Rallo L, Touzani A, Castaneda C, Serafin F, Trujillo I. Word catalogue of olive varieties. Madrid: International Olive Oil Council; 2000. p. 360. [Google Scholar]
- Bartolini G, Prevost G, Messeri C, Carignani G, Menini U. Olive germplasm. Cultivars and world-wide collections. Rome: FAO; 1998. [Google Scholar]
- Belaj A, Trujillo I, de la Rosa R, Rallo L, Gime’nez MJ. Polymorphism and discriminating capacity of randomly amplified polymorphic markers in an olive germplasm bank. J Am Soc Hortic Sci. 2001;126:64–71. [Google Scholar]
- Belaj A, Caballero JM, Barranco D, Rallo L, Trujillo I. Genetic characterization and identification of new accessions from Syria in an olive germplasm bank by means of RAPD markers. Euphytica. 2003;134:261–268. doi: 10.1023/B:EUPH.0000004954.93250.f5. [DOI] [Google Scholar]
- Belaj A, Satovic Z, Ciprian G, Baldoni L, Testolin R, Rallo L, Trujillo I. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and their effectiveness in establishing genetic relationship in olive. Theor Appl Genet. 2003;107:736–744. doi: 10.1007/s00122-003-1301-5. [DOI] [PubMed] [Google Scholar]
- Belaj A, Satovic Z, Ismaeli H, Panajoti D, Rallo L, Trujillo I. RAPD genetic diversity of Albanian olive germplasm and its relationships with other Mediterranean countries. Euphytica. 2003;130:387–395. doi: 10.1023/A:1023042014081. [DOI] [Google Scholar]
- Belaj A, Satovic Z, Trujillo I, Rallo L. Genetic relationships of spanish olive cultivars using RAPD markers. Hort Sci. 2004;39:916–1156. [Google Scholar]
- Ben-Ali S, Ben-Mohamed M, Guasmi F, Triki T, Kamel N. Genetic variability of Tunisian olive cultivars by ISSR and SSR markers. J N Sci Agric Biotechnol. 2015;24:1098–1107. [Google Scholar]
- Bendini A, Cerretani L, Carrasco-Pancorbo A, Gómez-Caravaca AM, Segura-Carretero A, Fernández-Gutiérrez A, Lercker G. Phenolic molecules in virgin olive oils: a survey of their sensory properties, health effects, antioxidant activity and analytical methods. An overview of the last decade. Molecules. 2007;12:1679–1719. doi: 10.3390/12081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Baradat P, Berville A. Genetic relationships in the olive (Olea europaea L.) reflects multilocal selection of cultivars. Theor Appl Genet. 2001;102:251–258. doi: 10.1007/s001220051642. [DOI] [Google Scholar]
- Besnard G, Garcıa-Verdugo C, Rubio de Casas R, Treier U, Galland N, Vargas P. Polyploidy in the olive complex (Olea europaea): evidence from flow cytometry and nuclear microsatellite analyses. Ann Bot. 2008;101:25–30. doi: 10.1093/aob/mcm275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton I, Doron-Faigenboim A, Jamwal M, Mani Y, Eshed R, Rosen A, et al. Development of a large set of SNP markers for assessing phylogenetic relationships between the olive cultivars composing the Israeli olive germplasm collection. Mol Breed. 2015;35:107–120. doi: 10.1007/s11032-015-0304-7. [DOI] [Google Scholar]
- Bogani P, Cavalieri D, Petruccelli R, Polsinelli L, Roselli G. Identification of olive tree cultivars by using random amplified polymorphic DNA. Acta Hort. 1994;356:98–101. doi: 10.17660/ActaHortic.1994.356.21. [DOI] [Google Scholar]
- Brake M, Migdadi H, Al-Gharaibeh M, Ayoub S, Haddad N, El Oqlah A. Characterization of Jordanian olive cultivars (Olea europaea L.) using RAPD and ISSR molecular markers. Sci Hort. 2014;176:282–289. doi: 10.1016/j.scienta.2014.07.012. [DOI] [Google Scholar]
- Brito G, Loureiro J, Lopes T, Rodriguez E, Santos C. Genetic characterization of olive trees from Madeira Archipelago using flow cytometry and microsatellite markers. Genet Resour Crop Evol. 2008;55:657–664. doi: 10.1007/s10722-007-9268-8. [DOI] [Google Scholar]
- Brousse G. Olive. In: Robbelen G, Downey RK, Ashri A, editors. Oil crops of the world, their breeding and utilization. New York: McGraw Hill; 1987. pp. 462–474. [Google Scholar]
- Campeol E, Flamini G, Chericoni S, Catalano S, Cremonini R. Volatile compounds from three cultivars of Olea europaea from Italy. J Agric Food Chem. 2001;49:5409–5411. doi: 10.1021/jf010455n. [DOI] [PubMed] [Google Scholar]
- Caraffa V, Giannettini J, Gambotti C, Maury J. Genetic relationships between cultivated and wild olives of Corsica and Sardinia using RAPD markers. Euphytica. 2002;123:263–271. doi: 10.1023/A:1014902210530. [DOI] [Google Scholar]
- Cipriani G, Marrazzo MT, Marconi R, Cimato A. Testolin R, Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet. 2002;104:223–228. doi: 10.1007/s001220100685. [DOI] [PubMed] [Google Scholar]
- Contento A, Ceccarelli M, Gelati MT. Diversity of Olea genotypes and the origin of cultivated olives. Theor Appl Genet. 2002;104:1229–1238. doi: 10.1007/s00122-001-0799-7. [DOI] [PubMed] [Google Scholar]
- Cordeiro AI, Sanchez-Sevilla JF, Alvarez-Tinaut MC, Gomez-Jimenez MC. Genetic diversity assessment in Portugal accessions of Olea europaea by RAPD markers. Biologia Plantarum. 2008;52(4):642–647. doi: 10.1007/s10535-008-0125-1. [DOI] [Google Scholar]
- Dabbou S, Rjiba I, Nakbi A, Gazzah N, Issaoui M, Hammami M. Compositional quality of virgin olive oils from cultivars introduced in Tunisian arid zones in comparison to Chemlali cultivars. Sci Hortic. 2010;124:122–127. doi: 10.1016/j.scienta.2009.12.017. [DOI] [Google Scholar]
- Di Giovacchino L, Costantini N, Serraiocco A, Surricchio G, Basti C. Natural antioxidants and volatile compounds of virgin olive oils obtained by two or three-phases centrifugal decamters. Eur J Lipid Sci Technol. 2001;103:279–285. doi: 10.1002/1438-9312(200105)103:5<279::AID-EJLT279>3.0.CO;2-I. [DOI] [Google Scholar]
- Erre P, Chessa I, Munoz-Diez C, Belaj A, Rallo L, Trujillo I. Genetic diversity and relationships between wild and cultivated olives (Olea europaea L.) in Sardinia as assessed by SSR markers. Genet Resour Crop Evol. 2010;57:41–54. doi: 10.1007/s10722-009-9449-8. [DOI] [Google Scholar]
- Essadki M, Ouazzani O, Lumaret R, Moumni M. ISSR variation in olive-tree cultivars from Morocco and other western countries of the Mediterranean Basin. Genet Resour Crop Evol. 2006;53:475–482. doi: 10.1007/s10722-004-1931-8. [DOI] [Google Scholar]
- Esselman EJ, Jianqiang L, Crawford DJ, Windusss JL, Wolfe AD. Clonal diversity in the rare Calamagrosis porteri ssp. insperata (Poaceae) comparative results for al-allozyme and random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. Mol Ecol. 1999;8:443–451. doi: 10.1046/j.1365-294X.1999.00585.x. [DOI] [Google Scholar]
- Flath RA, Forrey RR, Guadagni DG. Aroma components of olive oil. J Agric Food Chem. 1973;21(6):948–952. doi: 10.1021/jf60190a030. [DOI] [Google Scholar]
- Gallina-Toschi T, Cerretani L, Bendini A, BonoliCarbognin M, Lercker G. Oxidative stability and phenolic content of virgin olive oil: an analytical approach by traditional and high resolution techniques. J Sep Sci. 2005;28:859–870. doi: 10.1002/jssc.200500044. [DOI] [PubMed] [Google Scholar]
- Ganino T, Beghè D, Valenti S, Nisi R, Fabbri A. RAPD and SSR markers for characterization and identification of ancient cultivars of Olea europaea L. in the Emilia region, Northern Italy. Genet Resour Crop Evol. 2007;54(7):1531–1540. doi: 10.1007/s10722-006-9145-x. [DOI] [Google Scholar]
- Gomes S, Martins-Lopes P, Lima-Brito J, Meirinhos J, Lopes J, Martins A, Guedes-Pinto H. Evidence of clonal variation in olive Verdeal-Transmontana cultivar using RAPD, ISSR and SSR markers. J Hortic Sci Biotechnol. 2008;83(4):395–400. doi: 10.1080/14620316.2008.11512397. [DOI] [Google Scholar]
- Gomes S, Martins-Lopes P, Lopes J, Guedes-Pinto H. Assessing genetic diversity in Olea europaea L. using ISSR and SSR markers. Plant Mol Biol Rep. 2009;27:365–373. doi: 10.1007/s11105-009-0106-3. [DOI] [Google Scholar]
- Gomez-Rico A, Salvador MD, La Greca M, Fregapane G. Phenolic and volatile compounds of extra virgin olive oil (Olea europaea L. cv. Cornicabra) with regard to fruit ripening and irrigation management. J Agric Food Chem. 2006;54:7130–7136. doi: 10.1021/jf060798r. [DOI] [PubMed] [Google Scholar]
- Green PS. A revision of Olea L. (Oleaceae) Kew Bull. 2002;57:91–140. doi: 10.2307/4110824. [DOI] [Google Scholar]
- Haddada F, Krichène D, Manai H, Oueslati I, Daoud D, Zarrouk M. Analytical evaluation of six monovarietal virgin olive oils from Northern Tunisia. Eur J Lipid Sci Technol. 2008;110:905–913. doi: 10.1002/ejlt.200700251. [DOI] [Google Scholar]
- Hagidimitriou M, Katsiotis A, Menexes G, Pontikis C, Loukas M. Genetic diversity of major greek olive cultivars using molecular AFLPs and RAPDs markers and morphological traits. J Am Soc Hort Sci. 2005;130(2):211–217. [Google Scholar]
- Hakim IR, Grati-Kamoun N, Makhloufi E, Rebaï A. Discovery and potential of SNP markers in characterization of Tunisian olive germplasm. Diversity. 2010;2:17–27. doi: 10.3390/d2010017. [DOI] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4(1): 9 pp
- Hassawi DS, Hadeib T. Genetic analysis of olive genotypes (Olea europaea L.) using random amplified polymorphic DNA (RAPD) J Genet Breed. 2004;58:141–148. [Google Scholar]
- Hegazi S, Hegazi A, Tawfik A, Sayed H. Molecular characterization of local and imported olive cultivars grown in Egypt using ISSR technique. J Hort Sci Ornam Plants. 2012;4(2):148–154. [Google Scholar]
- Heywood VM. Flowering plants in the world. Oxford: Oxford University Press; 1978. [Google Scholar]
- Ipek M, Seker M, Ipek A, Gul MK. Identification of molecular markers associated with fruit traits in olive and assessment of olive core collection with AFLP markers and fruit traits. Genet Mol Res. 2015;14:2762–2774. doi: 10.4238/2015.March.31.6. [DOI] [PubMed] [Google Scholar]
- Irshad F, Raza I, Ali I, Naeem R, Jabeen Z. Genetic diversity of olive germplasm using RAPD markers. J Bio- Mol Sci. 2014;2(4):93–99. [Google Scholar]
- Issaoui M, Gharbi I, Flamini G, Cioni P, BendiniToschi T, Hammami M. Aroma compounds and sensory characteristics as biomarkers of quality of differently processed Tunisian virgin olive oils. Int J Food Sci Technol. 2015;50:1764–1770. doi: 10.1111/ijfs.12830. [DOI] [Google Scholar]
- Kalua CM, Allen MS, Bedgood DR, Bishop AG, Prenzler PD, Robards K. Olive oil volatile compounds, flavour development and quality: a critical review. Food Chem. 2007;100:273–286. doi: 10.1016/j.foodchem.2005.09.059. [DOI] [Google Scholar]
- Karp A, Kresovich S, Bhat KV, Ayad W G, Hodgkin T (1997) Molecular tools in plant genetic resources conservation: a guide to the technologies. In: IPGRI Technical Bulletin No. 2. International Plant genetic Resources Institute, Rome
- Kaya E. ISSR Analysis for Determination of genetic diversity and relationship in eight Turkish olive (Olea europaea L) cultivars. Not Bot HortiAgrobo. 2015;43(1):96–99. [Google Scholar]
- Khadari B, Breton C, Moutier N, Roger JP, Besnard G, Bervillé A, Dosba F. The use of molecular markers for germplasm management in a French olive collection. Theor Appl Genet. 2003;106:521–529. doi: 10.1007/s00122-002-1079-x. [DOI] [PubMed] [Google Scholar]
- Khierallah H, Bader S, Baum M, Hamwieh A. Genetic diversity of Iraqi Date Palms revealed by Microsatellite Polymorphism. J Am Soc Hort Sci. 2011;136:282–287. [Google Scholar]
- Lazzez A, Perri E, Caravita MA, Khlif M, Cossentini M. Influence of olive maturity stage and geographical origin on some minor components in virgin olive oil of the Chemlali variety. J Agric Food Chem. 2008;56:982–988. doi: 10.1021/jf0722147. [DOI] [PubMed] [Google Scholar]
- Leon L, Beltran G, Aguilera MP, Rallo L, Barranco D, De la Rosa R. Oil composition of advanced selections from an olive breeding program. Eur J Lipid Sci Technol. 2011;113:870–875. doi: 10.1002/ejlt.201000535. [DOI] [Google Scholar]
- Linos A, Nikoloudakis N, Katsiotis A, Hagidimitriou M. Genetic structure of the Greek olive germplasm revealed by RAPD, ISSR and SSR markers. Sci Hort. 2014;175:33–43. doi: 10.1016/j.scienta.2014.05.034. [DOI] [Google Scholar]
- Longobardi F, Ventrella A, Casiello G, Sacco D, Tasioula-Margari M, Kiritsakis AK. Characterization of the geographical origin of Western Greek virgin olive oils based on instrumental and multivariate statistical analysis. Food Chem. 2012;133:169–175. doi: 10.1016/j.foodchem.2011.09.130. [DOI] [PubMed] [Google Scholar]
- Lopes MS, Mendoça D, Sefc KM, Gil FS, Camara- Machado A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. Hort Sci. 2004;39:1562–1565. [Google Scholar]
- Lopez-Cortes I, Salazar-García DC, Velázquez-Martí B, Salazar DM. Chemical characterization of traditional varietal olive oils in East of Spain. Food Res Int. 2013;54:1934–1940. doi: 10.1016/j.foodres.2013.04.035. [DOI] [Google Scholar]
- Loukas M, Krimbas CB. History of olive cultivars based on their genetic distances. J Hortic Sci. 1983;58:121–127. doi: 10.1080/00221589.1983.11515099. [DOI] [Google Scholar]
- Lumaret R, Ouazzani N. Plant genetics. Ancient wild olives in Mediterranean forests. Nature. 2001;413(6857):700. doi: 10.1038/35099680. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Martins-Lopes P, Lima-Brito J, Gomes S, Meirinhos J, Santos L, Guedes-Pinto H. RAPD and ISSR molecular markers in Olea europaea L.: genetic variability and cultivar identification. Genet Resour Crop Evol. 2007;54:117–128. doi: 10.1007/s10722-005-2640-7. [DOI] [Google Scholar]
- Martins-Lopes P, Gomes S, Lima-BritoJ Lopes J, Guedes-Pinto H. Assessment of clonal genetic variability in Olea europaea L. Cobrançosa by molecular markers. Scientia Horticulturae. 2009;123(1):82–89. doi: 10.1016/j.scienta.2009.08.001. [DOI] [Google Scholar]
- Matos L, Cunha S, Amaral J, Pereira JA, Andrade P, Seabra R. Chemometric characterization of three varietal olive oils (Cvs. Cobranc, osa, Madural and Verdeal Transmontana) extracted from olives with different maturation indices. Food Chem. 2006;102:406–414. doi: 10.1016/j.foodchem.2005.12.031. [DOI] [Google Scholar]
- Morales MT, Aparicio R, Rios JJ. Dynamic headspace gas chromatographic method for determining Volatiles in virgin olive oil. J Chromatogr. 1994;668:455–462. doi: 10.1016/0021-9673(94)80139-8. [DOI] [Google Scholar]
- Muzzalupo I, Perri E. Genetic diversity in olive tree cultivars from Italy and other countries of the Mediterranean basin as revealed by RAPD and SSR molecular marker. Adv Hort Sci. 2009;23(4):263–275. [Google Scholar]
- Newmark H. Squalene, olive oil, and cancer risk: a review and hypothesis. Cancer Epidemiol Blornarkers Prev. 1997;6:1101–1103. [PubMed] [Google Scholar]
- Nieto ML, Hodaifa G, Lozano-Peña JL. Changes in phenolic compounds and Rancimat stability of olive oils from varieties of olives at different stages of ripeness. J Sci Food Agr. 2010;90:2393–2398. doi: 10.1002/jsfa.4097. [DOI] [PubMed] [Google Scholar]
- Noormohammadi Z, Sheidai M, Dehghani A, Parvini F, Mazinani S. Inter-population genetic diversity in Oleacuspidata subsp. cuspidata revealed by SSR and ISSR markers. Acta Biologica Szegediensis. 2012;56(2):155–163. [Google Scholar]
- Noormohammadi Z, Trujillo I, Belaj A, Ataei SH, Osseini-Mazinan M. Genetic structure of Iranian olive cultivars and their relationship with Mediterranean’s cultivars revealed by SSR markers. SciHort. 2014;178:175–183. [Google Scholar]
- Ostlund RE. Phytosterols and cholesterol metabolism. Curr Opin Lipidol. 2004;15(1):37–41. doi: 10.1097/00041433-200402000-00008. [DOI] [PubMed] [Google Scholar]
- Pardo JE, Cuesta MA, Alvarruiz JD, Granell M, Álvarez-Ortí M. Evaluation of potential and real qualities of virgin olive oil from the designation of origin (DO) “Aceite Montes de Alcaraz” (Albacete, Spain) Food Chem. 2010;124:1684–1690. doi: 10.1016/j.foodchem.2010.07.068. [DOI] [Google Scholar]
- Pasqualone A, Caponio F, Blanco A. Inter-simple sequence repeat DNA markers for identification of drupes from different Olea europaea L. cultivars. Eur Food Res Technol. 2001;213:240–243. doi: 10.1007/s002170100367. [DOI] [Google Scholar]
- Pasqualone A, Montemurro C, di Rienzo V, Summo C, Paradiso VM, Caponio F. Evolution and perspectives of cultivar identification and traceability from tree to oil and table olives by means of DNA markers. J Sci Food Agric. 2016;96(11):3642–3657. doi: 10.1002/jsfa.7711. [DOI] [PubMed] [Google Scholar]
- Penner GA. RAPD analysis of plant genome. In: Jauhar PP, editor. Methods of genome analysis in plants. Boca Raton: CRC Press; 1996. pp. 251–268. [Google Scholar]
- Perez AG, León L, Pascual M, Romero-Segura C, Sánchez-Ortiz A, de la Rosa R, Sanz C. Variability of virgin olive oil phenolic compounds in a segregating progeny from a single cross in Olea europaea L. and sensory and nutritional quality implications. PLoS One. 2014;9(3):e92898. doi: 10.1371/journal.pone.0092898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AG, Rosa RD, Pascual M, Sánchez-Ortiz A, Romero-Segura C, León L, Sanz C. Assessment of volatile compound profiles and the deduced sensory significance of virgin olive oils from the progeny of Picual × Arbequina cultivars. J Chromatogr. 2016;1428:305–315. doi: 10.1016/j.chroma.2015.07.055. [DOI] [PubMed] [Google Scholar]
- Poljuha D, Sladonja B, Šetić E, Milotić A, Bandelj D, Jakše J, Javornik B. DNA fingerprinting of olive varieties in Istria (Croatia) by microsatellite markers. Sci Hortic. 2008;115:223–230. doi: 10.1016/j.scienta.2007.08.018. [DOI] [Google Scholar]
- Ranalli A, De Mattia G, Patumi M, Proietti P. Quality of virgin olive oil as influenced by origin area. Grasas Aceites. 1999;50:249–259. doi: 10.3989/gya.1999.v50.i4.663. [DOI] [Google Scholar]
- Reale S, Doveri S, Díaz A, Angiolillo A, Lucentini L, Pilla F, et al. SNP-based markers for discriminating olive (Olea europaeaL.) cultivars. Genome. 2006;49:1193–1205. doi: 10.1139/g06-068. [DOI] [PubMed] [Google Scholar]
- Reboredo-Rodríguez P, González-Barreiro C, Cancho-Grande B, Simal-Gándara J. Effects of sedimentation plus racking process in the extra virgin olive oil aroma fingerprint obtained by DHS–TD/GC–MS. Food Bioprocess Technol. 2013;6(5):1290–1301. doi: 10.1007/s11947-011-0751-z. [DOI] [Google Scholar]
- Ridolfi M, Terenziani S, Patumi M, Fontanazza G. Characterization of the lipoxygenases in some olive cultivars and determination of their role in volatile compounds formation. J Agric Food Chem. 2002;50(4):835–839. doi: 10.1021/jf0109118. [DOI] [PubMed] [Google Scholar]
- Rugini E, Lavee S. Biotechnology of perennial fruit crops. Wallingford: CAB International; 1992. Olive; pp. 371–382. [Google Scholar]
- Salimia RB, Awad MK, Kalaitzis PK. Genetic fingerprinting of Palestinian olive (Olea europaea L.) cultivars using SNP markers. Jordan J Agric Sci. 2009;5:282–294. [Google Scholar]
- Sanz-Cortes F, Badenes ML, Paz S, Iiguez A, Ljcer G. Molecular characterization of olive cultivars using RAPD markers. J Am Soc Hortic Sci. 2001;126:7–12. [Google Scholar]
- Sefc KM, Lopes MS, Mendonça D, Rodrigues Dos Santos M, Da Câmara Laimer, Machado M, Da Câmara Machado A. Identification of microsatellite loci in olive (Olea europaea L.) and their characterization in Italian and Iberian olive trees. Mol Ecol. 2000;9:1171–1173. doi: 10.1046/j.1365-294x.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- Servili M, Montedoro G. Contribution of phenolic compounds to virgin olive oil quality. Eur J Lipid Sci Technol. 2002;104:602–613. doi: 10.1002/1438-9312(200210)104:9/10<602::AID-EJLT602>3.0.CO;2-X. [DOI] [Google Scholar]
- Tapiero KD, Tew G, Nguyen Ba, Mathé G. Polyphenols: do they play a role in the prevention of human pathologies? Biomed Pharmacother. 2002;56:200–207. doi: 10.1016/S0753-3322(02)00178-6. [DOI] [PubMed] [Google Scholar]
- Vargas P, Kadereit JW. Molecular fingerprinting evidence (ISSR, inter-simple sequence repeats) for a wild status of Olea europaea L. (Oleaceae) in the Eurosiberian North of the Iberian Peninsula. Flora. 2001;196:142–152. doi: 10.1016/S0367-2530(17)30029-4. [DOI] [Google Scholar]
- Vekiari SA, Papadopoulou P, Kiritsakis A. Effects of processing methods and commercial storage conditions on the extra virgin olive oil quality indexes. Grasas Aceites. 2007;58:237–242. doi: 10.3989/gya.2007.v58.i3.178. [DOI] [Google Scholar]
- Zarrouk W, Faouzia H, Baccouri B, Oueslati I, Taamalli W, Fernandez X. Characterization of virgin olive oil from Southern Tunisia. Eur J Lipid Sci Technol. 2008;110:81–88. doi: 10.1002/ejlt.200700085. [DOI] [Google Scholar]
- Zohary D, Hopf M. Domestication of plants in the Old World. 2. Oxford: Clarendon Press; 1994. [Google Scholar]
- Zohary D, Spiegel-Roy P. Beginnings of fruit growing in old world. Science. 1975;187:319–327. doi: 10.1126/science.187.4174.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.