Abstract
Fine particulate matter (PM2.5) exposure, especially to its organic components, induces adverse health effects on the respiratory system. However, the molecular mechanisms have still not been fully elucidated. Long non-coding RNA (lncRNA) is involved in various physio-pathological processes. In this study, the roles of lncRNA were investigated to reveal the toxicology of PM2.5. Organic extracts of PM2.5 from Nanjing and Shanghai cities were adopted to treat human bronchial epithelial cell lines (BEAS-2B and A549). RNA sequencing showed that the lncRNA functioned as antisense RNA, intergenic RNA and pre-miRNA. The mRNA profiles were also altered after exposure. PM2.5 from Nanjing showed a more serious impact than that from Shanghai. In detail, higher expression of n405968 was positively related to the elevated mRNA levels of inflammatory factors (IL-6 and IL-8). Increasing levels of metastasis associated lung adenocarcinoma transcript 1 (MALAT1) were positively associated with the induced epithelial-mesenchymal transition (EMT) process. Similar response was observed between both cell lines. The higher content of polycyclic aromatic hydrocarbons (PAHs) is likely to contribute to higher toxicity of PM2.5 from Nanjing than that from Shanghai. Antagonism of aryl hydrocarbon receptor (AHR) or inhibition of CYP1A1 diminished the effects stimulated by PM2.5. Our results indicated that lncRNAs could be involved in the toxicology of PM2.5 through regulating the inflammation and EMT process.
Introduction
Serious air pollution, especially fine particulate matter (PM2.5, particles with an aerodynamic diameter of 2.5 μm or less), is a major health concern1–3. PM2.5 has adverse impacts on the respiratory system. Epidemiological studies have shown that PM2.5 is associated with abnormal lung function4, lung infection5, and lung cancer6. Long-term improvement of air quality was associated with better lung-function growth in children7. By contrast, toxicological data have been largely insufficient for understanding the molecular mechanisms. PM2.5 is composed of organic carbon (OC), elemental carbon (EC), water-soluble inorganic ions and metal elements. OC consists of various organic compounds, including PAHs, PCBs, phthalate esters, aldehydes, ketones, benzene and so on. Crustal elements (K, Fe, Mg, Al, Ca) and toxic heavy metals (Hg, Cr, Cu, Pb, Zn) are both present in PM2.5. water-soluble inorganic ions contains NO3 −, SO4 2−, NH4 +, Cl−. The constituents of PM2.5 are regional and time specific8–10. The components of PM2.5 largely affected its toxic outcome11. The particle contributed to the toxicityin a way which is similar to that of nanomaterials12, 13. In addition, organic chemicals adsorbed on the surface possess appreciable toxicity14, 15.
Long noncoding RNA (lncRNA), which has more than 200 nucleotides in length, is commonly involved in physiological and pathological process16. Growing numbers of lncRNAs are being continuously discovered, and they play important roles in cancers and other diseases17–19. The expression levels of MALAT1 (metastasis associated lung adenocarcinoma transcript 1) and AFAP1-AS1 (actin filament associated protein 1 antisense RNA 1) were both significantly up-regulated, whereas TUG1 (taurine-upregulated gene 1) and HMlincRNA 717 were both down-regulated in non-small cell lung cancer (NSCLC)20. The usefulness of lncRNA as a non-invasive marker is expected in lung disease21.
LncRNA functions in response to environmental stimuli. MALAT1 and HOTAIR were both involved in the epithelial-mesenchymal transition (EMT) induced by cigarette smoke extract22, 23. Bisphenol-A and diethylstilbestrol could alter the expression of HOTAIR in vitro and in vivo 24. LncRNA could be a potential indicator of the toxicity induced by chemical exposure25.
To the best of our knowledge, lncRNA has not yet been considered in toxicological studies of PM2.5. In this study, organic extracts of PM2.5 from Nanjing and Shanghai cities were both adopted to treat both BEAS-2B and A549 cells. These two cell lines have been widely used in toxicological studies of pollutants on the respiratory system26–28. Differentially expressed lncRNAs were co-analyzed with their corresponding mRNAs. The expressions and phenotypes of several lncRNA-mRNA pairs related to the pathology of lung cancer were further studied upon exposure. The mediating role of the AHR signaling pathway was shown in the study and was also evidenced by the presence of 16 priority polycyclic aromatic hydrocarbons (PAHs) in PM2.5.
Results
Differentially expressed lncRNAs after exposure to PM2.5
To ensure that transcriptional changes induced by exposure were related to RNA expression rather than cell viability, the effect of PM2.5 on cell viability was first determined by MTT assay (supplementary information 1, Figure S1). The results showed that the dose used in our study (5 μg/mL) did not have a significant influence on the cell viability of BEAS-2B or A549 cells. RNA sequencing was conducted to detect the profiles of RNA transcripts from BEAS-2B cells after exposure at 5 μg/mL. A total of 6,624, 6,977 and 6,055 noncoding transcripts were obtained from control, NJ-exposed and SH-exposed BEAS-2B cells, respectively. We then distinguished the differentially expressed lncRNAs between groups (Fig. 1). In total, the expression levels of 618 lncRNAs were up-regulated, and 593 lncRNAs were down-regulated after exposure to Nanjing PM2.5, compared to controls. The numbers were 496 and 518 for the SH-exposed group, which was smaller than the NJ group. These two groups shared 381 common lncRNAs with the same expression patterns. Most of the differentially expressed lncRNAs were novel, and the known lncRNAs numbered relatively few. In contrast, A total of 356 and 326 coding genes were altered at the mRNA level after exposure to PM2.5 from NJ and SH, respectively. The mRNA transcripts of 137 genes were altered after exposure to PM2.5 from both cities. Detailed information on individual lncRNAs and mRNAs are available in supplementary information 2.
Figure 1.
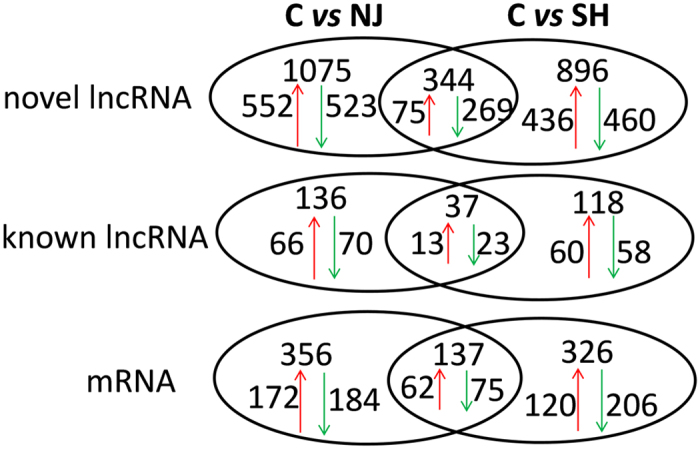
Comparison of the differentially expressed RNAs. C: control group, NJ: organic extract of PM2.5 from Nanjing city, SH: organic extract of PM2.5 from Shanghai city.
Functional prediction of differentially expressed lncRNAs
The predicted functions of differentially expressed genes were further categorized (Table 1). A total of 1,633 lncRNAs were predicted to be the antisense of coding transcripts. In contrast, the numbers were 253 and 4,340 for pre-miRNA and intergenic lncRNA, respectively. Most of the differentially expressed lncRNAs were predicted to be intergenic genes. They are located upstream or downstream of the coding genes. The numbers of antisense lncRNAs were 33 and 23 in the NJ and SH PM2.5-treated groups, respectively. They could form the antisense lncRNA-mRNA duplex. Novel pre-miRNA was also discovered from these differentially expressed lncRNAs.
Table 1.
Classification of differentially expressed lncRNA.
| C vs NJ | C vs SH | common | |
|---|---|---|---|
| Antisense | 33 | 23 | 12 |
| Up/downstream | 377 | 320 | 117 |
| PremiRNA_known | 5 | 7 | 2 |
| PremiRNA_novel | 43 | 33 | 12 |
| Family | 1 | 1 | 1 |
Antisense: antisense to related coding genes; Up/down stream: upstream lncRNA, overlaps with the promoter regions or other cis-regulatory elements of their co-expressed coding genes; downstream lncRNA, the 3’UTRs or downstream regions of protein-coding genes.
Functional analysis of differentially expressed mRNAs
The mRNA profiles were also studied after exposure. GO analysis showed that differentially expressed genes possessed various functions (supplementary information 1 Figure S2). The significantly enriched KEGG pathways are listed in supplementary information 1 Table S2. Both treatments shared the majority of pathways. They were mainly related to human disease, environmental information processing, and metabolism.
LncRNA-mRNA pairs and their phenotypes
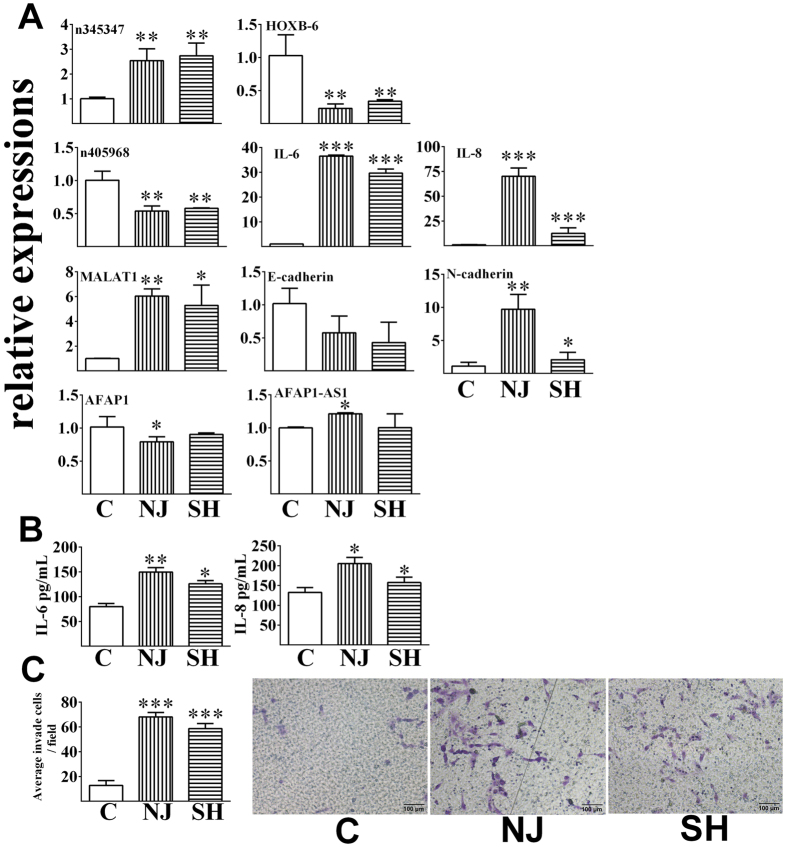
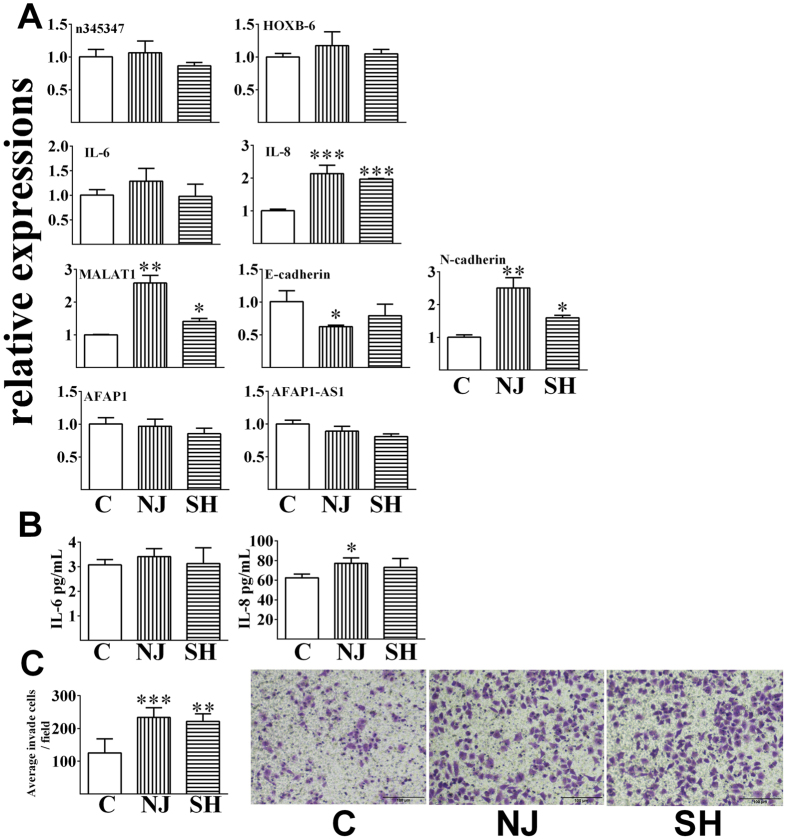
The levels of several lncRNA and mRNA pairs were further confirmed by RT-PCR (Fig. 2A). In BEAS-2B cells, the expression level of n345347 was significantly up-regulated upon exposure to PM2.5 from both cities. Its antisense gene (homeobox B6, HOXB-6) showed a reverse response to exposure, which decreased significantly upon exposure. The expression of n405968 was significantly down-regulated following either exposure. Abnormal expression of n405968 was related to the inflammatory response. The mRNA levels of interleukins (IL-6 and IL-8) were both significantly up-regulated upon exposure to PM2.5 from either city. Secreted IL-6 and IL-8 were also elevated in the cell culture media (Fig. 2B). The transcript of MALAT1 was markedly induced following either exposure. This increasing trend was positively related to the EMT process. Decreased levels of E-cadherin and elevated levels of N-cadherin were observed after exposure. The cell invasion ability was enhanced upon exposure (Fig. 2C). The expression level of AFAP1-AS1 increased, whereas the level of its sense gene AFAP1 decreased, following treatment with PM2.5 from Nanjing. In contrast, PM2.5 from Shanghai did not significantly affect the expression levels of this gene pair. The expression levels of related genes were also detected in A549 cells upon exposure (Fig. 3). Neither of the levels of n345347 and HOXB-6 was significantly influenced after exposure to PM2.5 from either city (Fig. 3A). The level of n405968 was not detectable in A549 cells. PM2.5 exposure did not significantly affect the level of IL-6. By contrast, the level of IL-8 significantly increased at both mRNA and protein levels which was similar to that occurred in BEAS-2B cells (Fig. 3B). Responses of MALAT1, E-cadherin and N-cadherin in A549 cells were also similar between these two cell types. The invasive ability of A549 cells was enhanced compared to control group upon exposure (Fig. 3C).
Figure 2.
The relative expression levels of lncRNA-mRNA pairs and the related phenotypes of BEAS-2B cells upon exposure. (A) Relative expression levels after exposure for 48 h by real-time RT-PCR; (B) The levels of IL-6 and IL-8 in the cell culture media after exposure for 48 h, measured by ELISA; (C) The number of invaded cells after exposure measured using a transwell assay. All data are presented as the means ± SDs (n = 3). Independent-samples t-tests were used for data analysis, *p < 0.05, **p < 0.01, ***p < 0.001.
Figure 3.
The relative expression levels of lncRNA-mRNA pairs and the related phenotypes of A549 cells upon exposure. (A) Relative expression levels after exposure for 48 h by real-time RT-PCR; (B) The levels of IL-6 and IL-8 in the cell culture media after exposure for 48 h, measured by ELISA; (C) The number of invaded cells after exposure measured using a transwell assay. All data are presented as the means ± SDs (n = 3). Independent-samples t-tests were used for data analysis, *p < 0.05, **p < 0.01, ***p < 0.001.
Determination of PAH levels
The levels of sixteen priority PAHs were determined, and their total concentrations are presented in Table 2. Total PAH levels of PM2.5 in Nanjing (21.687 ng/m3) were higher than those in Shanghai (15.695 ng/m3). Total BaP equivalent concentrations in Nanjing were 53% higher than those in Shanghai.
Table 2.
PAH contents of PM2.5 from Nanjing and Shanghai.
| ng/m3 | NJ | SH |
|---|---|---|
| Total PAHs | 21.687 | 15.695 |
| Total BaP equivalence | 3.623 | 2.614 |
Total PAHs were the sums of 16 priority PAHs. Total BaP equivalence was the sum of the equivalences of individual PAHs.
Mediating roles of the AHR signaling pathway on the effects of PM2.5
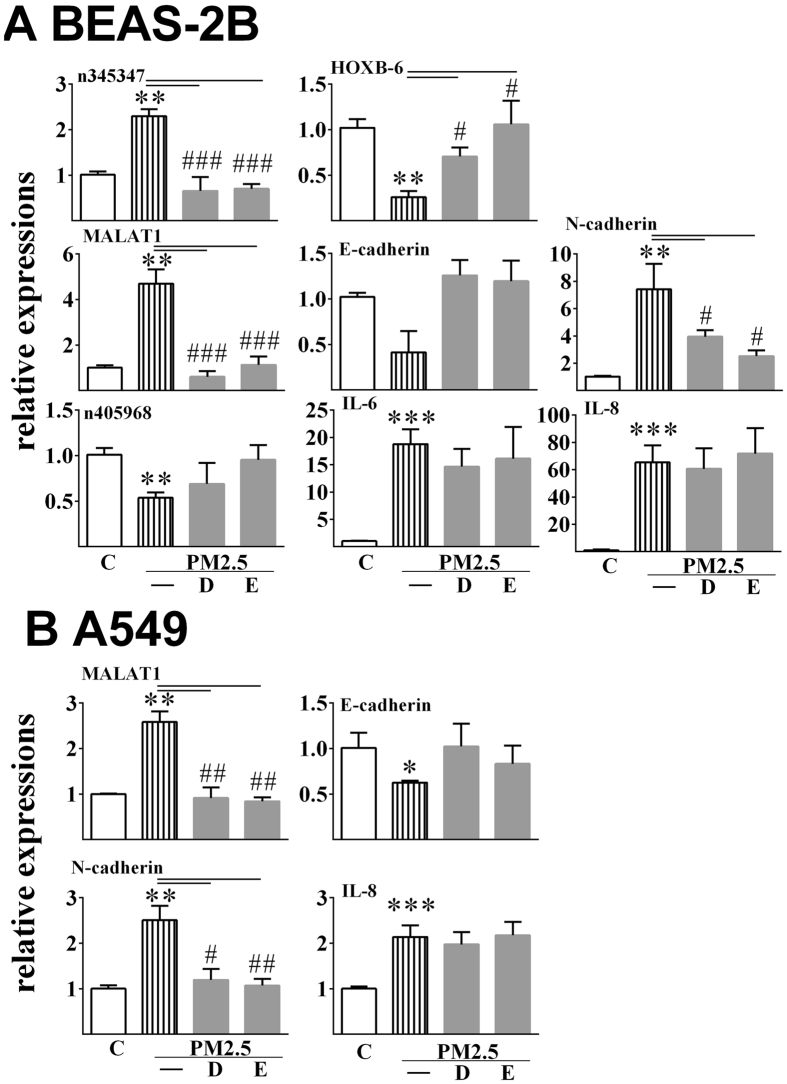
Cells were co-treated with Nanjing PM2.5 and AHR antagonist or CYP1A1 inhibitor (Fig. 4). Elevated levels of MALAT1 stimulated by PM2.5 significantly fell back to the level which was equal to that of control group after co-treatment in both cells. In accordance, the mRNA level of E-cadherin was higher in the co-exposed group than that in the single PM2.5-exposed group. The mRNA level of N-cadherin in the co-exposed group was significantly lower than in the single PM2.5-exposed group. Antagonism of AHR and inhibition of CYP1A1 both significantly diminished the stimulated effects of n345347 in BEAS-2B cells exposure to PM2.5. In the case of HOXB-6, both treatments led to the recovery of its expression in BEAS-2B cells. Neither of the chemicals affected the responses of n405968 and AFAP1-AS1 to PM2.5 exposure in both cells. Notably, the co-treatment did not significantly diminish the stimulatory effects of PM2.5 on the levels of both IL-6 and IL-8 in both cells. Similar trends were observed in co-treatment with PM2.5 from Shanghai.
Figure 4.
Effects of the antagonist on the relative expression levels of lncRNA and its related coding genes induced by PM2.5 in BEAS-2B cells (A) and A549 cells (B). All data are presented as the means ± SDs (n = 3). Independent-samples t-tests were used for data analysis, **p < 0.01 vs control group; # p < 0.05, ## p < 0.01, ### p < 0.001, PM2.5 single exposed group vs PM2.5/antagonist co-exposed group. DMF (D), antagonist to AHR; ellipticine (E), inhibitor of CYP1A1.
Discussion
The roles of lncRNAs in the toxicity of chemicals have been receiving attention recently. In this study, we detected the responses of lncRNAs to organic extracts of PM2.5 exposure in human bronchial epithelial cells. The lncRNA landscape in bronchial epithelial cells was obtained by RNA sequencing. LncRNA was widely involved in the toxicity of PM2.5. Previous studies of the toxicology of PM2.5 were all conducted at the level of DNA, mRNA, miRNA or protein. Our results provided a new layer of lncRNA to facilitate the understanding of PM2.5 toxicology. Differentially expressed lncRNAs were widely detected following exposure to PM2.5 from either city. Additionally, abnormal expression of lncRNA also occurred at the clinical level. Dysregulated lncRNA was widely observed in patients with lung diseases29.
In our study, two types of lung epithelial cells (BEAS-2B and A549) were adopted to determine the effects of organic components of PM2.5 on lung cells. PM2.5 has no significant toxicity to the viability of both cells at lower dose of 5 μg/mL. Inflammation and EMT are both involved in the etiology of lung cancer. LncRNA n405968 is the host gene of microRNA-155 (MIR155), which participates in inflammation30. In our study, the levels of IL-6 and IL-8 both increased in BEAS-2B cells after PM2.5 exposure. PM2.5 exposure also triggered the release of IL-8 in A549 cells. However, the level of IL-6 stayed at low levels after exposure. Regulation of n405968 might be one of the ways by which PM2.5 triggers the inflammatory response in BEAS-2B cells rather than in A549 cells. MALAT1 (also known as NEAT2) is involved in the regulation of cell cycle, cell migration, and cancer metastasis22. In our study, elevated levels of MALAT1 were positively associated with EMT processes, as indicated by the down-regulation of E-cadherin, up-regulation of N-cadherin, as well as the invasive ability of cells. PM2.5 likely triggers the EMT process by enhancing the expression of MALAT1. AFAP1-AS1 and its sense transcript AFAP1 showed reverse transcript patterns upon NJ PM2.5 exposure in BEAS-2B cells. LncRNA could function as an antisense transcript of the coding genes. In our study, AFAP1-AS1 and its antisense transcript AFAP1 showed reverse expression patterns upon PM2.5 exposure. A large amount of antisense lncRNA was predicted in our study. LncRNA (n345347) is the antisense to HOXB. HOXB acts as the transcription factor, and it is involved in lung development and related diseases, such as small cell lung cancers and bronchopulmonary sequestration31, 32.
Diverse sources of PM2.5 differ in their constituents and health outcomes33, 34. PM2.5 from Nanjing is predominantly from coal combustion whereas the major source is diesel and gasoline exhaust in Shanghai (as demonstrated by the local environmental protection bureaus: Nanjing: http://hbj.nanjing.gov.cn/; Shanghai: http://www.sepb.gov.cn). Concentrations of total PAHs and the corresponding benzopyrene (BaP) equivalences were higher in Nanjing than in Shanghai. Although the content in PM is very small, PAHs were regarded as the primary toxic constituents of PM and contributed to a higher risk of lung cancer35–37. In accordance, more differentially expressed lncRNAs were detected upon exposure to PM2.5 from Nanjing than from Shanghai. Levels of secreted inflammatory factors and their invasion ability were higher after treatment with PM2.5 from Nanjing than that from Shanghai. PAHs functioned through AHR receptor, which is an important mediator of the toxicity of organic chemicals. In this study, we found that this receptor-mediated effect also existed in the regulation of lncRNA expressions by PM2.5. Antagonism of AHR or inhibition of CYP1A1 obviously diminished the influence of PM2.5 on lncRNA-mRNA expression pairs. AHR mediated the carcinogenic effect stimulated by pollutants38.
In conclusion, our study provided new information on the toxic mechanism by which PM2.5 exerted its adverse health impacts through affecting the expressions of lncRNAs. LncRNA was involved in the inflammatory reaction and in the EMT process of lung epithelial cells. The AHR-CYP1A1 pathway mediated the effects of PAH-bound PM2.5 on lncRNA. This molecule could be a valuable target for revealing the toxicological mechanism of PM2.5.
Materials and Methods
Sample collection and extraction
PM2.5 was collected from Nanjing city (Gulou Campus, Nanjing University, 15 m height, 32° 055′ N, 118° 776′ E) and Shanghai city (Shanghai Academy of Environmental Sciences, 18 m height, 31°172′N, 121°425′ E) in the Yangtze River Delta region of China in Autumn 2014. The sampling period was on weekdays from November 6th to 17th. The climate parameters were as follows: average temperature (Nanjing, 21.3 ± 2.7 °C; Shanghai, 21.0 ± 2.9 °C) and barometric pressure (Nanjing, 765.1 ± 2.1 mm Hg; Shanghai, 765.4 ± 2.5 mm Hg). PM2.5 was captured on quartz filters (22.5 cm × 18.2 cm, Whatman Company, Kent, UK) at a flow rate of 1.05 m3/min by a large-size volume active sampler (HiVol 3000 air sampler, Ecotech, Australia). The average mass of PM2.5 in each membrane was 135.5 ± 37.6 mg and 121.2 ± 33.9 mg for Nanjing and Shanghai, respectively. One eighth of each membrane was used for solvent extraction of organic components. The remainder was dedicated to chemical analysis. For each city, samples from eight days were randomly pooled together for extraction. The membrane was cut into pieces, and then soxhlet extracted for 24 h in dichloromethane. The extract was dried under nitrogen sweeping. Then, the extracts were re-suspended at a stock concentration of 50 mg/mL in dimethyl sulfoxide (DMSO) to facilitate dissolution in cell media.
Cell culture and treatment
A human bronchial epithelial cell line (BEAS-2B) was obtained from the China Center for Type Culture Collection (CCTCC). The cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium) (HyClone, Logan, UT) supplemented with 10% FBS in a 5% CO2 humidified chamber at 37 °C. The human lung adenocarcinoma A549 cells (the Cell Bank of Type Culture Collection of Chinese Academy of Sciences) were maintained in DME/F12 (1:1) medium (HyClone, Logan, UT) supplemented with 10% FBS (fetal bovine serum) under the same condition. Both cells were exposed to organic extract of PM2.5 from both cities at the designated doses. The control group received the same extract from a null membrane. The dose of organic extract (5 μg/mL) equaled 10–20 μg/m3 of total PM2.5. These PM2.5 concentrations are in the regimen of the World Health Organization (WHO) annual standard39. In addition, this dose did not induce significant cellular toxicity as measured by MTT assay (Supplementary Figure 1). 3′,4′-dimethoxyflavone (DMF, AHR antagonist) and ellipticine (CYP1A1 inhibitor) were both co-incubated with PM2.5. Their final concentrations were both 10 μM.
MTT assay
Cell viability was determined by MTT (Sigma-Aldrich, St Louis, MO, USA) colorimetric assay. The cells were seeded in 96-well plates at a density of 6,000 per well. The serial exposure concentrations were 0, 5, 10, 25, 50, 75 and 100 μg/mL. After an exposure period of 48 h, MTT was added, and the optical density (OD) was measured at 550 nm by a SpectraMAX M5 microplate reader (Molecular Devices, CA, USA). Cell viability was calculated as the relative value of the exposed group vs the control group.
RNA isolation
Total RNA was obtained from cells by an RNA extraction kit (Omega Bio-Tek, Norcross, USA), according to the manufacturer’s instructions. RNA quality and quantity were determined on a nanophotometer (IMPLEN, GmbH, Germany). Then, equal amounts from each of the four replicates were pooled together for RNA sequencing.
RNA sequencing and bioinformatics analysis
The noncoding RNA and mRNA were enriched from total RNA of BEAS-2B cells by removing rRNA. Then, they were fragmented into short fragments (approximately 200–500 nt). The fragments were used to synthesize first-strand cDNA by a random hexamer primer. PCR amplification was then performed to construct a library. The library was sequenced using the Illumina HiSeqTM 2000 at the Beijing Genomics Institute. Raw sequencing data from the control, Nanjing and Shanghai PM2.5-exposed groups were assembled and predicted independently. Then, all of the data were merged together to remove the redundant transcripts and to optimize the structure of the transcripts. Transcript assemblies were generated using Cufflinks 40. Cuffcompare was utilized to distinguish known and novel lncRNA transcripts, based on UCSC hg19 and NONCODE V3.041. RNA expression levels were compared between different groups by Cuffdiff. RNAplex was adopted to predict antisense lncRNA-mRNA duplexes42. Pre-miRNA was predicted by miRBase43. The lncRNA family was predicted by the Rfam database44.
Real-time RT-PCR
First strand cDNA was synthesized from total RNA using the SYBR Premix Ex TaqTM kit (Takara, Dalian, China). The second PCR reaction was performed as follows: the thermal cycle of an initial denaturation step at 95 °C for 30 s, then 40 cycles at 95 °C for 5 s and 60 °C for 20 s on a Roche 480 instrument. A dissociation step was adopted to confirm the correct amplification of the target gene. Three replicates of each PCR reaction were performed for each tested gene. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the expression levels of target genes. The relative expression levels of the tested genes were analyzed using the 2−ΔΔCt method45. The primer sequences are listed in supplementary information 1 Table S1.
Enzyme-linked immunosorbent assay (ELISA)
The culture medium was collected for ELISA after treatment of cells with PM2.5. Concentrations of both interleukin-6 (IL-6) and IL-8 were measured by ELISA following the manufacturer’s instructions (R&D systems, Minneapolis, MN).
Cell invasion
Transwell assay was used to examine the cell invasion using a Matrigel invasion chamber with polycarbonate membranes (8-μm pores, Corning) coated with Matrigel (BD Biosciences). The invaded cells were fixed, stained, and counted by averaging ten fields under an Olympus CKX41 inverted microscope (Olympus, Tokyo, Japan).
Determination of total PAHs and benzo(a)pyrene (BaP) toxic equivalents
The concentrations of sixteen priority PAHs were determined in PM2.5 by GC-MS/MS following our previous study46. The total BaP equivalent concentration was obtained by totaling the equivalence values of individual PAHs. The individual equivalence was calculated by multiplying the concentration by its BaP coefficient, according to the previous study47.
Electronic supplementary material
Acknowledgements
The work was supported by the National Natural Science Foundation of China (41390240, 21477123, 21477124, and U1405235) and by the Institute of Urban Environment, Chinese Academy of Sciences (IUEMS201405).
Author Contributions
Qiansheng Huang developed the overall hypothesis and study design, analyzed the data and wrote the paper. Yulang Chi performed the cell experiments. Junjun Deng measured the concentrations of PAHs. Yiyao Liu performed the RT-PCR experiments. Yanyang Lu conducted the transwell assay. Jinsheng Chen provided the PM2.5 sample and advised the study. Sijun Dong was the principal investigator. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09818-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinsheng Chen, Email: jschen@iue.ac.cn.
Sijun Dong, Email: sjdong@iue.ac.cn.
References
- 1.Ma ZW, et al. Satellite-Based Spatiotemporal Trends in PM2.5 Concentrations: China, 2004–2013. Environ Health Persp. 2016;124:184–192. doi: 10.1289/ehp.1409481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiao L, et al. PM2.5 constituents and hospital emergency-room visits in Shanghai, China. Environ Sci Technol. 2014;48:10406–10414. doi: 10.1021/es501305k. [DOI] [PubMed] [Google Scholar]
- 3.Xu P, Chen Y, Ye X. Haze, air pollution, and health in China. Lancet. 2013;382 doi: 10.1016/S0140-6736(13)62693-8. [DOI] [PubMed] [Google Scholar]
- 4.Gehring U, et al. Air pollution exposure and lung function in children: the ESCAPE project. Environ Health Perspect. 2013;121:1357–1364. doi: 10.1289/ehp.1306770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacIntyre EA, et al. Air pollution and respiratory infections during early childhood: an analysis of 10 European birth cohorts within the ESCAPE Project. Environ Health Perspect. 2014;122:107–113. doi: 10.1289/ehp.1306755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamra GB, et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ Health Perspect. 2014;122:906–911. doi: 10.1289/ehp/1408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gauderman WJ, et al. Association of improved air quality with lung development in children. N Engl J Med. 2015;372:905–913. doi: 10.1056/NEJMoa1414123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, et al. Seasonal variations and chemical characteristics of PM2.5 in Wuhan, central China. Sci Total Environ. 2015;518:97–105. doi: 10.1016/j.scitotenv.2015.02.054. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, et al. Pollution characteristics, sources and lung cancer risk of atmospheric polycyclic aromatic hydrocarbons in a new urban district of Nanjing, China. J Environ Sci (China) 2017;55:118–128. doi: 10.1016/j.jes.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Ming L, et al. PM2.5 in the Yangtze River Delta, China: Chemical compositions, seasonal variations, and regional pollution events. Environ Pollut. 2017;223:200–212. doi: 10.1016/j.envpol.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Lippmann M. Toxicological and epidemiological studies of cardiovascular effects of ambient air fine particulate matter (PM2.5) and its chemical components: Coherence and public health implications. Crit Rev Toxicol. 2014;44:299–347. doi: 10.3109/10408444.2013.861796. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Ying GG, Jones KC, Martin FL. Real-world carbon nanoparticle exposures induce brain and gonadal alterations in zebrafish (Danio rerio) as determined by biospectroscopy techniques. Analyst. 2015;140:2687–2695. doi: 10.1039/C4AN02227K. [DOI] [PubMed] [Google Scholar]
- 13.Li J, et al. Low-dose carbon-based nanoparticle-induced effects in A549 lung cells determined by biospectroscopy are associated with increases in genomic methylation. Sci Rep. 2016;6 doi: 10.1038/srep20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh SM, Kim HR, Park YJ, Lee SY, Chung KH. Organic extracts of urban air pollution particulate matter (PM2.5)-induced genotoxicity and oxidative stress in human lung bronchial epithelial cells (BEAS-2B cells) Mutat Res-Gen Tox En. 2011;723:142–151. doi: 10.1016/j.mrgentox.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Osornio-Vargas AR, et al. Proinflammatory and cytotoxic effects of Mexico City air pollution particulate matter in vitro are dependent on particle size and composition. Environ Health Perspect. 2003;111:1289–1293. doi: 10.1289/ehp.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Xing Z, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao H, et al. Emerging role of long noncoding RNAs in lung cancer: Current status and future prospects. Resp Med. 2016;110:12–19. doi: 10.1016/j.rmed.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Vencken SF, Greene CM, McKiernan PJ. Non-coding RNA as lung disease biomarkers. Thorax. 2015;70:501–503. doi: 10.1136/thoraxjnl-2014-206193. [DOI] [PubMed] [Google Scholar]
- 22.Lu L, et al. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;289:276–285. doi: 10.1016/j.taap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;282:9–19. doi: 10.1016/j.taap.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Bhan A, et al. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrone AK, Beland FA, Pogribny IP. Noncoding RNA response to xenobiotic exposure: an indicator of toxicity and carcinogenicity. Expert Opin Drug Metab Toxicol. 2014;10:1409–1422. doi: 10.1517/17425255.2014.954312. [DOI] [PubMed] [Google Scholar]
- 26.Ekstrand-Hammarstrom B, et al. Human primary bronchial epithelial cells respond differently to titanium dioxide nanoparticles than the lung epithelial cell lines A549 and BEAS-2B. Nanotoxicology. 2012;6:623–634. doi: 10.3109/17435390.2011.598245. [DOI] [PubMed] [Google Scholar]
- 27.Courcot E, et al. Xenobiotic Metabolism and Disposition in Human Lung Cell Models: Comparison with In Vivo Expression Profiles. Drug Metab Dispos. 2012;40:1953–1965. doi: 10.1124/dmd.112.046896. [DOI] [PubMed] [Google Scholar]
- 28.Fischer BM, et al. Use of high-throughput RT-qPCR to assess modulations of gene expression profiles related to genomic stability and interactions by cadmium. Arch Toxicol. 2016;90:2745–2761. doi: 10.1007/s00204-015-1621-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White NM, et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532:1–12. doi: 10.1016/j.gene.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Flagiello D, Poupon MF, Cillo C, Dutrillaux B, Malfoy B. Relationship between DNA methylation and gene expression of the HOXB gene cluster in small cell lung cancers. Febs Letters. 1996;380:103–107. doi: 10.1016/0014-5793(96)00017-8. [DOI] [PubMed] [Google Scholar]
- 32.Volpe MV, Archavachotikul K, Bhan I, Lessin MS, Nielsen HC. Association of bronchopulmonary sequestration with expression of the homeobox protein Hoxb-5. J Pediatr Surg. 2000;35:1817–1819. doi: 10.1053/jpsu.2000.19266. [DOI] [PubMed] [Google Scholar]
- 33.Sarnat SE, Winquist A, Schauer JJ, Turner JR, Sarnat JA. Fine particulate matter components and emergency department visits for cardiovascular and respiratory diseases in the St. Louis, Missouri-Illinois, metropolitan area. Environ Health Perspect. 2015;123:437–444. doi: 10.1289/ehp.1307776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laden F, Neas LM, Dockery DW, Schwartz J. Association of fine particulate matter from different sources with daily mortality in six U.S. cities. Environ Health Perspect. 2000;108:941–947. doi: 10.1289/ehp.00108941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue HF, Yun Y, Gao R, Li GK, Sang N. Winter Polycyclic Aromatic Hydrocarbon-Bound Particulate Matter from Peri-urban North China Promotes Lung Cancer Cell Metastasis. Environ Sci Technol. 2015;49:14484–14493. doi: 10.1021/es506280c. [DOI] [PubMed] [Google Scholar]
- 36.Pozzoli L, et al. Polycyclic aromatic hydrocarbons in the atmosphere: Monitoring, sources, sinks and fate. I: Monitoring and sources. Ann Chim-Rome. 2004;94:17–32. doi: 10.1002/adic.200490002. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Tao S, Shen H, Ma J. Inhalation exposure to ambient polycyclic aromatic hydrocarbons and lung cancer risk of Chinese population. Proc Natl Acad Sci U S A. 2009;106:21063–21067. doi: 10.1073/pnas.0905756106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Safe S, Lee SO, Jin UH. Role of the Aryl Hydrocarbon Receptor in Carcinogenesis and Potential as a Drug Target. Toxicol Sci. 2013;135:1–16. doi: 10.1093/toxsci/kft128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO. World Health Organization air quality guidelines for particulate matter, ozone, nitrogen dioxide and sulfur dioxide. http://apps.who.int/iris/bitstream/10665/69477/1/WHO_SDE_PHE_OEH_06.02_eng.pdf (2006). [PubMed]
- 40.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bu D, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tafer H, Hofacker IL. RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-[Delta][Delta]CT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Hong Y, et al. Effects of urbanization on gaseous and particulate polycyclic aromatic hydrocarbons and polychlorinated biphenyls in a coastal city, China: levels, sources, and health risks. Environ Sci Pollut Res Int. 2015;22:14919–14931. doi: 10.1007/s11356-015-4616-2. [DOI] [PubMed] [Google Scholar]
- 47.Tsai PJ, et al. Assessing and predicting the exposures of polycyclic aromatic hydrocarbons (PAHs) and their carcinogenic potencies from vehicle engine exhausts to highway toll station workers. Atmos Environ. 2004;38:333–343. doi: 10.1016/j.atmosenv.2003.08.038. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.