Abstract
Nuclear factor Y (NF-Y) is a heterotrimeric transcription factor with three distinct NF-YA, NF-YB and NF-YC subunits. It plays important roles in plant growth, development and stress responses. We have reported earlier on development of gain-of-function mutants in an indica rice cultivar, BPT-5204. Now, we screened 927 seeds from 70 Ac/Ds plants for salinity tolerance and identified one activation-tagged salt tolerant DS plant (DS-16, T3 generation) that showed enhanced expression of a novel ‘histone-like transcription factor’ belonging to rice NF-Y subfamily C and was named as OsNF-YC13. Localization studies using GFP-fusion showed that the protein is localized to nucleus and cytoplasm. Real time expression analysis confirmed upregulation of transcript levels of OsNF-YC13 during salt treatment in a tissue specific manner. Biochemical and physiological characterization of the DS-16 revealed enhanced K+/Na+ ratio, proline content, chlorophyll content, enzymes with antioxidant activity etc. DS-16 also showed transcriptional up-regulation of genes that are involved in salinity tolerance. In-silico analysis of OsNF-YC13 promoter region evidenced the presence of various key stress-responsive cis-regulatory elements. OsNF-YC13 subunit alone does not appear to have the capacity for direct transcription activation, but appears to interact with the B- subunits in the process of transactivation.
Introduction
Rice (Oryza sativa L.) is a major staple food crop consumed globally by more than three billion people. Various abiotic stresses such as extreme temperature, water deficiency, high salinity and submergence adversely affect rice crop growth and productivity. Soil salinity, in addition to other abiotic stresses, is one of the most severe problems in agriculture and partly responsible for lower rice productivity1. Increasing soil salinization in irrigated areas has necessitated the identification of genes that confer resistance to salinity either by conventional breeding or genetic engineering2.
Salinity is a complex trait, which affects almost every aspect of the physiology and biochemistry of plants both at whole plant and cellular levels through osmotic and ionic stresses3, 4. Absorption of excessive salt inhibits both root and shoot growth, reduces reproductive activity and affects viability of plants. To counter salinity stress, plant cells have several defense mechanisms. These include secretion of toxic levels of sodium ions from the cytosol to the apoplast or into the vacuole mediated by the Na+/H+ exchanger, SOS1 located at the plasma membrane and the vacuolar Na+/H+ antiporter NHX1, respectively5.
Recent molecular genetic studies of rice have also revealed several factors involved in mechanisms that ensure plant growth under saline conditions particularly root using loss-of-function mutants6, 7. Development of salinity tolerant rice varieties through transgenic approach is an efficient option to overcome salinity stress. Several studies reported that overexpression of stress-induced/transcription factor/transporter genes from rice or other plant species conferred salinity tolerance in rice. Some of these genes conferring enhanced salt tolerance in rice are OsLEA4 8, OsPP1a 9, OsGly1 10, OsJRL 11. Although transgenic plants targeting the modification of a single gene have shown improved tolerance to salinity, progress in this area of research is hampered due to lack of complete understanding of the molecular basis of salt tolerance, which involves a complex network of genes operating in close coordination12, 13. Hence, there is still the possibility of identifying further novel genes through functional genomic approaches, which facilitate the identification of novel genes and their functions that would greatly strengthen our efforts in the development of stress tolerance (both biotic/abiotic) in rice. In order to identify novel candidate gene(s), an efficient gain-of-function mutagenesis strategy called ‘activation-tagging’ (AT) was employed in an indica rice variety. AT is a powerful tool used for generating huge numbers of independent transformed lines with the possibility of gain-of-function mutagenesis and identification of novel genes. High throughput profiling of these activation-tagging lines provides useful resources to identify genes that are involved in regulatory or biosynthesis pathways14. This system uses tetramer copies of cauliflower mosaic virus (CaMV) 35 S enhancers15 and the enhancers upon integration in the recipient plant genome can function in either orientation at the place of insertion, thereby causing transcriptional activation of nearby genes resulting in dominant gain-of-function mutations16. There are several reports citing this technique with the development of AT lines in Arabidopsis 17, 18, japonica rice16, 19, indica rice20, tomato21, 22, poplar23, strawberry, potato24, barley25 and sorghum26.
Transcription factors (TFs) are a group of regulatory proteins that regulate the expression of genes under different environmental stress conditions27, 28. These TFs bind to specific cis-elements in the promoter regions of stress-responsive genes and initiate complex signaling cascades in various biosysnthetic pathways including those in response to abiotic stresses29. Nuclear factor Y (NF-Y), also known as Heme activator protein (HAP) or CCAAT binding factor (CBF) is a heterotrimeric complex transcription factor with high affinity and sequence specificity to a cis-element, CCAAT box in eukaryote promoters and has been reported to play roles in the activation of diverse genes30. This heterotrimeric complex is composed of three subunits: NF-YA, NF-YB and NF-YC. A unique pattern of heterotrimer formation of NF-Y complex happens first in cytoplasm with NF-YB and NF-YC forming a heterodimer, which subsequently translocates to the nucleus, where the dimer interacts with NF-YA subunit to form a mature NF-Y transcription factor31, 32. It is binding to a specific sequence (CCAAT) in a promoter results in either positive or negative transcriptional regulation of genes under its control33.
In most eukaryotic genomes, the NF-Y is encoded by a single gene, whereas in plant genomes, each subunit is encoded by a family of genes34. Genome wide survey of NF-Ys in Arabidopsis revealed 10 NF-YA, 13 NF-YB, and 13 NF-YC genes35–37. In wheat, 37 NF-Y and Dr1 genes (10 NF-YA, 11 NF-YB, 14 NF-YC and 2 Dr1) were identified38. Recently, several groups have identified NF-Y genes in different plant species such as 68 NF-Ys in soybean39, 59 NF-Ys in tomato40, 27 in Prunus mume 41, 34 in Vitis vinifera 42. In rice, 28 NF-Y genes were identified including 10 NF-YAs, 11 NF-YBs, 7 NF-YCs43. However, Yang and co-workers44 reported the identification of some other genes in the Rice Genome Annotation Project (RGAP) database, which were included in the NF-Y family enlarging the total to 40 putative NF-Y genes (11 NF-YA, 13 NF-YB, and 16 NF-YC). Reports suggest that NF-Y gene encoded proteins undertake combinatorial and functional diversity like plant growth, development and stress responses. However, the exact molecular mechanism of NF-Y towards plant development and stress response is unclear, which needs to be explored45, 46.
The objective of the current investigation is to identify salt-tolerant rice lines among the activation-tagged indica rice (variety BPT 5204, Samba Mahsuri) lines and characterize the flanking sequences on either side of the integration of Ds element carrying multiple enhancers and study the expression of genes in order to identify the tagged genes. In this communication, we report on the identification and characterization of a novel nuclear factor Y gene (NF-YC13) in one of the identified salt tolerant AT lines that has been shown to impart salt tolerance in rice.
Results
Screening and confirmation of identified salt-tolerant DS lines
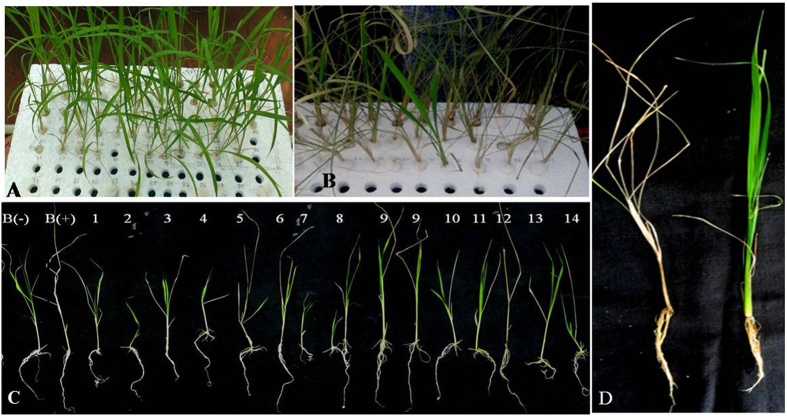
The rice variety BPT 5204 was transformed with pSQ5 vector, which carries the Ac element with the hph gene for plant selection, Ds element carrying multiple 35S enhancers and RFPgene47 and the transgenic plants carrying both Ac/Ds elements and stable Ds lines were developed20. A total of 1828 seeds from 70 transgenic Ac/Ds element carrying plants in T3 generation were inoculated in ½ MS medium. About 927 seeds germinated into seedlings that were subsequently transferred to Yoshida’s culture solution for 3–5 d in the transgenic greenhouse (Fig. 1A). At two or three-leaf stage (15–20 d), seedlings were treated with 150 mM sodium chloride at pH 5.8 for two weeks. In the salt treatment, fresh solution was replaced every three days to maintain the pH 5.8. Along with the transgenic plants, seedlings of wild type BPT-5204 were also transferred and treated in the same tray. On the third day of salt stress, the older leaves started rolling and drying up. During the stress treatment, some of the stable Ds plants showed tolerance where the leaves remained green without the burning symptoms, whereas the susceptible transgenic plants along with the controls were completely dried and died (Fig. 1B and C). After stress treatment (15 d), the tolerant plants were transferred to fresh Yoshida’s culture solution and allowed for 1–2 weeks for recovery from the salt stress treatment (Fig. 1D). Out of 927 seedlings (70 lines), 59 showed salt tolerance to varying levels and recovered. After complete recovery of the 59 plants, 35 plants showed normal growth in vegetative as well as reproductive stages, while the remaining 24 plants showed retarded growth.
Figure 1.
Hydroponic screening of Ds plants for salinity tolerance. (A) Two weeks old seedlings were in Yoshida’s solution before stress treatment. (B) Seedlings in Yoshida’s solution supplemented with 150 mM NaCl. (C) Salt tolerant Ds plants after NaCl treatment. (D) Plants after recovery (Represented line). B(−): WT-BPT without stress; B( + ): WT-BPT with NaCl stress treatment; 1–14: Ds lines showing tolerance after two weeks of NaCl stress.
The 35 salt tolerant transgenic plants were screened for the presence of Ac/Ds elements through PCR. In this procedure, the amplification with hph (Hygromycin phospho transferase) gene specific primers indicates the presence of the Ac element. Similarly, amplification with RFP (Red fluorescent protein) gene specific primers indicates the presence of Ds element in the corresponding plant. From PCR analysis 30 out of 35 plants were found to be stable Ds element carrying plants whereas the remaining 5 plants represented Ac-Ds plants (supplementary Fig. 1A).
Identification of insertion site and flanking sequences on the chromosome
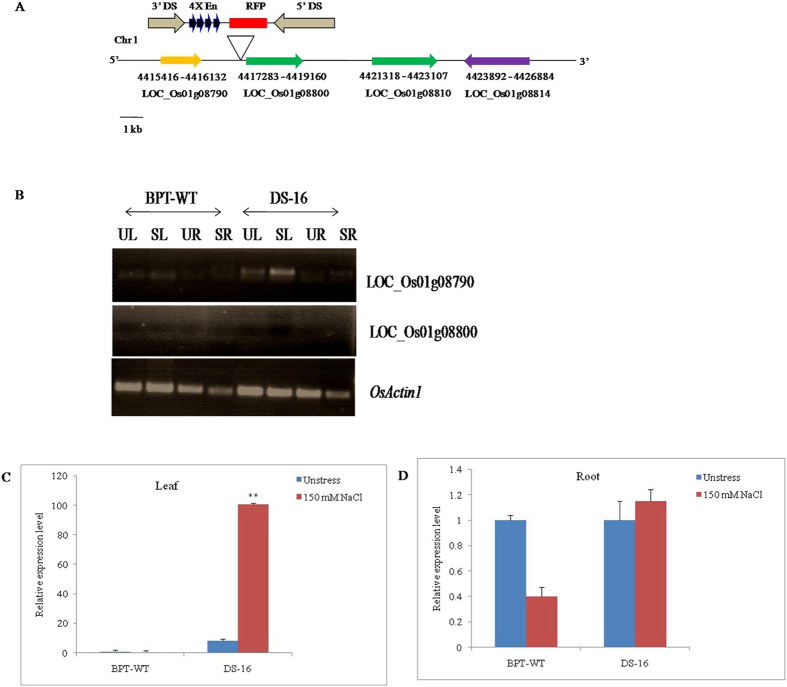
Twenty best salt tolerant plants were selected out of the 30 selected Ds lines and genomic DNA was isolated followed by a three-step TAIL PCR to identify the flanking sequences. Twenty transgenic plants gave specific TAIL-PCR products above 500 to 750 bp (supplementary Fig. 1B), which were subsequently gel purified, cloned and sequenced. The sequences were analyzed through the rice annotation sites at Michigan State University (http://rice.plantbiology.msu.edu), The Institute of Genomics Research (http://blast.jcvi.org/euk-blast/) and RAP-DB (http://rapdb.dna.affrc.go.jp/). All the transgenic plants showed independent Ds insertion events with the Ds element getting inserted at different loci on the rice chromosomes (Supplementary Table 2). Apart from mapping the insertion of Ds element on the corresponding rice chromosome, a 10 kb region on either side of the insertion site was also mapped, which aided in the identification of the genes flanking the Ds element insertions (Supplementary Fig. 2). The plant DS-16 showed the insertion on chromosome 1, and the BLAST results showed that the insertion on chromosome 1 was at an intergenic region near LOC_Os01g08800 encoding a putative cytochrome P450 gene (Fig. 2A). A 10 kb region on either side of the Ds insertion, carried the genes histone-like transcription factor and archeal histone (LOC_Os01g08790), cytochrome P450 (LOC_Os01g08810), an expressed protein (LOC_Os01g08814). Similarly, the insertion sites of other different salt-tolerant DS lines and details of nearby genes located are shown (Supplementary Table 2). Further, the left boarder (LB) flanking sequence analysis was performed for both PDS-16 plant (Parent of DS-16, with both Ac/Ds elements) and Ac plant (after transposition of Ds element from T-DNA) to confirm the stability of the Ac element. Results revealed that the T-DNA was located at a non-coding region on chromosome number 12 (position at approximately 24871372). Nearby gene was identified as expressed protein (LOC_Os12g40180) with unknown function towards downstream of integration loci. There was no positional variation in T-DNA, before and after the transposition.
Figure 2.
Physical mapping of insertion site of Ds-Enhancer (DS-En) elements and expression analysis of NF-YC13 in salt tolerant activation-tagged DS-16 line. (A) Ds element was inserted in the intergenic region on chromosome 1 and location of neighboring four putative genes (up to 20 kb region) near the insertion site. (B) Semi-quantitative RT-PCR analysis of activation-tagged DS-16 line. Two weeks old seedlings grown in Yoshida’s solution supplemented with 150 mM NaCl for 15 d and those without NaCl supplementation used as control. One of the genes, LOC_Os01g08790 shows expression during control and NaCl stress conditions while no expression in leaf and root of WT-BPT and LOC_Os01g08800 respectively. (C,D) Quantitative real time-PCR analysis of expression of NF-YC13 in the activation-tagged DS-16 line in leaf and root tissues. OsActin1 was used as a reference gene for quantitative RT-PCR. Means of three independent samples and standard errors are presented. ** and * indicates significant difference at P < 0.01 and P < 0.05.
Enhanced expression of tagged gene in DS-16 and analysis of other stress-responsive genes
Out of all salt tolerant Ds plants, the plant DS-16 was selected based on its salt tolerance and the insertion of the enhancer carrying Ds element in the intergenic region without disturbing any genes, whereas the insertion site was in the intergenic region in other Ds plants. Also the tagged gene at the locus, LOC_Os01g08790 was yet to be reported/characterized for abiotic stress. Hence, the DS-16 plant was analyzed for the expression profile of the neighboring genes identified near the insertion site of the Ds element for the possible identification of the activated gene(s), if any. Analysis of the RNA samples of both plants, control and NaCl stressed conditions of DS-16 and WT-BPT respectively was carried out using semi-quantitative and qRT-PCR. In DS-16, four putative genes, LOC_Os01g08790, LOC_Os01g08800, LOC_Os01g08810 and LOC_Os01g08814 were located within a 20 kb region near the Ds insertion site on either side (Fig. 2A). The Ds element was identified at a site 636 bp (4416647–4417283) upstream of the of the LOC_Os01g08800 gene, which encodes a putative Cytochrome P450 on chromosome 1 (Fig. 2A). At the same time, LOC_Os01g08790 gene (putative, histone-like transcription factor) was located 1.4-kb downstream of the Ds insertion. To check the enhancer effect, we performed semi-quantitative PCR analysis of DS-16 and WT root and leaf tissues and the result showed the activation of only LOC_Os01g08790 gene, which was expressed in both root and leaf tissues whereas its expression was not detected in WT (Fig. 2B). At the same time, we also checked the 4X enhancer effect on other three neighboring genes. We did not detect any expression in all the three genes (LOC_Os01g08800, LOC_Os01g08810 and LOC_Os01g08814) (data not shown). Further, we analyzed the transcript level of LOC_Os01g08790 by a real-time analysis and observed higher expression (8-fold) of LOC_Os01g08790 gene during control condition in the leaves of DS-16 with respect to WT. However, the level of transcript was enhanced 100-fold during NaCl stress treatment (Fig. 2C) as compared to WT. Similarly, in roots, it was observed also under control condition with the transcript level being quite low but with a little increased (1.5-fold) transcript level during salt stress treatment (Fig. 2D). At the same time, we checked transcript levels of the other upstream genes (LOC_Os01g08800, LOC_Os01g08810 and LOC_Os01g08814), which showed no detectable expression pattern in both leaf and root tissues of DS-16 plant (data not shown). Therefore, only one gene at the locus, LOC_Os01g08790 in DS-16 plant was activated by CaMV35S 4X enhancer elements, which also showed higher expression during abiotic stress treatments, particularly salt stress in this study.
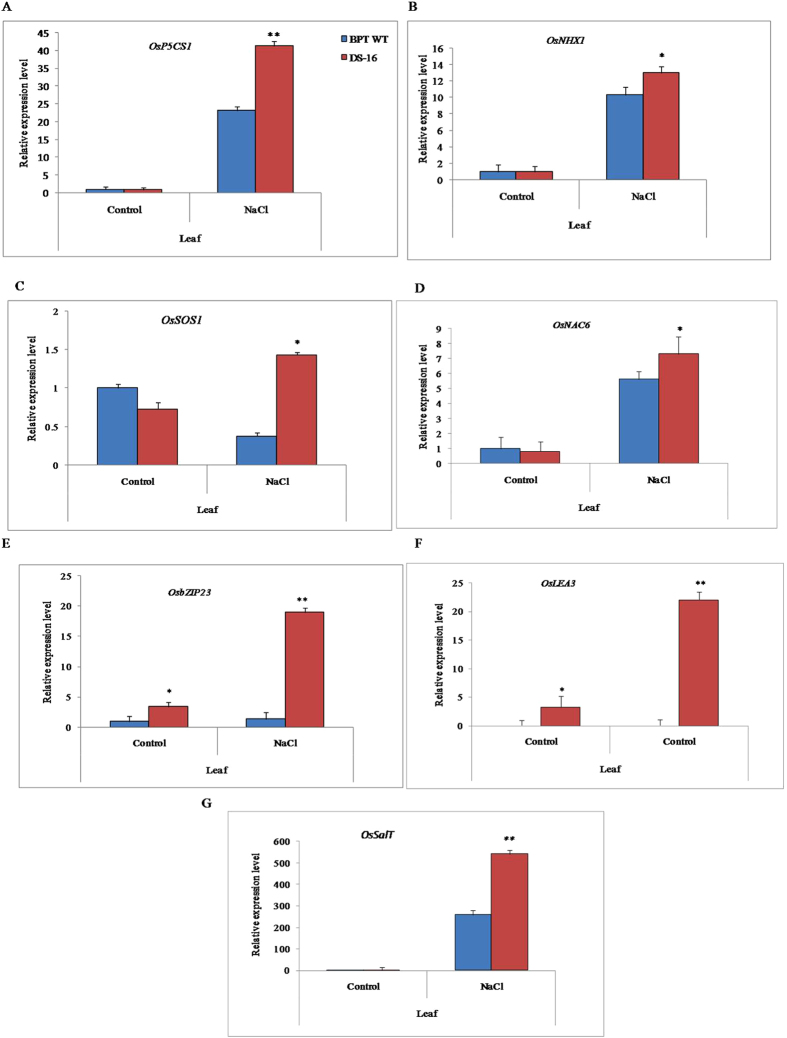
Since we have observed enhanced expression of tagged gene during NaCl treatment, we analyzed the expression of a few selected stress-responsive genes such as OsP5CS1, OsNAC6, OsLEA3, OsSOS1, OsZIP23, OsNHX1 and OsSalT in the leaf tissues of DS-16. Interestingly, quantitative analysis revealed a significantly enhanced expression of these genes under NaCl stress as compared to the WT and as well as under control conditions (Fig. 3A–G). Though there was some up-regulation of some of these genes in the stressed WT plants also, the expression particularly of OsSOS1, OsbZIP23 and OsSalT was significantly higher. The results clearly suggested that the tagged gene, LOC_Os01g08790 has a role in salt stress conditions.
Figure 3.
Analysis of transcript levels of stress responsive marker genes in activation tagged DS-16 line in comparison with WT-BPT under NaCl stress treatment. (A) OsP5CS1; (B) OsNHX1; (C) OsSOS1; (D) OsNAC6; (E) OsbZIP23; (F) OsLEA3; (G) OsSalT. Two weeks old seedlings grown in Yoshida’s solution supplemented with 150 mM NaCl for 15 d and those without NaCl supplement used as control. OsActin1 was used as a reference gene for quantitative real-time RT-PCR. Means of three independent samples and standard errors are presented. ** and * indicates significant difference at P < 0.01 and P < 0.05.
LOC_Os01g08790 identified as OsNF-YC13 and phylogenetic analysis of the gene
Previous reports showed that there are 28 NF-Y genes identified in the rice genome34, 43. However, a recent report included some new genes as new OsNF-Y genes. As a result, 40 putative NF-Y genes in rice were annotated, which included 11 NF-YA, 13 NF-YB, and 16 NF-YC44. We searched gene at the locus LOC_Os01g08790 that got activation-tagged in the present investigation in the Plant Transcription factor database (PlantTFDB) (http://planttfdb.cbi.pku.edu.cn). Apart from the earlier annotated 7 NF-YC transcription factors, 5 new members viz., OsNF-YC9 to OsNF-YC12 48 were included in the PlantTFDB. We found the tagged gene LOC_Os01g08790 in the PlantTFDB and the protein has been predicted to possess a coiled coil region (from 3 to 47 AA) and NF-YC domain (from 119 to 180 AA) with a score of 45.4 and 1.8e-14 E-value (Supplementary Fig. 3A). Hence, we named LOC_Os01g08790 as OsNF-YC13. The OsNF-YC13 gene has an uninterrupted ORF of 717 bp, encoding a 239-amino acid protein with the polypeptide mass of 25.146 KDa and an isoelectric point (pI) of 4.21.
To construct the phylogenetic tree, a total of 16 unique NF-YCs from the O. sativa genome (http://planttfdb.cbi.pku.edu.cn) were retrieved for performing the BLASTp analysis. The OsNF-YC13 showed similarity to TFs present in the wild rice genotypes. Further, the phylogenetic analysis of all the members of NF-YCs family in O. sativa with those of the wild rice accessions, S.bicolor, S.italica and G.max indicated that OsNF-YC13 is separately grouped to the TFs of wild rice and other monocot species (Supplementary Fig. 3B). Multiple sequence alignment of OsNF-YC13 with the wild rice accessions indicated a 100% identity with NF-YC TFs of O.rufipogan, 99% with O.nivara, 56% with Sorghum bicolor and 55% with Setaria italica and very less homology with the members of A.thaliana (20%) and Glycine max (17%) (Supplementary Fig. 3C and Supplementary Table 3).
Expression of OsNF-YC13 under salt stress
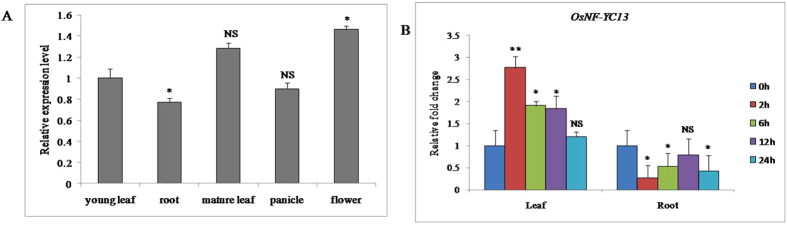
The transcripts of OsNF-YC13 were detected in different organs with higher level in mature leaf and flower of control WT-BPT (Fig. 4A). The response of OsNF-YC13 expression under NaCl (150 mM) was monitored in a real time analysis. The OsNF-YC13 transcripts in shoot got gradually decreased after 6–24 h of NaCl treatment respectively (Fig. 4B). Similarly, we observed the expression pattern of OsNF-YC13 in roots where the transcript level showed a ≤ 2-fold change in NaCl stress (Fig. 4B). The result indicated that OsNF-YC13 transcription factor was differentially regulated in salt stress treatments with shoot specific expression.
Figure 4.
Expression pattern of OsNF-YC13 in wild-type BPT 5204. (A) Relative expression profile of OsNF-YC13 gene in different organs of wild-type rice plants. The transcript level of OsNY-YC13 in young leaf as standard. (B) Relative expression of OsNF-YC13 under NaCl stress (150 mM) treatment in leaf and root. Two week-old rice seedlings grown in Yoshida’s solution were collected for gene expression analysis of different time intervals. Total RNA was prepared from 2-week-old seedlings of wild-type rice after the above treatments and then cDNA was prepared. Real-time PCR was performed using OsActin1 was used as an internal control. qRT-PCR was performed with OsNF-YC13 specific primers. Data represent means and standard errors of three biological replicates are shown.
Subcellular localization of OsNF-YC13
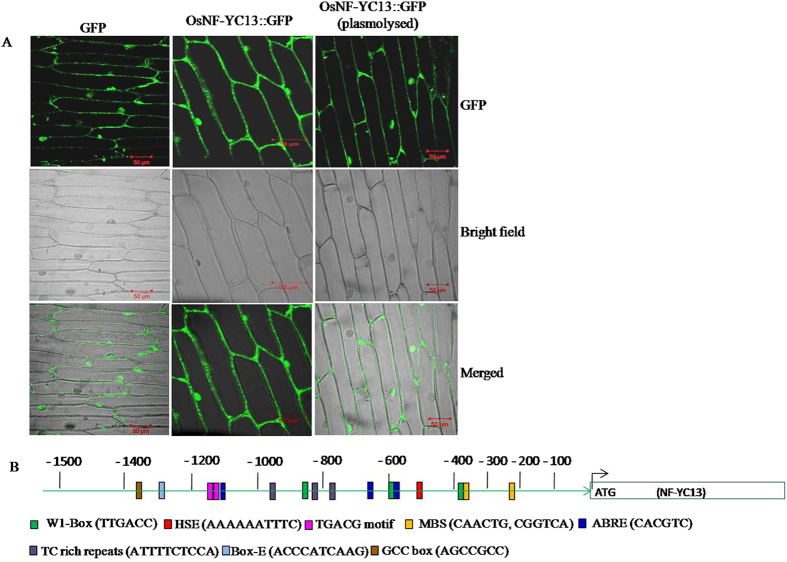
To determine the subcellular localization of the OsNF-YC13, we generated OsNF-YC13::GFP fusion constructs under the control of CaMV 35S promoter. These constructs were then expressed in the onion epidermal cells. We observed that OsNF-YC13::GFP protein fluorescence was localized to the nucleus and in the peripheral region of the transformed onion cells (Fig. 5A). Further, the transformed onion cells were subjected to plasmolysis, which showed that the GFP was present in nucleus as well as cytoplasm (Fig. 5A).
Figure 5.
Subcellular localization of OsNF-YC13 in onion epidermal cells and graphical representation of various cis-regulatory elements in 1.5 kb upstream regulatory region of OsNF-YC13. (A) The coding region of OsNF-YC13 was fused to the N-terminus of green fluorescence protein (GFP) and transformed into onion epidermal cells through Agrobacterium-infiltration using GFP as control. The GFP fluorescence was detected under confocal laser scanning microscope. (B) Physical mapping of key cis-elements involved in different stresses. A 1.5 kb upstream region of OsNF-YC13 was extracted from rice genome database and searched for various stress related elementsusing online tool, PlantCare.
Yeast transactivation assay of OsNF-YC13 and possible interacting proteins
To check the transactivation activity of OsNF-YC13 subunit, we performed transcriptional activation assay using yeast cells showed that OsNF-YC13 did not possess transactivational activity alone (Supplementary Fig. 4A). In order to find out the possible other protein interaction network with OsNF-YC13, we used STRING database (https://string-db.org/). The result showed that OsNF-YC13 might have interacted with OsNF-Y B subunits such as NF-YB3, B4, B6, B7 and others (Supplementary Fig. 4B).
Cis-elements in 1.5-kb promoter region of OsNF-YC13
Because of the up-regulation of OsNF-YC13 gene in other abiotic stresses, we investigated for the presence of stress responsive cis-elements in the putative promoter region upstream to the coding region. We extracted 1500-bp upstream sequence from rice database and analyzed using PlantCARE database49. We found various stress responsive cis-elements in the 1.5 kb region, which were physically mapped (Fig. 5B). Major stress-responsive cis-elements such as abscisic acid responsive elements (1 ABRE: CACGTC), heat stress elements (1 HSE: AAAAAATTTC), low temperature responsive elements (2 LTR: CCGAAA), MYB binding site (2 MBS: CAACTG, CGGTCA) which is involved in drought-inducibility, methyl jasmonate responsive elements (2 TGACG motif, 1 CGTCA motif), TC-rich repeats (2 ATTTTCTCCA) involved in defense and stress reponse, GCC box (1 AGCCGCC), Box-W1 motif (3 TTGACC), which are responsive to fungal elicitors were found in the 1.5-kb upstream regulatory region.
Increased growth and biomass of activation-tagged line DS-16
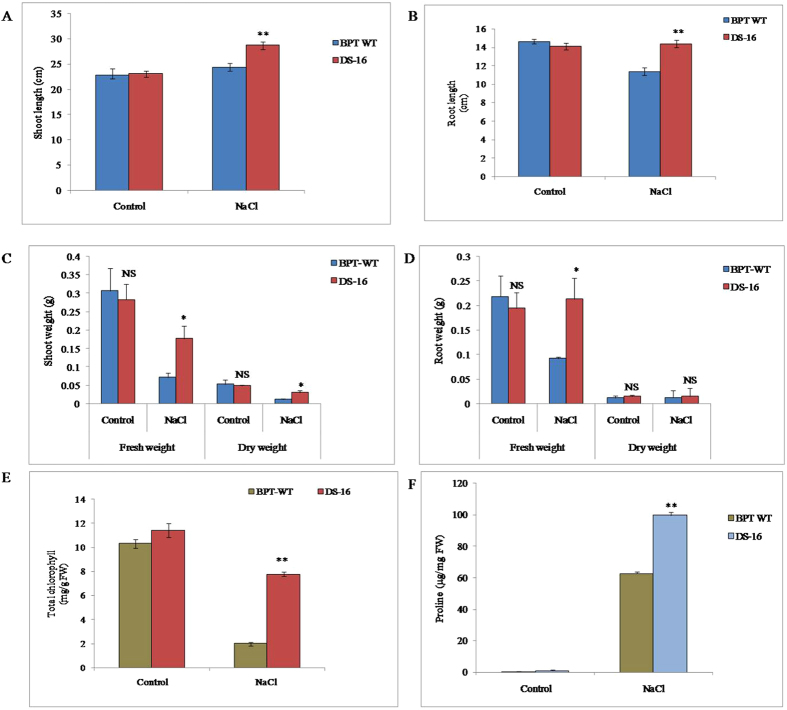
The seedling growth and biomass were measured before and after NaCl stress treatments in the salt tolerant DS-16 line. The shoot and root length of both WT and DS-16 were similar in length during control condition whereas both shoot and root length were increased significantly as compared to that of WT seedlings under sodium chloride stress (Fig. 6A and B).
Figure 6.
Evaluation of activation tagged DS-16 plants for salinity tolerance and chlorophyll and proline content in the activation tagged DS-16 line. (A,B) Shoot and root length. (C,D) Fresh and dry weight of shoot and root. E: Total chlorophyll content (F) proline content. Two weeks old rice seedlings from DS-16 and WT-BPT were treated with 150 mM NaCl stress treatment for 2-weeks. Experiment was carried out before and after stress treatment of DS-16 and WT. A total of 5–10 rice seedlings were measured and two independent biological repeats were performed for each treatment with error bar. ** indicates significant differences in comparison with the WT at P < 0.01, *P < 0.05, NS - non significant.
Similarly, the weight of the plants has not been affected by salt stress. Fresh and dry weight of DS-16 shoots under salt stress treatment was significantly increased compared with that of WT. However, no significant changes were observed in both the tagged line and WT without stress. In addition to shoot biomass, root biomass was also measured after stress treatment (Fig. 6C). During stress condition, the root fresh weight of WT was negatively affected and decreased by 10% compared to its control condition. But, in the tagged line, the root fresh weight was not affected during NaCl stress condition. With no change in the fresh root mass of DS-16, the dry weight of tagged line was on par with WT root (Fig. 6D). Overall, the tagged line DS-16 was appeared to be tolerant to 150 mM NaCl stress treatment compared with the WT.
Estimation of total chlorophyll and proline content
Leaf of DS-16 and WT was collected freshly before and after NaCl stress treatment for measuring the total chlorophyll and proline contents. Under control condition, there were no significant differences in the chlorophyll content between WT and DS-16 plants. However, after two weeks of NaCl stress condition, the chlorophyll content of WT plants was significantly lower than that of DS-16. The mean chlorophyll contents of DS-16 plants were found a 3.8-fold higher (7.7 mg g−1FW) under NaCl stress as compared with WT (2.0 mg g−1 FW) (Fig. 6E). In addition to total chlorophyll content estimation, proline content was also measured. Under control condition, there were no significant differences in proline content between DS-16 and WT. However, after two weeks of NaCl stress treatment (150 mM), the proline content in DS-16 and WT plants was measured as 99.8 µg mg−1 FW and 62.6 µg mg−1 FW respectively. A 1.5-fold elevated proline content was observed in DS-16 plants as compared to the WT (Fig. 6F). The results of higher chlorophyll and proline content in DS-16 are consistent with the salt tolerance phenotype in DS-16.
Accumulation of Na+ and K+ content in leaf and root of DS-16 during salt stress
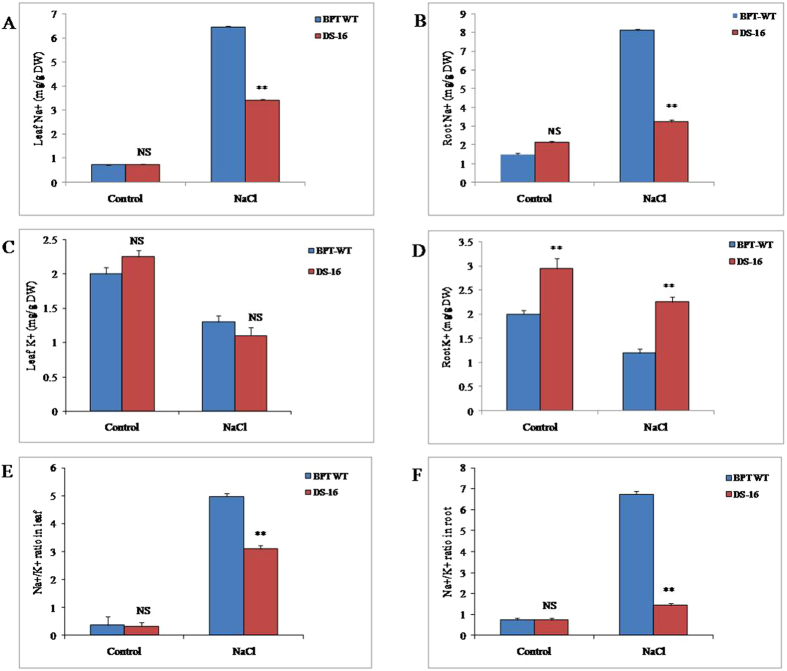
Accumulation of sodium and potassium ions in the seedlings of DS-16 and WT under control and NaCl stress conditions was investigated. As shown in Fig. 7A, the Na+ concentration in leaves of DS-16 and WT control plants showed no significant changes under control condition. In contrast, the concentration of Na+ in leaves of WT increased by ~9-fold under 150 mM NaCl stress (from 0.74 to 6.46 mg g−1 DW) in comparison with its control conditions (no stress). At the same time, the DS-16 line showed only a five-fold increase in Na+ concentration (from 0.74 to 3.41 mg g−1 DW) compared to its control condition. Relatively, the DS-16 plants maintained a ~1.8-fold lower Na+ concentration under NaCl stress conditions compared to the WT. Similarly, under control and NaCl stress conditions, no significant changes in K+ accumulation were observed in both WT and DS-16 (Fig. 7C).
Figure 7.
Estimation of Sodium and potassiumcontent of salt tolerant activation tagged DS-16 line. (A) Leaf Na+, (B) root Na+, (C) leaf K+, (D) root K+, (E) Leaf Na+/K+ ratio, (F) root Na+/K+ ratio. Two weeks old rice seedlings from DS-16 and WT-BPT were treated with 150 mM NaCl stress for two weeks. Ion accumulation was estimated before and after stress treatment in DS-16 and WT. Three rice seedlings were used in each treatment and three independent biological repeats were performed for each treatment with error bar. * indicates significant differences in comparison with the WT at P < 0.05, **P < 0.01, NS - non significant.
Further, we measured the Na+ and K+ concentrations in the roots. There were no significant differences in the accumulation of Na+ in roots in both types in the control condition. However, NaCl stress resulted in a 2.5-fold decreased Na+ in roots of DS-16 plants compared to WT (Fig. 7B). Besides the presence of a lower level of Na+ in DS-16, a 1.4- and 1.8-fold higher K+ accumulation was observed in the roots of DS-16 during control and NaCl stress conditions respectively compared to the WT (Fig. 7D). Further, we calculated Na+/K+ ratio in leaf and root of DS-16 and WT control under both conditions. During control condition, no significant differences were observed in the ratio in leaf and root of both types of plants, whereas under NaCl stress, the ratio decreased to 1.5-fold in leaf and 4.7-fold in root of DS-16 as compared with that of WT (Fig. 7E and F). This analysis clearly indicated that the DS-16 line retained lower Na+ and higher K+ concentration in roots and lower Na+ in leaf under salt stress conditions for maintaining ion homeostasis.
Analysis of antioxidant enzyme activity
Antioxidant enzymes such as superoxide dismutase (SOD), peroxidase (POX) and catalase (CAT) are involved in scavenging ROS and protecting the cells from the oxidative damage and eventually leading to enhanced stress tolerance. In the present study, these three enzymes were analyzed at the seedling stage and they were significantly increased before and after NaCl stress in DS-16 plants as compared to WT. SOD activity increased1.5-fold under NaCl stress conditions in both shoot and root of DS-16 plants as compared with that of WT (Fig. 8A). Similarly, enhanced POX (1.7-fold) and CAT (2-fold) activities were observed in shoots of DS-16 plants (Fig. 8B and C), while in the root, the POX and CAT activity increased by a 2.2- and a 1.6-fold respectively (Fig. 8B and C). The increased content of SOD, POX and CAT would result in reduced oxidative damage to cells caused by salt stress treatment and enhances tolerance in DS-16 to 150 mM NaCl stress.
Figure 8.
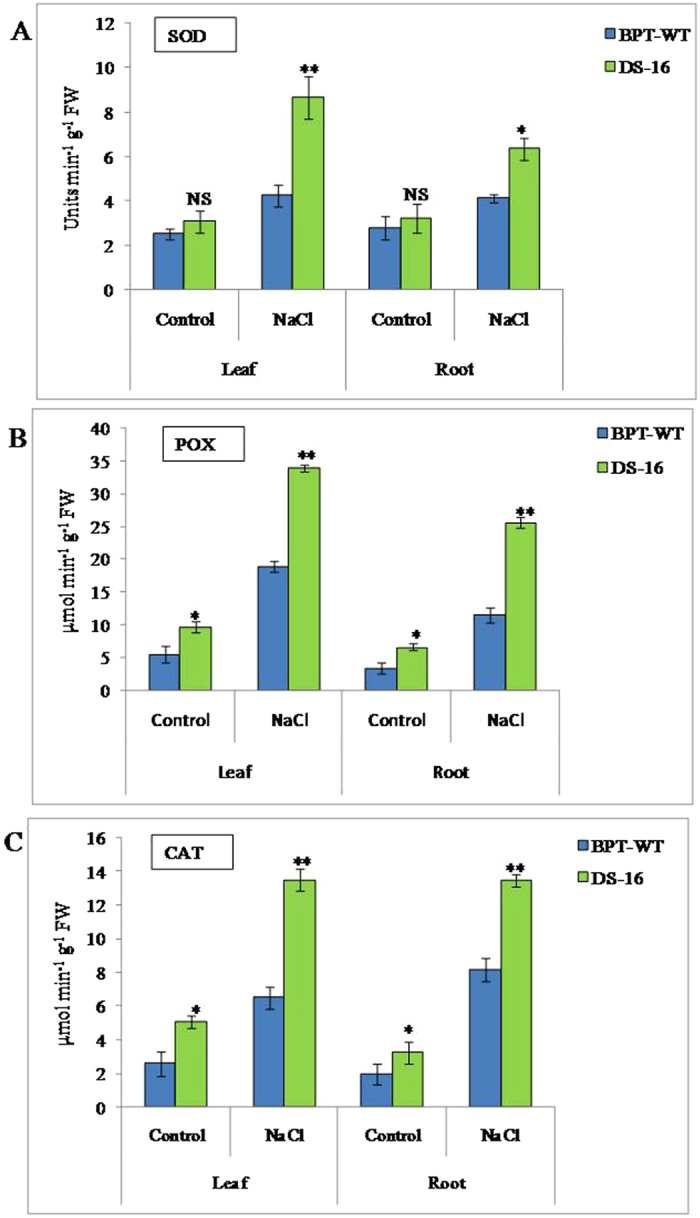
Analysis of antioxidant enzymes activity in activation tagged DS-16 and WT before and after NaCl treatment. (A) super oxide dismutase (SOD), (B) peroxidase (POX), (C) catalase (CAT). Two weeks old DS-16 seedlings and WT-BPT were treated with 150 mM NaCl for twoweeks. Three rice seedlings were used in each treatment and three independent biological repeats were performed for each treatment with error bar. * indicates significant differences in comparison with the WT at P < 0.05, **P < 0.01, NS - non significant.
Discussion
Plants are constantly exposed to various abiotic stresses causing yield losses. Salinity is one of the major abiotic stresses that reduces the plant growth and production especially in food crops50. To reduce the yield loss by increasing soil salinization, breeders have developed various salt-tolerant rice varieties by introgressing QTL/genes conferring salt tolerance from other related genetic resources and subsequent identification based on marker-assisted breeding. On the other hand, deploying the novel gene(s) or altering the expression of existing genes to increase the degree of salt tolerance by generation of transgenic plants is an option that needs thorough investigations51.
Rice (Oryza sativa L.) crop is more sensitive to increased soil salinization at seedling and reproductive stages52, 53. Development of salinity tolerant rice varieties is an efficient option to overcome salinity stress and enhance productivity under stress. Activation-tagging (AT) system has been recently used for developing gain-of-function mutants in a widely cultivated indica rice variety (BPT 5204, Samba Mahsuri) to identify novel genes20. Many research groups utilized this system earlier for developing large mutant populations in different plants species (Arabidopsis, japonica type Rice, Tomato, Potato, Strawberry, Poplar, Barley, Sorghum) as resources for functional genomics.
In the present study, we have screened activation-tagged indica rice lines on 150 mM sodium chloride and developed an activation-tagged rice line showing enhanced expression of a previously uncharacterized gene, LOC_Os01g08790 under salt stress treatment. Further, the phenotype of tagged salt tolerant line was characterized by physiological, biochemical and molecular expression analyses. Sequence analysis of the corresponding activation-tagged loci in selected salt tolerant rice Ds plants revealed that the integration of the Ds-Enhancer elements was in intragenic region (95%) of most of the plants screened and the remaining (5%) were in the intergenic regions on the chromosomes. This result was consistent with other earlier reports where most of the insertions were maximally in intragenic (coding region, promoter, 3′ UTR), while it was lesser in intergenic regions22, 54. From the twenty salt tolerant Ds plants, DS-16 was selected for further analysis because the tagged gene was intergenic, with a novel/uncharacterized gene encoding ‘Histone-like transcription factor’ (LOC_Os01g08790) getting activated by the CaMV 35S 4X enhancers during NaCl stress conditions. Expression analysis of DS-16 plant by qRT-PCR revealed that the tagged gene histone-like transcription factor (LOC_Os01g08790), where the Ds-4X Enhancer positioned between LOC_Os01g08790 and LOC_Os01g08800 (Cytochrome P450) was expressed at a 8-fold enhanced level under control condition while the transcript level was enhanced to a 100-fold during NaCl stressed condition.
Previous reports suggested that one or two genes can be activated at a time by the multiple CaMV enhancers in different Ds lines20, 47. It has also been reported that the degree of activation and distance of the Ds insertion to the gene has no correlation with the enhancer function16, 47. In our investigations on the DS-16 line, only one gene (LOC_Os01g08790) was activated by the CaMV 35S 4X enhancers and the transcript level was elevated during salt stressed condition. As the LOC_Os01g08790 gene showed higher transcript level during NaCl stress condition, we analyzed the expression level of a few selected stress-responsive genes (OsP5CS1, OsNAC6, OsLEA3, OsSOS1, OsZIP23, OsNHX1 and OsSalT) based on the previous reports, which established that these stress-responsive genes were up-regulated during abiotic stress condition particularly in salinity stress55, 56. Interestingly, these stress-responsive genes were also up-regulated in DS-16 plants with significantly higher expression level during salt stressed conditions and also associated with significantly up-regulated LOC_Os01g08790 gene expression. Theses result indicates that the gene LOC_Os01g08790 has a significant role in abiotic stress condition, particularly in salt stress.
The newly identified gene LOC_Os01g08790 encodes a putative histone-like transcription factor that belongs to nuclear factor Y-C subfamily and we named it as OsNF-YC13 with the data presented in the Results section. The Nuclear factor Y (NF-Y), is a heterotrimeric complex transcription factor consisting of three subunits, namely NF-YA (also called HAP2 or CBF-B), NF-YB (HAP3 or CBF-A), and NF-YC (HAP5 or CBF-C)43. A total of 40 putative NF-Y genes including 11 NF-YAs, 13 NF-YBs and 16 NF-YCs were found in rice database44.
Further, we have studied the localization of the transcription factor activity through transient expression of OsNFYC13::GFP fusion protein construct in transformed onion epidermal cells, which indicated that the gene is localized to nucleus and cytoplasm, which is consistent with the previous reports on CdtNF-YC1 from Bermudagrass57 and OsNF-YB1 58. Localization of OsNF-YC13::GFP in both nucleus and cytoplasm of onion cells suggests that the NF-YC13 may interact with other unknown OsNF-YB subunits to from dimers that translocate from cytoplasm to nucleus. Similar to our result, localization of OsNF-YB1::GFP was observed in both nucleus and cytoplasm of root cells whereas the protein confined to nucleus of aleurone layers cells58. In the present study, we could not observe transcriptional activation of OsNF-YC13 alone as no colonies were observed on the selection medium, which indicate transactivation. The gene might require additional protein interactions to regulate transactivation and modulate the expression of downstream targets. Similar to our result, Xu and co-workers58 found no transcriptional activation of OsNF-YB1 alone and it requires additional partner like OsERF#115 factor. Analysis using the STRING online tool showed that the NF-YC13 might interact with NF-YB subunits B3, B4, B6, B7 etc.
NF-Ys have diverse functions in plant growth, development and also in stress responses34, 59. Several groups worked on different individual NF-Ys in model systems and characterized their role in development and physiological process34, 46. For instance, one of the reports demonstrated that AtNF-YB9 (LEC1) plays a key role in embryo development60. Similarly, a rice nuclear factor Y, OsNF-YB1 is specifically expressed in the aleurone layer of developing endosperm regulating grain filling and endosperm development58. Zhu and co-workers61 identified a new uncharacterized gene OsHAPL1 (NF-YC like) with involvement in spikelet heading date in rice. After characterization, they proposed that OsHAPL1 functions as a transcriptional regulator and forms a complex with DTH8, Hd1, which influences the heading date in rice61. The above reports suggest that NF-Y proteins play a crucial role in plant developmental process.
Although NF-Y proteins play crucial roles in plant development, they also appear to play roles in biotic/abiotic stresses. Recently, Chen and co-workers57 have investigated the expression profile of some rice NF-YC genes under dehydration, salt and ABA stress treatments. Genome wide expression analysis of soybean with 68 NF-Y (21 GmNF-YA, 32 GmNF-YB, 15 GmNF-YC and 13 NC2) genes demonstrated their differential expression in response to drought stress39. In our case, the identified OsNF-YC13 gene was differentially regulated in salt stress (NaCl) treatment.
Many individual NF-Y subfamily genes were characterized separately through transgenic approach and they appear to be involved in different plant physiological process including abiotic stress condition. Overexpression of AtNF-YB1 in Arabidopsis and maize (Zea mays) was shown to significantly improve drought resistance and yield under drought stress conditions62. Overexpression of AtNF-YA5 increased drought tolerance in Arabidopsis plants63. Similarly, overexpression of OsNF-YA2 (OsHAP2E) in rice also showed resistance to Magnaporthe oryzae or Xanthomonas oryzae pv. oryzae with tolerance to salinity and drought64. Ectopic expression of Bermudagrass NF-YC (Cdt-NF-YC1) in rice showed elevated tolerance level to drought, salt and sensitive to ABA57. Overexpression of garlic NF-YC (AsNF-YC8) in tobacco conferred tolerance to drought and salinity stress65.
Since the OsNF-YC13 was induced during salt stress condition, we tried to focus on various stress responsive cis-regulatory elements such as ABRE, HSE, LTR, MBS, methyl jasmonate responsive elements, TC-rich repeats, GCC box, Box-W1 motif in the 1.5 kb putative promoter region of OsNF-YC13. The presence of various stress responsive elements in the promoter region of the gene supported our observations on the enhanced expression of the novel NF-YC13 in rice under abiotic stress particularly salinity condition. As the stress responsive genes were up-regulated during NaCl treatment in DS-16 plants, we searched for the presence of CCAAT elements in the promoter region of OsP5CS1, OsNHX1, OsLEA3, OsbZIP23, OsSOS1 and OsSalT genes. Interestingly, we observed the presence of 1–2 CCAAT cis-elements in the promoters of all the genes except OsNHX1 and OsbZIP23 (Supplementary Table 4). Similar to our results, Alam et al.64 found 1–5 CCAAT cis-elements in the promoter regions of ABA induced marker genes that got up-regulated under salt and drought stress treatments in rice transformed with Cdt-NF-YC1. The up-regulation of selected stress responsive genes in DS-16 plants correlated with the presence of respective stress responsive elements in the promoters of most the stress related genes suggests that OsNF-YC13 holds a role during salt stress condition through directly or indirectly activating the ABA-dependent signaling pathway.
Because of OsNF-YC13 gene was induced during NaCl treatment, the DS-16 was further characterized for salinity tolerance by using physiological and biochemical analyses. As rice seedlings are very sensitive to salinity stress, the seedlings (15 d old) of DS-16 after salt stress treatment showed significantly better growth (shoot and root length), biomass and higher chlorophyll content than its WT. Our result was consistent with the over-expression of OsNF-YA2 (OsHAP2E) in rice, which resulted in enhanced shoot, root length, weight and higher chlorophyll content under salinity treatment64. Increased accumulation of free proline in plants has been shown increase plant tolerance to osmotic stress and the free proline not only acts as free radical scavenger but also protects from cell damage during stress condition66. Also DS-16 showed significantly higher free proline content accumulation compared to the WT under NaCl stress treatment.
Plants exposed to high salinity experience both ionic and osmotic stresses, which restrict Na+ uptake and entry into their cells. Increased Na+ levels are toxic to cells and as the Na+ ion competes for binding sites with K+ as it has similar physiochemical properties and this leads to metabolic disturbances causing severe cellular damage67, 68. Thus, plants either adapt to or evade salt stress by excluding Na+ from the roots thereby maintaining a lower level of Na+ in the leaves69. Salt-tolerant plants accumulate salt in the vacuoles thereby controlling lower salt concentration and maintaining higher K+/Na+ ratio in the cytoplasm of the cells70. There are several studies reporting that transgenic or salt-tolerant plants accumulate less Na+ and more K+ maintaining a lower Na+/K+ ratio than the control or susceptible plants52, 71, 72. Consistent with these reports, DS-16 plants showed significantly lower level of Na+ in leaves and roots under salt stress condition whereas no significant accumulation level of K+ was observed in leaves. However, the roots of DS-16 plants accumulated higher level of K+ accumulation than its WT control. As a result of this, they maintained lower level of Na+/K+ ratio in leaves and roots with better ion homeostasis that prevents cell damage.
During abiotic stress condition, plants should maintain low level of reactive oxygen species (ROS) to minimize the cellular damage caused by osmotic stress73. Plant defense system possesses ROS-detoxification antioxidant enzymes like SOD, CAT and POX for scavenging ROS and reduced oxidative damage74. In our study, we have checked the antioxidant enzyme activities in DS-16 plants, which showed higher activities of SOD, POX and CAT in comparison with WT under salt stress treatment. Evidence supports reduced oxidative damage and increased salinity tolerance in plants are associated with efficient antioxidant defence system in plants75, 76. Similarly, overexpression of garlic NF-YC, (AsNF-YC8) in tobacco showed higher antioxidant enzymes activity (SOD, POX, CAT, APX) than the wild type tobacco65.
Overall, with the results from above experiments, we identified an activation-tagged line DS-16 for salt tolerance at 150 mM NaCl and studied the expression of a novel and previously uncharacterized gene OsNF-YC13, which was identified a candidate gene for salt tolerance. This novel gene was induced under different abiotic stress conditions. The activation-tagged DS-16 plants exhibited salinity tolerance, which was supported by physiological, biochemical and expression analysis of several stress responsive genes. From this present study, we conclude that the novel OsNF-YC13 gene has a significant role in salinity stress in rice. Further studies are being made in rice to demonstrate its role during plant developmental/physiological process.
Materials and Methods
Plant material, seed germination and salt stress treatment
Seeds of rice plants in T3 generation carrying both Ac and Ds (with multiple 35S enhancers) elements and wild-type BPT 5204 (WT) were surface sterilized with 70% ethanol and 0.1% HgCl2 solution. The sterilized mature seeds were inoculated on hormone free ½ MS basal medium. The plates were incubated under light for 15 d for germination. Two week old transgenic rice seedlings were directly transferred to Yoshida’s culture solution77 for 3–4 d followed by salt stress treatment in Yoshida’s solution supplemented with 150 mM NaCl (pH 5.8) for two weeks. The solution was prepared freshly and replaced after every three days to maintain pH 5.8. Two week after salt stress treatment, the tolerant transgenic rice plants were recovered and transplanted to pots and maintained in the green house for further characterization.
To examine the expression pattern of OsNF-YC13 gene, which got activated by CaMV 35S 4X enhancers in salt stress treatment, two week old rice seedlings (WT) were transferred to Yoshida’s culture solution supplemented with sodium chloride (NaCl; 150 mM) for salt treatment. After treatment, shoot and root samples were collected separately at 2 h, 6 h, 12 h and 24 h intervals. Treatment with water served as non-stressed control (0 h). RNA isolation and gene expression analysis under various stress treatments was performed using qRT-PCR.
PCR confirmation of salt tolerant activation-tagged rice transgenic plants
The DNA samples isolated from leaves of salt tolerant rice mutants and non-transformed control plants were used in PCR analyses78. PCR amplification was performed with 50 ng of template DNA, using primers for hph (hygromycin phospho-transferase) and RFP (Red fluorescent protein)-Nos (nopaline synthase termination signal) using a Thermal Cycler (Eppendorf, Germany). The PCR reaction mixture (10 μL) was prepared with genomic DNA, 1 × Taq assay buffer (containing 1.5 mmol/L MgCl2), 125 μmol/L dNTPs, 0.4 μmol/L each of forward and reverse primers and 1 U of Taq DNA polymerase (KAPA Biosystem, USA). The PCR profile was set as follows: 95 °C for 5 min, 95 °C for 30 s + 58 °C for 30 s + 72 °C for 1 min (35 cycles) + 72 °C for 7 min. The PCR products were resolved on 1% TAE agarose gels. Gene specific primers used in this study are listed in Supplementary Table 1.
Identification of DS insertion site by TAIL-PCR and flanking sequence analysis
Thermal Asymmetric Interlaced Polymerase Chain Reaction (TAIL - PCR)79 was performed using Ex-Taq DNA polymerase (Takara, Japan) in Eppendrof thermal cycler (Eppendrof, Germany) following Moin et al.20. Genomic DNA isolated from salt tolerant rice plants was subjected to three separate PCR runs using a degenerate primer and three nested primers specific for the 5′ end of the Ds element for the identification of the insertion in the rice genome and analysis of flanking sequences. The details of degenerate and nested primers have been detailed in earlier report20. The final products were run on 1% agarose gels and the expected products were sliced out for purification and sequencing. Sequences were checked using the BLAST against the rice annotation sites at MSU (http://rice.plantbiology.msu.edu), TIGR (http://blast.jcvi.org/euk-blast/) and RAP-DB (http://rapdb.dna.affrc.go.jp/).
RNA isolation and Real-time PCR analysis of activation-tagged line
Total RNA from leaf and root tissues of activation-tagged transgenic and wild-type BPT 5204 (both control and stressed conditions) was isolated according to the manufacturer’s instructions (RNAiso Plus, Takara, Japan) and PCR was performed as per Manimaran et al.80. Two microgram of total RNA was used for first-strand cDNA synthesis using oligo d(T) primers (SMART MMLV RT, Takara, Japan). The cDNA after RNaseH treatment was mixed with 12.5 µL of 2X KAPA SYBR® FAST qPCR mix universal (KAPA Biosystem, USA), 2 µM each of forward and reverse primers in a final volume of 20 µL. PCR with no template control (NTC) was also performed for each primer pair. The real-time PCR was performed in Mastercycler® ep realplex (Eppendorf, Germany). The conditions for qRT-PCR were as follows. 3 min at 95 °C, 35 cycles of 95 °C - 3 s and 60 °C - 20 s in 96-well optical reaction plates (Axygen, USA). The amplicon specificity was verified by melt curve analysis from 60 to 95 °C after completion of 35 cycles. Each sample was replicated thrice. The comparative threshold cycle (CT) method was used to quantify the relative expression levels in real-time PCR. OsActin1 was used as a reference gene to normalize gene expression. The ΔCT for BPT-WT (control condition) was used as a control for the calculation of relative changes in expression in activation-tagged transgenic line (control), WT (NaCl stressed) and transgenic line (NaCl stressed). Relative expression level of genes was calculated using the ΔΔCTT formula81. Primers used for gene expression studies and stress-responsive genes (OsP5CS1, OsNAC6, OsLEA3, OsSOS1, OsZIP23, OsNHX1 and OsSalT) are listed in Supplementary Table 1.
Phylogenetic analysis of the OsNF-YC13
Oryza sativa NF-YC family protein sequences were obtained from Plant Transcription factor database (http://planttfdb.cbi.pku.edu.cn). Additionally, NF-YC family protein sequences of the wild rice species were obtained from NCBI/UniProt. Multiple sequence alignment was performed in ClustalW. The phylogenetic tree was constructed by Neighbor-joining method in MEGA 6.0 with default parameters of 1000 bootstraps.
Isolation, cloning and subcellular localization of OsNF-YC13:GFP in onion epidermal cells
As OsNF-YC13 has uninterrupted coding region, the coding sequence of OsNF-YC13 was amplified directly from genomic DNA and fused in frame to the coding region of the N-terminus of GFP in the vector pRT35S-GFP vector to create the OsNF-YC13::GFP fusion protein under the control of the 35 S promoter. The OsNF-YC13::GFP construct was mobilized into Agrobacterium strain EHA105. We performed Agrobacterium-mediated onion epidermal peel transformation for studying the subcellular localization82. After 48 h co-cultivation, GFP localization was detected using a confocal laser scanning microscope (Carl Zeiss LSM880). The primers used for OsNF-YC13 subcellular localization are given in Supplementary Table S1.
Transactivation assay for NF-YC13 in yeast system
To assay the transactivation capacity of the NF-YC13 subunit, the coding region of OsNF-YC13 was amplified by PCR, subcloned into pGBKT7 (Clonetech, USA) and fused with the BD domain. The plasmid was confirmed by restriction enzyme analysis. The confirmed plasmid OsNF-YC13-pGBKT7 and pGBKT7 (control) were transformed into yeast strain PJ69–4A and plated on SD/-Trp medium for selection of positive colonies. Some colonies were picked and plated on SD/-Trp/-His and SD/-Trp/-Ade medium to check for the transactivation property of the subunit. At the same time, a serial dilution was performed by using liquid culture. For this, a colony was picked and inoculated in -Trp liquid medium. At 1.0 O.D, the cultures were serially diluted upto 10−5 and spotted on SD/-Trp, SD/-Trp/-His and SD/-Trp/-Ade plates for examining the transactivating property of OsNF-YC13 protein subunit. Empty vector was used as a control. Sequences of primers used are listed in Supplementary Table S1.
In-silico analysis of upstream regulatory sequence of OsNF-YC13
To identify the presence of various cis-regulatory elements in OsNF-YC13 gene, we extracted 1.5 kb upstream region from rice database RAP-DB (http://rapdb.dna.affrc.go.jp/) and analyzed using the online program PlantCare at bioinformatics.psb.ugent.be/webtools/plantcare49.
Physiological parameters of activation-tagged line for salt stress treatment
Growth and weight of salt tolerant activation-tagged line
Growth parameters like shoot and root length and total dry weight were recorded after two weeks under control and NaCl stress treatment conditions. After recording the growth parameters, fresh weight of the seedlings was rapidly measured. Fresh samples were then dried in a hot air oven at 80 °C for 72 h and dry weights were measured separately.
Total chlorophyll content
Total chlorophyll content was estimated and calculated according to Lichtenthaler and Wellburn83. Twenty five mg samples of fresh leaves (150 mM NaCl treated) were incubated in 10 ml of 80% acetone and kept in dark for 48 h. The absorbance was measured at 663 and 647 nm for chl a and chl b respectively.
Estimation of proline content
The proline estimation was carried according to Bates et al.84. One gram of fresh leaf material was ground with 20 ml of 3% sulfosalicyclic acid (W/V) and the homogenate was centrifuged at 10,000 rpm for 10 min. The supernatant was used for the estimation of free proline. The reaction mixture comprised 2.0 ml of glacial acetic acid, 2.0 ml ninhydrin reagent and 2.0 ml of supernatant. The whole reaction mixture was boiled for one hour at 110 °C. After cooling the liquid to room temperature, 4 ml of toluene was added and mixed vigorously for 30 s on a cyclomixer. The chromophore (toluene) aspirated from the aqueous phase was taken and its absorbance was measured at 520 nm. Proline concentration in the samples was determined by a standard curve and expressed in terms of micro gram proline per one gram fresh weight.
Estimation of Na+ and K+ ion content
For Na+ and K+ analyses, weighed dried samples of leaf and root (100 mg) were taken in digestion tubes containing 5 ml of tri-acid mixture, which consisted of nitric, perchloric and sulfuric acids (10:4:1). The tubes were subjected to 1 h digestion at 300 °C followed by overnight cooling. The digested liquid was then filtered through Whatmann No. 2 filter paper and the volume was made up to 50 ml with deioinized water. Sodium and potassium levels were analyzed using a flame photometer (ELICO-CL378, Type-223, India). The quantitative measurement of sodium and potassium ions was made using a standard curve.
Extraction and estimation of antioxidant enzymes
Assay of antioxidant enzyme activities: The activity of antioxidant enzymes (superoxide dismutase (SOD), peroxidase (POX) and catalase (CAT)) were estimated in both leaf and root samples after two weeks of imposition of 150 mM NaCl stress.
Enzyme extraction
The extraction procedure for SOD, POD and CAT was similar. Freshly weighed (100 mg) of leaf or root samples were frozen in liquid nitrogen to prevent proteolytic activity. The frozen tissues were ground using 5 ml of extraction buffer containing 100 mM phosphate buffer (pH 7.5) and 0.5 mM EDTA. The extract was centrifuged for 15 min at 12,000 rpm at 4 °C and the supernatant was used in the enzyme analyses.
Superoxide dismutase activity (EC 1.15.1.1)
The SOD activity was measured according to Dhindsa et al.85. The reaction mixture comprised 3 ml each of methionine (200 mM), nitroblue tetrazolium chloride (NBT) (2.25 mM), EDTA (3.0 mM), riboflavin (60 μM), sodium carbonate (1.5 M), phosphate buffer (100 mM, pH 7.8) and the sample. The tube without enzyme was considered as control and the tube with enzyme considered as test. These tubes were kept under light (15 W) for 15 min followed by addition of enzyme and the absorbance measurement at 560 nm. The enzyme activity was expressed in [unit g−1(FW) min−1]. One unit of activity is the quantity of enzyme required to inhibit 50% initial reduction of NBT under light.
Peroxidase (POD) activity (EC 1.11.1.7)
Peroxidase assay was carried out according to Castillo et al.86. The reaction mixture comprised 1.0 ml of phosphate buffer (pH 6.1), 0.5 ml guaicol, 0.5 ml H2O2, 0.1 ml of sample and the final volume was made up to 3 ml with double distilled water. Increase in the absorbance due to the formation of tetra-guaicol was recorded at 470 nm and it was expressed in μmol (guaicol reduced) g−1(FW) min−1.
Catalase (CAT) activity (EC 1.11.1.6)
The catalase activity was measured according to Aebi87. The reaction mixture consisted of 1.5 ml phosphate buffer pH 7.0, 0.5 ml H2O2, 0.05 ml enzyme and the final volume was made upto 3 ml with distilled water. The Decrease in the absorbance was recorded at 240 nm and it was expressed in μmol H2O2 oxidized g−1(FW) min−1. All the observations on the absorbance were recorded in a double beam UV-VIS spectrophotometer (Spectrascan UV 2600, Chemito, India).
Statistical analysis
All the recorded data were analyzed using Student’s t-test for statistical significance. **P < 0.01 and *P < 0.05 represent significant differences at 1 and 5% level respectively compared with the wild-type control.
Electronic supplementary material
Acknowledgements
The material used in the present investigation for analysis has been generated under a Research Grant (DBT No: BT/PR13105/AGR/02/684/2009) to PBK. Authors acknowledge the Head, Dept of Plant science for access to the research facilities provided by DST-FIST, DBT-CREBB and UGC-SAP to the Dept of Plant sciences, University of Hyderabad. PM thank the Dept. of Biotechnology, Govt. of India for the Post Doctoral Research Associateship. Authors thank the Project Director, IIRR for providing facilities. MM and SVR acknowledge DBT and CSIR-UGC Junior Research Fellowships respectively.
Author Contributions
P.M., S.M.B. and P.B.K. conceived and designed the experiments. P.M. and S.V.R. performed screening, cloning, PCR, RNA isolation, qRT-PCR experiments, in-silico analysis, yeast transactivation assay. M.M. provided the seed material and performed TAIL-PCR & sequence analysis. P.M., M.R.R. and P.Y. performed ion estimation, antioxidant enzyme assay and physiology experiments. S.S.M. performed phylogenetic and statistical analyses. PBK supervised the research. P.M. and P.B.K. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
P. Manimaran and S. Venkata Reddy contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10022-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
P. Manimaran, Email: p.maran@gmail.com
P. B. Kirti, Email: pbkirti@uohyd.ac.in
References
- 1.Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 2.Kumari S, et al. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomics. 2009;9:109–123. doi: 10.1007/s10142-008-0088-5. [DOI] [PubMed] [Google Scholar]
- 3.Cuartero J, Bolarin MC, Asins MJ, Moreno V. Increasing salt tolerance in tomato. J Exp Bot. 2006;57:1045–1058. doi: 10.1093/jxb/erj102. [DOI] [PubMed] [Google Scholar]
- 4.Murphy KST, Durako MJ. Physiological effects of short-term salinity changes on Ruppia maritima. Aquat Bot. 2003;75:293–309. doi: 10.1016/S0304-3770(02)00206-1. [DOI] [Google Scholar]
- 5.Deinlein U, et al. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa D, Morita H, Hattori T, Takeda S. Molecular characterization of the rice protein RSS1 required for meristematic activity under stressful conditions. Plant Physiol Biochem. 2012;61:54–60. doi: 10.1016/j.plaphy.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Toda Y, et al. Rice salt sensitive3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell. 2013;25:1709–1725. doi: 10.1105/tpc.113.112052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu T, et al. Overexpression of OsLEA4 enhances drought, high salt and heavy metal stress tolerance in transgenic rice (Oryza sativa L.) Environ Exp Bot. 2016;123:68–77. doi: 10.1016/j.envexpbot.2015.10.002. [DOI] [Google Scholar]
- 9.Liao YD, Lin KH, Chen CC, Chiang MC. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol Breed. 2016;36 doi: 10.1007/s11032-016-0446-2. [DOI] [Google Scholar]
- 10.Zeng Z, et al. Overexpression of a glyoxalase gene, OsGly I, improves abiotic stress tolerance and grain yield in rice (Oryza sativa L.) Plant Physiol Biochem. 2016;109:62–71. doi: 10.1016/j.plaphy.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 11.He X, et al. A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice. Plant Biol J. 2017;19:257–267. doi: 10.1111/plb.12514. [DOI] [PubMed] [Google Scholar]
- 12.Bohnert HJ, Gong Q, Li P, Ma S. Unraveling abiotic stress tolerance mechanisms–getting genomics going. Curr Opin Plant Biol. 2006;9:180–188. doi: 10.1016/j.pbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Sreenivasulu N, Sopory SK, Kishor PK. Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene. 2007;388:1–13. doi: 10.1016/j.gene.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Seki H, et al. Hairy root-activation tagging: a high-throughput system for activation tagging in transformed hairy roots. Plant Mol Biol. 2005;59:793–807. doi: 10.1007/s11103-005-1008-x. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi H, Czaja I, Lubenow H, Schell J, Walden R. Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Science. 1992;258:1350–1353. doi: 10.1126/science.1455228. [DOI] [PubMed] [Google Scholar]
- 16.Jeong DH, et al. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tani H, et al. Activation tagging in plants: a tool for gene discovery. Funct Integr Genomics. 2004;4:258–266. doi: 10.1007/s10142-004-0112-3. [DOI] [PubMed] [Google Scholar]
- 19.Kolesnik T, et al. Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J. 2004;37:301–314. doi: 10.1046/j.1365-313X.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 20.Moin M, et al. Activation tagging in indica rice identifies ribosomal proteins as potential targets for manipulation of water-use efficiency and abiotic stress tolerance in plants. Plant Cell Environ. 2016;39:2440–2459. doi: 10.1111/pce.12796. [DOI] [PubMed] [Google Scholar]
- 21.Mathews H, et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell. 2003;15:1689–1703. doi: 10.1105/tpc.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter JD, Pereira A, Dickerman AW, Veilleux RE. An active Ac/Ds transposon system for activation tagging in tomato cultivar m82 using clonal propagation. Plant Physiol. 2013;162:145–56. doi: 10.1104/pp.113.213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fladung M, Polak O. Ac/Ds-transposon activation tagging in poplar: a powerful tool for gene discovery. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu N, et al. Transposon based activation tagging in diploid strawberry and monoploid derivatives of potato. Plant Cell Rep. 2014;33:1203–16. doi: 10.1007/s00299-014-1610-y. [DOI] [PubMed] [Google Scholar]
- 25.Lindemose S, O’Shea C, Jensen MK, Skriver K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013;14:5842–5878. doi: 10.3390/ijms14035842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayliffe MA, et al. A barley activation tagging system. Plant Mol Biol. 2007;64:329–347. doi: 10.1007/s11103-007-9157-8. [DOI] [PubMed] [Google Scholar]
- 27.Antre HA, et al. An Ac/Ds-mediated Insertional Mutagenesis System for Functional Genomics in Sorghum. Molecular Plant Breeding. 2013;4:220–227. [Google Scholar]
- 28.Fang H, et al. Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol Biol. 2015;87:441–58. doi: 10.1007/s11103-015-0294-1. [DOI] [PubMed] [Google Scholar]
- 29.Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell. 2008;20:3122–3135. doi: 10.1105/tpc.108.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/S0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 31.Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle. 2004;3:217–222. doi: 10.4161/cc.3.2.654. [DOI] [PubMed] [Google Scholar]
- 32.Kahle J, Baake M, Doenecke D, Albig W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol Cell Biol. 2005;25:5339–5354. doi: 10.1128/MCB.25.13.5339-5354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceribelli M, et al. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008;28:2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petroni K, et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012;24:4777–4792. doi: 10.1105/tpc.112.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riechmann JL, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 36.Gusmaroli G, Tonelli C, Mantovani R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene. 2002;283:41–48. doi: 10.1016/S0378-1119(01)00833-2. [DOI] [PubMed] [Google Scholar]
- 37.Siefers N, et al. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephenson TJ, McIntyre CL, Collet C, Xue GP. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol. 2007;65:77–92. doi: 10.1007/s11103-007-9200-9. [DOI] [PubMed] [Google Scholar]
- 39.Quach TN, et al. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol Genet Genomics. 2015;290:1095–115. doi: 10.1007/s00438-014-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, et al. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics. 2016;17:1–16. doi: 10.1186/s12864-015-2294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Wan XL, Guo C, Zhang JW, Bao MZ. Identification and expression analysis of nuclear factor Y families in Prunus mume under different abiotic stresses. Biol. Plant. 2016;60:419–426. doi: 10.1007/s10535-016-0624-4. [DOI] [Google Scholar]
- 42.Ren C, Zhang Z, Wang Y, Li S, Liang Z. Genome-wide identification and characterization of the NF-Y gene family in grape (vitis vinifera L.) BMC Genomics. 2016;17 doi: 10.1186/s12864-016-2989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genomics. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- 44.Yang W, Lub Z, Xionga Y, Yaoa J. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J. 2017;5:21–31. doi: 10.1016/j.cj.2016.06.014. [DOI] [Google Scholar]
- 45.Swain, S., Myers, Z. A., Siriwardana, C. L., & Holt, B. F. III. The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim Biophys Acta, doi:10.1016/j.bbagrm.2016.10.012 (2016). [DOI] [PubMed]
- 46.Zanetti ME, Rípodas C, Niebel A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbagrm.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Qu S, Desai A, Wing R, Sundaresan V. A versatile transposon-based activation tag vector system for functional genomics in cereals and other monocot plants. Plant physiol. 2008;146:189–199. doi: 10.1104/pp.107.111427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue LJ, Zhang JJ, Xue HW. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. Plos one. 2012;7 doi: 10.1371/journal.pone.0031081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–50. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi T, Blumwald E. Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 2005;10:615–620. doi: 10.1016/j.tplants.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Ghaffari A, Gharechahi J, Nakhoda B, Salekdeh GH. Physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at the vegetative stage. J Plant Physiol. 2014;171:31–44. doi: 10.1016/j.jplph.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 53.Domingo C, Lalanne E, Catalá MM, Pla E, Reig-Valiente JL. & Talón, M. Physiological Basis and Transcriptional Profiling of Three Salt-Tolerant Mutant Lines of Rice. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuromori T, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313X.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 55.Amir Hossain M, et al. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol. 2010;72:557–566. doi: 10.1007/s11103-009-9592-9. [DOI] [PubMed] [Google Scholar]
- 56.Asano T, et al. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol Biol. 2011;75:179–191. doi: 10.1007/s11103-010-9717-1. [DOI] [PubMed] [Google Scholar]
- 57.Chen M, Zhao Y, Zhuo C, Lu S, Guo Z. Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol J. 2015;13:482–491. doi: 10.1111/pbi.12270. [DOI] [PubMed] [Google Scholar]
- 58.Xu JJ, Zhang XF, Xue HW. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J Exp Bot. 2016;67:6399–6411. doi: 10.1093/jxb/erw409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laloum T, De Mita S, Games P, Baudin M, Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18:157–166. doi: 10.1016/j.tplants.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Lee H, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis leafy cotyledon1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, S. et al. The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. J Exp Bot doi:10.1093/jxb/erw468 (2017). [DOI] [PMC free article] [PubMed]
- 62.Nelson DE, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. US. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li WX, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post transcriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alam MM, et al. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant BiotechnolJ. 2015;13:85–96. doi: 10.1111/pbi.12239. [DOI] [PubMed] [Google Scholar]
- 65.Sun X, Lian H, Liu X, Zhou S, Liu S. The garlic NF-YC gene, AsNF-YC8, positively regulates non-ionic hyperosmotic stress tolerance in tobacco. Protoplasma. 2016;254:1353–1366. doi: 10.1007/s00709-016-1026-3. [DOI] [PubMed] [Google Scholar]
- 66.Kishor PBK, Hong Z, Miao G, Hu C, Verma DPS. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline overproduction and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shabala S, Cuin TA. Potassium transport and plant salt tolerance. Physiol. Plant. 2008;133:651–669. doi: 10.1111/j.1399-3054.2007.01008.x. [DOI] [PubMed] [Google Scholar]
- 68.Wang M, Zheng Q, Shen Q, Guo S. The critical role of potassium in plant stress response. Int J Mol Sci. 2013;14:7370–7390. doi: 10.3390/ijms14047370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munns R, Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 70.Glenn EP, Brown JJ, Blumwald E. Salt tolerance and crop potential of halophytes. CRC Crit Rev Plant Sci. 1999;18:227–255. doi: 10.1016/S0735-2689(99)00388-3. [DOI] [Google Scholar]
- 71.Hoang TML, Williams B, Khanna H, Dale J, Mundree SG. Physiological basis of salt stress tolerance in rice expressing the anti-apoptotic gene SfIAP. Funct Plant Biol. 2014;41:1168–1177. doi: 10.1071/FP13308. [DOI] [PubMed] [Google Scholar]
- 72.Mekawy AMM, et al. Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol Biochem. 2015;87:17–25. doi: 10.1016/j.plaphy.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Foyer CH, Shigeoka S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011;155:93–100. doi: 10.1104/pp.110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–457. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 75.Wang G, et al. A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol Plant. 2016;158:45–64. doi: 10.1111/ppl.12444. [DOI] [PubMed] [Google Scholar]
- 76.Wei T, et al. Ectopic Expression of DREB Transcription Factor, AtDREB1A, Confers Tolerance to Drought in Transgenic Salvia miltiorrhiza. Plant Cell Physiol. 2016;57:1593–609. doi: 10.1093/pcp/pcw084. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida, S., Forno, D. A., Cock, J. H., & Gomez, K. A. Routine procedures for growing rice plants in culture solution. In: Cock J. H., Gomez K. A. Laboratory Manual for Physiological Studies of Rice. Los Banos, Philippines: IRRI: 61–66 (1976).
- 78.Manimaran P, et al. Infection of early and young callus tissues of indica rice BPT 5204 enhances regeneration and transformation efficiency. Rice Sci. 2013;20:415–426. doi: 10.1016/S1672-6308(13)60153-5. [DOI] [Google Scholar]
- 79.Liu YG, Mitsukawa M, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant j. 1995;3:457–463. doi: 10.1046/j.1365-313X.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 80.Manimaran P, et al. Identification of cis-elements and evaluation of upstream regulatory region of a rice anther-specific gene, OSIPP3, conferring pollen-specific expression in Oryza sativa (L.) ssp. indica. Plant Reprod. 2015;28:133–42. doi: 10.1007/s00497-015-0264-4. [DOI] [PubMed] [Google Scholar]
- 81.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 82.Sun W, Cao Z, Li Y, Zhao Y, Zhang H. A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia, Bratislava. 2007;62:529–532. doi: 10.2478/s11756-007-0104-6. [DOI] [Google Scholar]
- 83.Lichtenthaler HK, Wellburns AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983;11:591–592. doi: 10.1042/bst0110591. [DOI] [Google Scholar]
- 84.Bates LS, Waldran R, Teare ID. Rapid determination of free proline for water studies. Plant and Soil. 1973;39:205–208. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 85.Dhindsa RS, Plumb-dhindsa P, Thorpe TA. Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot. 1981;32:93–101. doi: 10.1093/jxb/32.1.93. [DOI] [Google Scholar]
- 86.Castillo FI, Penel I, Greppin H. Peroxidase release induced by ozone in Sedum album leaves. Plant Physiol. 1984;74:846–851. doi: 10.1104/pp.74.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–6. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.