Abstract
The hnRNP A1 protein and a shortened derivative (UP1) promote telomere elongation in mammalian cells. In support of a direct role for A1 in telomere biogenesis, we have shown that the recombinant UP1 protein binds to telomeric DNA sequences in vitro, and pulls down telomerase activity from a cell extract. Here we show that A1/UP1 can interact directly with the RNA component of human telomerase (hTR). A portion of A1/UP1 that contains RNA recognition motif 2 (RRM2) is sufficient for an interaction with the first 208 nt of hTR. Given that the portion of A1/UP1 that contains RRM1 is sufficient for binding to a telomeric DNA oligonucleotide, we have tested whether A1/UP1 can interact simultaneously with both nucleic acids. Using a chromatography assay, we find that A1/UP1 bound to hTR can interact with telomeric DNA. Notably, these interactions are sufficiently robust to withstand incubation in a cell extract. Our results suggest that hnRNP A1 may help recruit telomerase to the ends of chromosomes.
INTRODUCTION
The extremities of eukaryotic chromosomes, or telomeres, are organized into specialized structures that protect the ends from nucleolytic degradation and prevent their recognition as double-stranded breaks. However, to counteract the loss of telomeric sequences following conventional DNA replication, telomeres must remain accessible to telomerase, the ribonucleoprotein enzyme that carries out telomere elongation (1). How telomerase is recruited to telomeres is not known. In the yeast Saccharomyces cerevisiae, the Est1p protein is a component of telomerase that interacts with telomerase RNA (2). Because Est1p can also bind to G-rich telomeric DNA in vitro (3), Est1p may mediate telomerase access to chromosome termini (2,4). In mammals, the hTERT and mTERT proteins have been identified as the catalytic component of the human and mouse telomerases, respectively (5–10). Several proteins including TEP1 (11,12), the RNA binding proteins hStau and L22 (13), and the chaperone proteins p23 and Hsp90 (14) have been found to associate with human telomerase. Whether any of these proteins facilitate the interaction of mammalian telomerase with telomeric DNA has not been addressed. More recently, immunoprecipitation assays in a cell extract have shown that the hnRNP C1 and C2 proteins can interact with the RNA component of the human telomerase (15).
HnRNP A1 is one of the most abundant nuclear proteins in actively growing mammalian cells. A1 is involved in a variety of RNA-related processes including alternative RNA splicing (16–19) and mRNA transport (20). We have previously shown that a deficiency in A1 expression in a mouse erythroleukemic cell line is associated with short telomeres, and that restoring A1 expression increases the length of telomeres (21). The expression of a shortened version of A1 (UP1), lacking the glycine-rich domain, can also promote telomere elongation in mouse and human cells.
Although we do not yet know the mechanism by which A1/UP1 modulates telomere length, this activity may be mediated through the direct binding of A1/UP1 to single-stranded telomeric sequences. In mammals, some of the 50–150 nt extensions of the G-rich strand (22,23) may be sequestered into duplex structures (24), a process that should expose internal single-stranded G-rich repeats. UP1 was initially found to be part of complexes that assemble on telomeric oligonucleotides following incubation in a HeLa nuclear extract (25). A1/UP1 binds directly and specifically to vertebrate telomeric repeats in vitro (21,26). Although the recent structure of a co-crystal of UP1/telomeric DNA suggests that both RNA binding domains of UP1 (RRM1 and RRM2) can interact with telomeric DNA (27), our biochemical assays indicate that RRM1 is sufficient for strong and specific binding to single-stranded telomeric DNA in vitro (26). Consistent with the possibility that one function of A1/UP1 may be to shield and protect the ends of chromosomes, we have shown that the binding of A1/UP1 to telomeric DNA oligonucleotides prevents access to a variety of nucleases and polymerases (26).
Another observation that argues in favor of a direct role for A1/UP1 in telomere biogenesis is the ability of recombinant UP1 to interact with telomerase in a mammalian cell extract (21). Given that A1 is an RNA binding protein and that telomerase contains an RNA component (hTR in human cells), we sought to test whether A1/UP1 could interact directly with hTR. We report that A1/UP1 binds to hTR with specificity, and that this interaction requires the RRM2 domain. Because A1/UP1 can interact simultaneously with single-stranded telomeric DNA sequences, one function of hnRNP A1 may be to recruit telomerase to mammalian telomeres.
MATERIALS AND METHODS
Plasmids and oligonucleotides
The expression plasmids pGEX-UP1 and pGEX-A1 were described previously in LaBranche et al. (21). The construction of pGEX-UP1Δ1 and pGEX-UP1Δ2 have been described in Dallaire et al. (26). The pGem-T plasmid encoding the human RNA telomerase component was kindly provided by S.Bacchetti and R.Reddel. The telomeric oligonucleotide TS10 (TTAGGG)10 was used in the columns and competition experiments.
In vitro transcription and RNA purification
The plasmid pGem-T, containing the sequence of hTR, was digested with a variety of restriction enzymes (MscI, DdeI, BsaHI, XbaI, AvaII, NaeI or BglI). RNA molecules produced from these plasmids (hTR-M, hTR-D, hTR-Ba, hTR-X, hTR-A, hTR-F and Bg, respectively) were obtained by transcription in vitro with either SP6 or T7 RNA polymerase (Amersham Pharmacia Biotech) in the presence of cap analog and [α-32P]UTP (New England Nuclear). RNA purification was performed as described previously (28).
Gel mobility shift assay
The binding assay of recombinant A1 or UP1 to telomerase RNA or the TS10 DNA oligonucleotide was performed in 10 µl containing 75 mM KCl, 2.5 mM MgCl2, 3% Ficoll, 40 mM poly(dI-dC), 0.1 mM EDTA, 5 mM HEPES and 1 mM DTT. In the assay presented in Figure 1C, poly(dI-dC) was substituted for heparin (final concentration of 2.5 µg/µl). Nucleic acids were always added to the mixture before addition of the protein. The reaction mixtures were incubated on ice for 5 min before the addition of 2 µl of dye and migration on a non-denaturing 5% acrylamide gel (29:1 acrylamide/bis-acrylamide, 50 mM Tris pH 8.8, 50 mM glycine and 5% glycerol) run in Tris–glycine buffer (50 mM Tris pH 8.8 and 50 mM glycine).
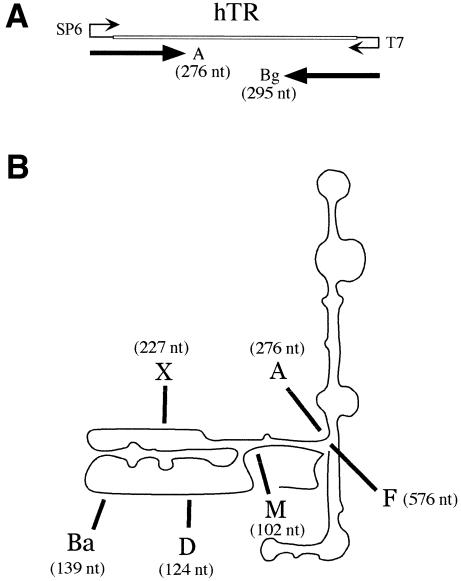
Figure 1.
UP1 binds specifically to hTR. (A) Diagram of the hTR portion (451 bp) of the pGem-T plasmid with the transcription start sites of the SP6 and T7 RNA polymerases. hTR-A RNA is 276 nt, the first 68 nt of which are plasmid sequences. Bg RNA is a control RNA derived from the complementary strand. (B) A schematic representation of hTR is shown and is based on the report by Chen et al. (34). The 3′ end of each transcript used in (C) is indicated and corresponds to the position used for plasmid linearization and run-off transcription with SP6 RNA polymerase. The lengths of the products obtained after transcription from the pGEM-T plasmid are indicated in parenthesis and include the 5′ portion derived from plasmid sequence. (C) The binding of UP1 to various portions of hTR. Mixtures containing recombinant UP1 and portions of hTR or control RNAs were fractionated in a 5% native polyacrylamide gel. Each set contains the RNA alone (1 fmol) and increasing amounts of UP1 protein (2.5, 5 and 10 pmol). This gel-shift assay was performed in the presence of heparin at a final concentration of 2 µg/µl. (D) UP1 binding in the presence of competitor RNAs. UP1 (5 pmol) and the 32P-labeled hTR-A RNA (1 fmol) were incubated in the presence of increasing amounts of competitor hTR-A RNA (1, 50, 100, 250, 500 and 1000 fmol) or Bg RNA (1, 50, 100, 500 and 1000 fmol). Because the unlabeled RNAs were synthesized in the presence of residual amounts of [32P]-UTP to facilitate purification and quantification, aliquots of ‘cold’ hTR-A and Bg RNAs alone were loaded as controls (lanes 9 and 15, respectively).
Protein expression and purification
Procedure I to purify recombinant GST-A1 and GST-UP1 has been described in Dallaire et al. (26). Purification of GST-A1, GST-UP1, GST-UP1Δ1 and GST-UP1Δ2 by procedure II was as follows. The expression plasmids encoding A1 or UP1 were transformed in Escherichia coli B21 strains and incubated overnight in 25 ml of L-broth medium containing 100 µg/µl of ampicillin. An aliquot of 10 ml of this culture was transferred to 500 ml of L-broth/ampicillin. Protein induction was realized by addition of IPTG to a final concentration of 100 µM when the culture reached 0.6–2.0 OD600. After 4 h of induction, the culture was centrifuged at 3500 r.p.m. at 4°C for 10 min. The supernatant was removed and the pellet washed and resuspended in washing buffer (50 mM piperazine pH 9.8, 0.5 M NaCl, 1 mM EDTA and 1 mM DTT). Cells were spun at 10 000 r.p.m. at 4°C for 10 min. The pellet was resuspended in lysis buffer (washing buffer containing 0.3 mg/ml of lysozyme, 0.5 mM PMSF, 1.6 mM benzamidine and 2 µM bacitracine). The resuspended pellet was sonicated 4 × 30 s. Triton X-100 was then added to a final concentration of 1% and the solution was incubated on a rotator at 4°C for 30 min. The mixture was centrifuged at 4°C for 10 min at 10 000 r.p.m. The pellet was washed, centrifuged again and resuspended in washing buffer. Gluthathione–Sepharose beads (500 µl) were then added to the solution and incubated for 1 h at 4°C. Beads were recovered and elution of GST-A1 and GST-UP1 was performed in washing buffer containing 200 mM piperazine and 20 mM gluthathione. This last step was repeated three times. The eluted fractions were pooled and dialyzed against buffer DN (20 mM HEPES pH 7.9, 100 mM KCl, 5% glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT and 0.1% NP-40).
Binding assay on agarose adipic acid hydrazide columns
The telomeric oligonucleotide TS10 was purified on a denaturing 10% acrylamide gel (38:2 acrylamide/bis-acrylamide, 20% formamide, 8 M urea, 90 mM Tris–borate and 2 mM EDTA). The purified oligonucleotide (10 nmol) was resuspended in water and coupled to 250 µl of agarose adipic hydrazide according to the manufacturer’s recommendation (Amersham Pharmacia Biotech). Fifty microliters (packed volume) of agarose beads coupled with TS10 were packed in a 200 µl pipetman tip. Each protein/RNA mixture (15 fmol RNA and 96 pmol of protein) was prepared, mixed with buffer DN and half was loaded onto the column (the other half was kept as the input fraction). Each column was washed several times with 200 µl (4 × 50 µl) of buffer DN. In some experiments the salt concentration of buffer DN (normally containing 100 mM KCl) was changed to 150 or 75 mM KCl, as indicated. Stepwise elution was accomplished with buffer DN containing increasing salt concentrations (250, 500 or 750 mM or 1 M KCl). For each salt concentration, elution was performed with a 200 µl vol (4 × 50 µl). Input, flow-through, wash and eluted fractions were split in half and each set was processed independently. For the first set, the protein profile was monitored by silver staining after fractionation by SDS–PAGE. For the second set, labeled hTR-A RNA was extracted with phenol/chloroform/isoamylalcohol, ethanol precipitated and fractionated on a 5% acrylamide–8 M urea gel.
The HeLa S100 extract (protein concentration of 5 µg/µl) was prepared as described previously (29). An aliquot of 15 fmol of hTR-A (with or without 75 pmol of A1) was added to 10 µl of S100 extract in a total volume of 100 µl in buffer DN. After a 10 min incubation at 4°C, the mixture was loaded onto a TS10 column.
RESULTS
UP1 interacts with human telomerase RNA
We showed previously that a column made with GST-UP1 can specifically recover telomerase activity from a cell extract (21). To test whether UP1 can interact directly with telomerase RNA, we performed a gel mobility shift assay using recombinant UP1 protein. As substrate, we used a battery of 32P-labeled RNAs transcribed from the gene encoding human telomerase RNA (hTR). These RNAs were produced by run-off transcription of pGem-T linearized at internal positions (Fig. 1A and B). As controls, we used a 295 nt RNA-containing sequence complementary to the 3′ end of hTR (Bg), and the 91 nt CE4 RNA, which contains a high affinity binding site for A1/UP1 (19). Mixtures were fractionated in a native polyacrylamide gel. As shown in Figure 1C, hTR-M, hTR-D and hTR-X were not significantly bound by UP1. In contrast, >50% of hTR-Ba, hTR-A and hTR-F existed in complexes at the highest concentration of UP1 tested (Fig. 1C, lanes 12, 20 and 24). Although the control antisense Bg RNA was bound at the highest concentration of UP1 (Fig. 1C, lane 28), <50% of the input RNA was in the complex. These results suggest that UP1 can bind to the 5′ half of hTR. Because hTR-Ba is the shortest RNA molecule bound by UP1, the binding site may reside within the first 71 nt of hTR, a region that includes the template region (+46 to +55). It is likely that the secondary structure of the RNA molecule plays an important role in the presentation of the binding site since UP1 did not bind to a longer RNA (hTR-X), but bound to a slightly longer version (hTR-A).
To address the specificity of the UP1/hTR interaction we performed the binding assay in the presence of molar excesses of competitor RNAs. As shown in Figure 1D, a gradual increase in the amount of cold hTR-A RNA compromised the formation of UP1/32P-hTR-A complexes (Fig. 1D, lanes 3–8). In contrast, these complexes were not affected by the addition of a 1000-fold excess of unlabeled Bg RNA (Fig. 1D, lanes 10–14). The above binding assays do not address whether UP1 binds specifically to one strong binding site in hTR or to multiple weaker binding sites, or to a combination thereof. This issue will be resolved by testing a battery of mutated hTR derivatives.
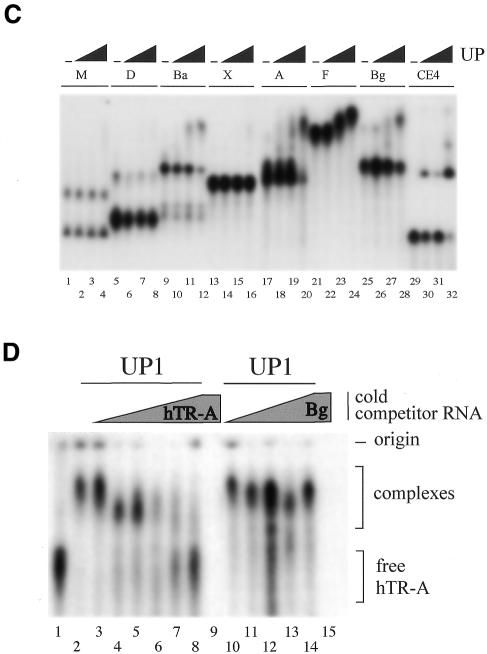
We reported previously that UP1 but not the full-length hnRNP A1 protein could recover telomerase activity from a mammalian extract (21). Thus, recombinant hnRNP A1 might not interact with hTR-A RNA. Indeed, we observed that a preparation of recombinant A1 protein interacted much less efficiently than UP1 with hTR-A (Fig. 2A), despite the fact that this A1 preparation could bind to a telomeric oligonucleotide with an efficiency similar to UP1 (data not shown). However, during the course of these investigations we modified the procedure used for the preparation of recombinant proteins (procedure II, see Materials and Methods). A1 preparations made according to procedure II interacted with hTR-A RNA as efficiently as UP1 (Fig. 2B). These results indicate that both recombinant A1 and UP1 can interact with telomerase RNA. The failure of early A1 preparations to interact with hTR may be due to an improper folding of the glycine-rich domain, a region that has been reported to be highly disordered (30), and therefore possibly more inclined to interfere with the activity of a nearby nucleic acid binding domain (see Discussion).
Figure 2.
A1 binds to hTR. Gel-shift assay showing the binding activity of different preparations of recombinant A1 to hTR-A. Procedure I (A) and procedure II (B) were used to prepare recombinant A1 and UP1 proteins. The binding assays were performed with 1 fmol of hTR-A and 2.5, 5 or 10 pmol of recombinant proteins in (A), and with 1 fmol of hTR-A and 1, 2, 4, 6, 8 or 10 pmol of recombinant proteins in (B).
The RRM2 domain is required for the interaction with telomerase RNA
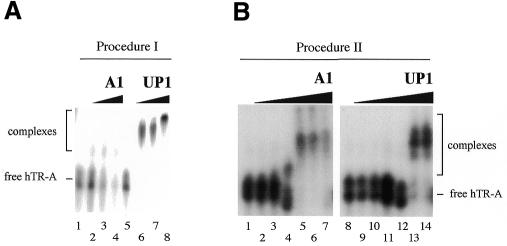
In a previous report we analyzed the role played by protein domains in the specific binding of UP1 to telomeric DNA sequences in vitro (26). Using a substrate oligonucleotide carrying 10 telomeric repeats (TS10), we found that a truncated version of UP1 that contains RRM1 and the linker region (UP1Δ2) is sufficient for strong and specific binding to telomeric sequences, as judged by gel-shift assays (26). To determine which portion of UP1 is required for the binding to hTR, we performed a gel-shift assay with two UP1 derivatives, UP1Δ1 and UP1Δ2. UP1Δ1 lacks most of RRM1 while UP1Δ2 lacks RRM2 (Fig. 3A). As shown previously (26), only UP1Δ2 can form complexes with TS10 at the concentrations tested (Fig. 3B). Interestingly, while UP1Δ1 forms a complex with hTR-A (Fig. 3C, lane 2), UP1Δ2 does not (Fig. 3C, lane 3). The interaction of UP1Δ1 with hTR-A appears specific since UP1Δ1 did not bind to Bg RNA (Fig. 3C, lane 4). These results suggest that the two RRMs of hnRNP A1 have distinct specificities: RRM1 binds to telomeric DNA sequences while RRM2 interacts with the RNA component of telomerase.
Figure 3.
UP1 binding to hTR requires RRM2. (A) Structure of the UP1 derivatives. UP1Δ1 lacks amino acids 13–134 while UP1Δ2 lacks amino acids 107–196. (B) Gel-shift assay using UP1Δ1 and UP1Δ2 and the labeled TS10 telomeric DNA oligonucleotide (10 fmol). Increasing amounts of the UP1 derivatives were used in each set (0.5, 1.0, 1.5 and 2.0 pmol). (C) Gel-shift assay with UP1Δ1 and UP1Δ2. Binding was performed using uniformly 32P-labeled hTR-A (lanes 1–3) or Bg RNA (lanes 4–6). (D) A telomeric oligonucleotide does not efficiently compete the binding of UP1 to hTR RNA. UP1 (5 pmol) and the 32P-labeled RNA (1 fmol) were incubated in the presence of increasing amounts of the telomeric oligonucleotide TS10 (1, 50, 100, 250, 500 and 1000 fmol).
If the binding to telomeric DNA and telomerase RNA occurs through distinct RRMs, an excess of one nucleic acid (e.g., telomeric DNA) should not interfere with the binding of UP1 to the other nucleic acid (e.g., telomerase RNA). Consistent with this prediction, the binding of UP1 to [32P]-hTR-A remained relatively insensitive to gradual increases in the amount of the telomeric oligonucleotide TS10 (Fig. 3D, lanes 3–8). While large amounts of TS10 promoted some dissociation of hTR-A (Fig. 3D, lanes 7 and 8), similar amounts of competitor hTR-A had led to near complete disruption of the complexes (see Fig. 1D, lanes 7 and 8). These results support the notion that the two RRMs bind to different nucleic acids. However, they do not address whether UP1 can interact simultaneously with telomeric DNA and telomerase RNA since, in the gel system used, the migration of a complex is determined more by the size and number of proteins than by the length and number of nucleic acids.
A1/UP1 can interact simultaneously with telomerase RNA and telomeric DNA
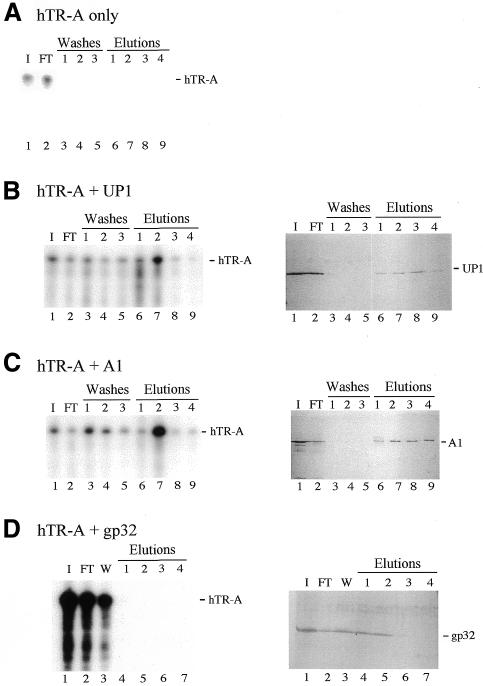
The ability of A1/UP1 to bind to telomeric sequences through the RRM1 domain and to interact with telomerase RNA through the RRM2 domain suggests that these interactions can occur simultaneously on the same A1/UP1 molecule. To address whether a ternary complex made up of A1/UP1, telomeric DNA and hTR RNA could form, we performed the following experiment. TS10 oligonucleotides were covalently coupled to an adipic acid hydrazide agarose column. The TS10 column was loaded with a mixture containing 32P-labeled hTR-A RNA and GST-UP1 or GST-A1. A control loading was performed with a mixture containing labeled hTR-A only. Loaded columns were washed extensively with buffer containing 100 mM KCl and the bound material was eluted in a stepwise manner using solutions containing 250, 500 or 750 mM or 1 M KCl. In the absence of protein, labeled hTR-A RNA was not significantly retained by the column, as noted by the presence of hTR-A in the flow-through fraction only (Fig. 4A, lane 2). In contrast, in the presence of UP1 or A1, hTR-A bound to the column and eluted principally in the 0.5 M KCl fraction (Fig. 4B and C, lane 7 in the left panels). The bulk of bound UP1 and A1 also began eluting at 0.5 M KCl (Fig. 4B and C, lanes 7–9 in the right panels). The fact that little hTR-A RNA remained in the 0.75 M KCl fractions (lane 8), which still contain A1/UP1, indicates that the RNA–protein interaction tolerates 250 mM KCl but does not resist 0.5 M KCl. These results demonstrate the existence of a complex containing recombinant A1/UP1, hTR-A and telomeric DNA.
Figure 4.
A1 and UP1 interact with telomeric DNA sequences and hTR simultaneously. Mixtures of [32P]-hTR-A and proteins were loaded onto TS10 columns. Left panels are acrylamide–urea gels exposed to visualize 32P-labeled hTR-A in various fractions, while right panels represent silver-stained protein gels. I, input; FT, flow-through fraction; W, wash. Washes 1, 2 and 3 are successive washes with each of 4 × 50 µl of loading buffer (buffer DN). Elutions 1, 2, 3 and 4 are successive elutions with each of 4 × 50 µl of buffer DN containing 250, 500 or 750 mM or 1 M KCl, respectively. Except for the left panel of D, in which all the I, FT and W fractions have been loaded (W is a pool of three successive washes with buffer DN containing 75 mM KCl), 10 times less of the I and FT fractions have been loaded. The positions of the proteins and hTR-A are indicated.
To address the specificity of this interaction, we carried out an interaction assay with the single-stranded nucleic acid binding protein gp32. gp32 can bind telomeric DNA oligonucleotides, as judged by gel-shift assays (26). While gp32 also binds to the TS10 column (Fig. 4D, lanes 4 and 5 in the right panel), bound gp32 did not retain hTR-A (Fig. 4D, lanes 4–7 in the left panel).
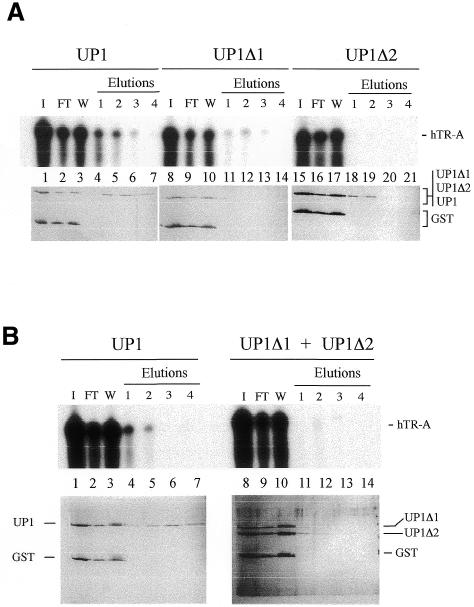
As additional controls, we tested the ability of GST-UP1Δ1 and GST-UP1Δ2 to retain hTR-A on the telomeric DNA column. Loading was performed at higher salt concentration (150 mM KCl), to minimize non-specific interactions, and in the presence of the protein GST as an internal control. In these conditions, GST-UP1 was specifically retained by the TS10 column (Fig. 5A, lanes 1–7, bottom panel), and the majority of hTR-A was recovered in the 250 mM and 0.5 M KCl washes (Fig. 5A, lanes 4 and 5, top panel). While UP1Δ2 could bind to the TS10 column, it eluted at a lower salt concentration than UP1 (Fig. 5A, lanes 18–21, bottom panel), consistent with our previous observation that UP1Δ2 binds to TS10 less efficiently than UP1 (26). Although UP1Δ2 bound to the column, it did not retain hTR-A (Fig. 5A, lanes 18–21, top panel). In contrast and consistent with our previous report, UP1Δ1 did not bind significantly to the telomeric DNA column (Fig. 5A, lanes 11–14, bottom panel). Consequently, little hTR-A was retained by the column (Fig. 5A, lanes 11–14, top panel). The residual amount of hTR-A may be due to the varying tendency of the recombinant proteins to form aggregates. Because UP1Δ1 can bind to hTR-A (Fig. 3C), this may explain why some hTR-A is found in the eluted fractions obtained with UP1Δ1. Performing the experiment with a mixture of GST-UP1Δ2 and GST-UP1Δ1 essentially led to the same conclusion: very little hTR-A was retained on the TS10 column (Fig. 5B, lanes 11–14, top panel). These results confirm the importance of RRM1 in telomeric DNA binding, and of RRM2 in hTR-A binding. Thus, simultaneous binding of UP1 to TS10 and hTR-A requires the presence of both RRMs.
Figure 5.
Simultaneous binding requires both RRMs. Mixtures containing [32P]-hTR-A, GST, recombinant UP1 or derivatives (UP1Δ1 and UP1Δ2) were loaded onto TS10 columns. Top panels are acrylamide–urea gels used to fractionate 32P-labeled hTR-A. Bottom panels are silver-stained protein gels. I, input; FT, flow-through fraction; W, wash (a pool of three washes with buffer DN containing 150 mM KCl). Half of the I, F, W and eluted fractions were loaded on the RNA gel and the other half on a protein gel. The positions of the proteins and hTR-A are indicated.
We have shown in a previous study using adipic acid columns that the recombinant A1 protein can interact with itself, and that this interaction requires the glycine-rich domain since it does not occur with UP1 (19). Because GST-UP1 but not free GST is retained on the column (Fig. 5), dimer formation through the GST domains is unlikely in our experimental conditions. Because a GST-A1 column can pull-down other GST-A1 molecules (data not shown), the possibility that recombinant molecules already exist as stable dimers appears unlikely. Although we cannot rule out that a GST-induced dimerization could conceivably be a problem in our assay, we favor the notion that the simultaneous binding of UP1 to telomerase RNA and telomeric DNA takes place on a single UP1 molecule, and not on a complex containing several UP1 proteins.
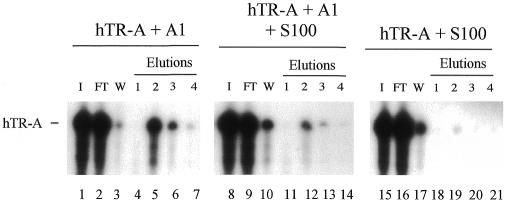
As the experiments described above were performed with purified components, we wished to address whether these interactions could occur in an environment that is more biologically relevant. Thus, a solution containing recombinant A1 and hTR-A was added to a HeLa S100 extract, which is a cellular preparation used to carry out conventional telomerase extension assays (31,32). The mixture was loaded onto a TS10 column to determine whether hTR-A could be retained in an A1-dependent manner. The results shown in Figure 6 indicate that complexes containing hTR-A/A1/TS10 can form in a HeLa S100 extract. Thus, the interactions between A1, hTR-A and TS10 are sufficiently strong to withstand incubation in a cell extract.
Figure 6.
The TS10/A1/hTR-A ternary complex resists incubation in a nuclear extract. Left, hTR-A and recombinant A1 were mixed and loaded onto the TS10 column. Middle, hTR-A and recombinant A1 were incubated together and mixed in a HeLa S100 extract. The mixture was then loaded onto a TS10 column. Right, as in the middle panel except that A1 was omitted. In all cases, RNA recovered from each fraction was fractionated on a denaturing polyacrylamide gel. I, input (total fraction); FT, flow-through fraction (total fraction); W, wash fractions (total fractions). Elutions 1, 2, 3 and 4 are successive elutions with buffer DN containing 250, 500 or 750 mM or 1 M KCl, respectively. The position of hTR-A is indicated.
DISCUSSION
We have shown that A1 and its shortened derivative UP1 can bind specifically to hTR in vitro. The first 214 nt of hTR (hTR-A) are sufficient for binding. This region corresponds to the minimal region of hTR that can rescue telomerase activity in an extract where endogenous hTR had been inactivated by micrococcal nuclease digestion (33). Moreover, an exhaustive phylogenetic comparative analysis suggests that this region forms a distinct motif containing a pseudoknot domain (34). The binding of UP1 to shorter versions of hTR suggests that the binding site may be located within the first 71 nt, a portion that contains the template region. We are currently using a variety of approaches to precisely map the binding site of A1/UP1 on the 5′ portion of hTR.
The binding of UP1 to a component of the telomerase was predicted from our previous demonstration that UP1 could specifically recover telomerase activity from a mouse cell extract (21). Recently we have shown that the first nucleic acid binding domain of UP1 (RRM1) is sufficient for strong binding to telomeric DNA sequences in vitro (26). Because the second nucleic acid binding domain of UP1 (RRM2) is not essential for binding to telomeric DNA, we reasoned that it could be involved in binding to telomerase RNA. This prediction was confirmed in vitro: the deletion of RRM2 compromised binding to hTR-A, and a derivative lacking most of RRM1 but containing a complete RRM2 bound specifically to the 5′ end portion of telomerase RNA.
Although the structure of a co-crystal of UP1/telomeric DNA indicates that both RRM1 and RRM2 interact with telomeric DNA (27), our biochemical assays indicate that RRM1 is sufficient for strong and specific binding to single-stranded telomeric DNA in vitro (26), while the C-terminal portion of UP1 that contains RRM2 is sufficient for binding to hTR-A. Given that the RRM2 portion has a weaker affinity for telomeric sequences than the RRM1 portion (26), the RRM2 portion may interact with a telomeric sequence when it is the only sequence available (as in the crystal), but may be capable of interacting with another nucleic acid molecule, like hTR.
Most importantly, we have shown that the interactions of A1 with telomeric DNA and hTR-A may occur simultaneously in vitro, and that these interactions can resist incubation in a cell extract. Although some proteins have the ability to bind to either DNA or RNA (e.g., gp32, ssB, TFIIIA), a protein that can interact simultaneously with DNA and RNA represents a unique situation in biological systems. Interestingly, the yeast protein Est1 has recently been shown to interact with the Tlc1 telomerase RNA (2). Although Est1 has been reported to interact with telomeric DNA with low affinity (3), it is not known whether these nucleic acid interactions can occur simultaneously on Est1. We have spent considerable efforts in trying to document the formation of a hTR/UP1/telomeric DNA oligonucleotide complex by native gel analysis. Despite observing binding of UP1 to either hTR or telomeric DNA, we have been unable to confirm simultaneous binding using this approach. The stable migration of a UP1 complex in an electrical field may require that both RRMs interact with hTR or the DNA oligonucleotide. Although the chromatography assay may be considered less stringent (no heparin, no electrical field), we have minimized low affinity interactions by performing the assay at higher salt concentrations (100–150 mM KCl). The fact that the UP1-dependent binding of hTR-A to the TS10 column was resistant to a 250 mM KCl wash also suggests that the assay monitored relatively strong interactions.
Another important finding concerns the ability of the complete hnRNP A1 protein to interact with hTR. We had observed initially that the recombinant A1 protein could not recover telomerase activity from a cell extract (21). We have shown here that the procedure used to prepare recombinant A1 is critical to confer the ability to bind hTR. The procedure used in the initial report did not yield A1 molecules that could interact with hTR. In contrast, the new procedure yielded A1 proteins that bound to hTR as efficiently as UP1. Although the reason for these differences remains unclear, it has been reported that the glycine-rich domain (GRD) of A1, which is absent from UP1, is largely disordered (30). Therefore, one possibility is that an unstructured GRD interferes with the binding activity of the closest RRM (RRM2), which binds to hTR. Because cell lysis in procedure II is performed at 0.5 M NaCl (as compared with 130 mM NaCl in procedure I), procedure II may yield a more structured GRD, allowing RRM2 to interact more stably with hTR-A.
Our initial observation had led us to suggest that the conversion of A1 into UP1 might be important to yield a protein capable of interacting with telomerase. However, based on the results presented in this study we would instead postulate that the structure of the GRD may modulate the interaction between A1 and hTR. Notably, phosphorylation of the GRD changes its conformation (30). Several kinases, including PKCζ, have been documented to phosphorylate the GRD in vitro and in vivo (30,35,36). Thus, recruitment of telomerase to chromosome ends may be modulated by kinases that target the GRD of A1.
Although the binding of hnRNP A1 to single-stranded extensions at the ends of mammalian chromosomes remains to be demonstrated in vivo, our results support a model in which hnRNP A1 plays a direct role in telomere biogenesis. Our previous report (26) suggests that the binding of A1 to telomeric DNA may help shield the ends of chromosomes from nucleolytic attack and from surveillance mechanisms that detect double-stranded DNA breaks. The recruitment of the telomerase ribonucleoprotein complex to chromosome ends may contribute to this protective role. In addition, the simultaneous interaction of A1 with telomeric DNA and hTR may help position telomerase in preparation for the extension of 3′ overhangs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Silvia Bacchetti and Roger Reddel for the hTR plasmid. This work was supported by a grant from the National Cancer Institute of Canada with funds from the Canadian Cancer Society. B.C. is a Research Scholar from the Fonds de la Recherche en Santé du Québec and is a member of the Sherbrooke RNA/RNP group supported by the Fonds pour la Formation des Chercheurs et l’Aide à la Recherche.
References
- 1.Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J., Hidaka,K. and Futcher,B. (2000) The est1 subunit of yeast telomerase binds the tlc1 telomerase RNA. Mol. Cell. Biol., 20, 1947–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virta-Pearlman V., Morris,D.K. and Lundblad,V. (1996) Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev., 10, 3094–3104. [DOI] [PubMed] [Google Scholar]
- 4.Evans S.K. and Lundblad,V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- 5.Harrington L., Zhou,W., McPhail,T., Oulton,R., Yeung,D.S., Mar,V., Bass,M.B. and Robinson,M.O. (1997) Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev., 11, 3109–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 7.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q., Bacchetti,S., Haber,D.A. and Weinberg,R.A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg R.A., Allsopp,R.C., Chin,L., Morin,G.B. and DePinho,R.A. (1998) Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene, 16, 1723–1730. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Rivera L., Herrera,E., Albar,J.P. and Blasco,M.A. (1998) Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl Acad. Sci. USA, 95, 10471–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington L., McPhail,T., Mar,V., Zhou,W., Oulton,R., Bass,M.B., Arruda,I. and Robinson,M.O. (1997) A mammalian telomerase-associated protein. Science, 275, 973–977. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama J., Saito,M., Nakamura,H., Matsuura,A. and Ishikawa,F. (1997) TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell, 88, 875–884. [DOI] [PubMed] [Google Scholar]
- 13.Le S., Sternglanz,R. and Greider,C.W. (2000) Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell., 11, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt S.E., Aisner,D.L., Baur,J., Tesmer,V.M., Dy,M., Ouellette,M., Trager,J.B., Morin,G.B., Toft,D.O., Shay,J.W., Wright,W.E. and White,M.A. (1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev., 13, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford L.P., Suh,J.M., Wright,W.E. and Shay,J.W. (2000) Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol., 20, 9084–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- 17.Mayeda A., Munroe,S.H., Cáceres,J.F. and Krainer,A.R. (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J., 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X., Bani,M.R., Lu,S.J., Rowan,S., Ben-David,Y. and Chabot,B. (1994) The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl Acad. Sci. USA, 91, 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchette M. and Chabot,B. (1999) Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J., 18, 1939–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izaurralde E., Jarmolowski,A., Beisel,C., Mattaj,I.W., Dreyfuss,G. and Fischer,U. (1997) A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J. Cell Biol., 137, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBranche H., Dupuis,S., Ben-David,Y., Bani,M.R., Wellinger,R.J. and Chabot,B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet., 19, 199–202. [DOI] [PubMed] [Google Scholar]
- 22.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 23.McElligott R. and Wellinger,R.J. (1997) The terminal DNA structure of mammalian chromosomes. EMBO J., 16, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa F., Matunis,M.J., Dreyfuss,G. and Cech,T.R. (1993) Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell Biol., 13, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallaire F., Dupuis,S., Fiset,S. and Chabot,B. (2000) Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J. Biol. Chem., 275, 14509–14516. [DOI] [PubMed] [Google Scholar]
- 27.Ding J., Hayashi,M.K., Zhang,Y., Manche,L., Krainer,A.R. and Xu,R.M. (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabot B. (1994) In Higgins,S.J. a.H.B.D. (eds), RNA Processing, A Practical Approach. IRL Press, Oxford, Vol. 1, pp. 1–29.
- 29.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idriss H., Kumar,A., Casas-Finet,J.R., Guo,H., Damuni,Z. and Wilson,S.H. (1994) Regulation of in vitro nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein protein A1 by reversible phosphorylation. Biochemistry, 33, 11382–11390. [DOI] [PubMed] [Google Scholar]
- 31.Morin G.B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- 32.Prowse K.R., Avilion,A.A. and Greider,C.W. (1993) Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl Acad. Sci. USA, 90, 1493–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Autexier C., Pruzan,R., Funk,W.D. and Greider,C.W. (1996) Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J., 15, 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J.L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 35.Cobianchi F., Calvio,C., Stoppini,M., Buvoli,M. and Riva,S. (1993) Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res., 21, 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Municio M.M., Lozano,J., Sanchez,P., Moscat,J. and Diaz-Meco,M.T. (1995) Identification of heterogeneous ribonucleoprotein A1 as a novel substrate for protein kinase C ζ. J. Biol. Chem., 270, 15884–15891. [DOI] [PubMed] [Google Scholar]