Abstract
Since pork accounts for about 40% of global meat consumption, the pig is an important economic animal for meat production. Pig is also a useful medical model for humans due to its similarity in size and physiology. Understanding the mechanism of muscle development has great implication for animal breeding and human health. Previous studies showed porcine muscle satellite cells (PSCs) are important for postnatal skeletal muscle growth, and Notch1 signaling pathway and miRNAs regulate the skeletal muscle development. Notch1 signal pathway regulates the transcription of certain types of miRNAs which further affects target gene expression. However, the specific relationship between Notch1 and miRNAs during muscle development has not been established. We found miR-34c is decreased in PSCs overexpressed N1ICD. Through the overexpression and inhibition of mi-34c, we demonstrated that miR-34c inhibits PSCs proliferation and promotes PSCs differentiation. Using dual-luciferase reporter assay and Chromatin immunoprecipitation, we demonstrate there is a reciprocal regulatory loop between Notch1 and miR-34c. Furthermore, injection of miR-34c lentivirus into mice caused repression of gastrocnemius muscle development. In summary, our data revealed that miR-34c can form a regulatory loop with Notch1 to repress muscle development, and this result expands our understanding of muscle development mechanism.
Introduction
Muscle satellite cells, also known as muscle stem cells, reside as quiescent cells beneath the basal lamina that surrounds muscle fibers, and function as myogenic precursors for muscle growth and repair1, 2. Quiescent muscle satellite cells are PAX7 +/MYF5 −, when exposed to stress or damage, they can respond and be activated3, 4. Activated muscle satellite cells are PAX7 +/MYF5 +, and after several rounds of replication, they will fuse to the nearby myofibers5. During these processes, the Notch1 signaling pathway is known to play a critical role6.
Notch1 is a receptor that mediates intercellular signaling through a pathway conserved across the metazoan7. After two cleavages, the intracellular domain of Notch1 (N1ICD) is released and translocated into the nucleus, where it interacts with the CSL (CBF-1, Suppressor of hairless, Lag) family and forms part of a CSL-N1ICD complex on the DNA binding site (GTGGGAA) to regulate genomic DNA transcription8, 9. Notch1 signaling is known to inhibit myogenic differentiation10 and has been proposed to promote the expansion of undifferentiated progenitors indirectly11. Rando et al. found that activation of Notch1 signaling stimulates the proliferation of satellite cells and leads to the expansion of proliferating myoblasts. And, inhibition of Notch1 signaling abolishes satellite cell activation and impairs muscle regeneration11. Also, recent studies found Notch1 is active in quiescent muscle satellite cells, and Notch1 signaling is critical for maintaining the quiescence of muscle satellite cells12, 13.
MicroRNA (miRNA) is a small non-coding RNA molecule (containing ~22 nucleotides) found in plants, animals, and some viruses14. MiRNA is encoded by nuclear DNA and usually transcribed by RNA polymerase II15, 16. As many as 40% of miRNA lie in the introns or even exons of other genes. Thus they usually are regulated together with their host genes. Other miRNAs are initially transcribed using their own promoter16. MiRNA exert their functions in RNA silencing by binding to complementary sequences within its target RNA17, 18. For example, in mice, miR-1 and miR-133 are clustered on the same chromosomal loci and transcribed together in a tissue-specific manner during development, but miR-133 enhances proliferation by repressing serum response factor, whereas miR-1 promotes myogenesis through repressing histone deacetylase 419.
The miR-34 family members (miR-34a, miR-34b, and miR-34c) were discovered computationally20 and later verified by experiment21, 22. In human, three miR-34 precursors are produced from two transcriptional units, miR-34a precursor is transcribed from chromosome 1, and miR-34b and miR-34c precursors are co-transcribed from a region on chromosome 1123. But only miR-34a and miR-34c are found in pig24, 25. A previous study indicated miR-34a inhibits mouse smooth muscle cell proliferation by directly targeting Notch1 26. Another miR-34 family member miR-34c also has been shown to inhibit rat vascular smooth muscle cell proliferation27. But, the role of miR-34 plays in pig skeletal muscle development has not been reported.
Our laboratory has demonstrated that Notch1 promotes PSCs proliferation28. We have also constructed an N1ICD overexpressing PSCs model for miRNA sequencing to examine satellite cell developmental mechanisms. From the RNA-seq data, we discovered that many miRNAs were differently expressed in the N1ICD overexpressing PSCs, including miR-34c29. However, there is no report regarding the role of miR-34c on PSCs development; therefore, the objective of this study was to define the role of miR-34c on PSCs development and ascertain whether there is a regulatory relationship between miR-34c and N1ICD.
In this study, we used miR-34c mimics and inhibitor to manipulate the miR-34c level in transfected PSCs and that elevated miR-34c not only inhibits PSCs proliferation but also promotes PSCs differentiation. Using dual-luciferase reporter assay and Chromatin immunoprecipitation (ChIP), we demonstrated there is a regulatory loop between Notch1 and miR-34c. Finally, through miR-34c lentivirus injection into mice gastrocnemius muscle, we confirmed miR-34c represses mice muscle development. Our results not only established Notch1 is the target gene of miR-34c but also discovered that Notch1, in turn, directly regulates miR-34c in PSCs. This reciprocal regulatory loop formed by miR-34c and Notch1 that controls skeletal muscle development is novel, and this information expands our understanding of the mechanisms involved in muscle development.
Results
Overexpressing N1ICD decreases miR-34c expression in vitro
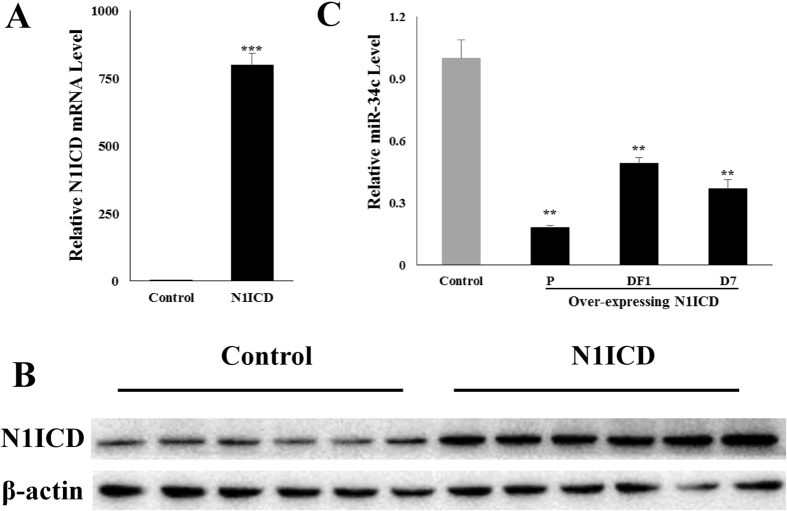
To explore the regulations of Notch1 signaling in PSCs development, we constructed the constitutively activated N1ICD PSCs. Both the N1ICD mRNA and protein levels were increased (p < 0.001); Figs 1A and B) in the N1ICD overexpressed PSCs. The PSCs cell samples were collected on proliferation day 1 (P), differentiation day 1 (DF1) and differentiation day 7 (DF7). The expression of miR-34c was decreased in the N1ICD overexpressed cells (p < 0.01; Fig. 1C). These results demonstrate we have successfully overexpressed N1ICD in PSCs and N1ICD decreased miR-34c expression in all three periods of PSCs development.
Figure 1.
Overexpressing N1ICD decreases miR-34c expression during PSCs development. (A) PSCs treated with either pEGFP-N1ICD (N1ICD) or pEGFP-N1 (control). N1ICD mRNA level was increased in N1ICD overexpressing PSC on proliferation day 1 (P). (B) N1ICD protein level was increased in N1ICD overexpressing PSC on proliferation day 1. (C) qRT-PCR analysis of miR-34c on proliferation day 1 (P), differentiation day 1 (DF1) and differentiation day 7 (DF7), during the development of N1ICD-overexpressed PSCs. *Indicates a difference between treatment and control. **p < 0.01; ***p < 0.001. The results are presented as Mean ± S.E.M. of three replicates for each group. The full-length blot images are presented in Supplementary Figure 1.
Overexpressing miR-34c inhibits PSCs proliferation in vitro
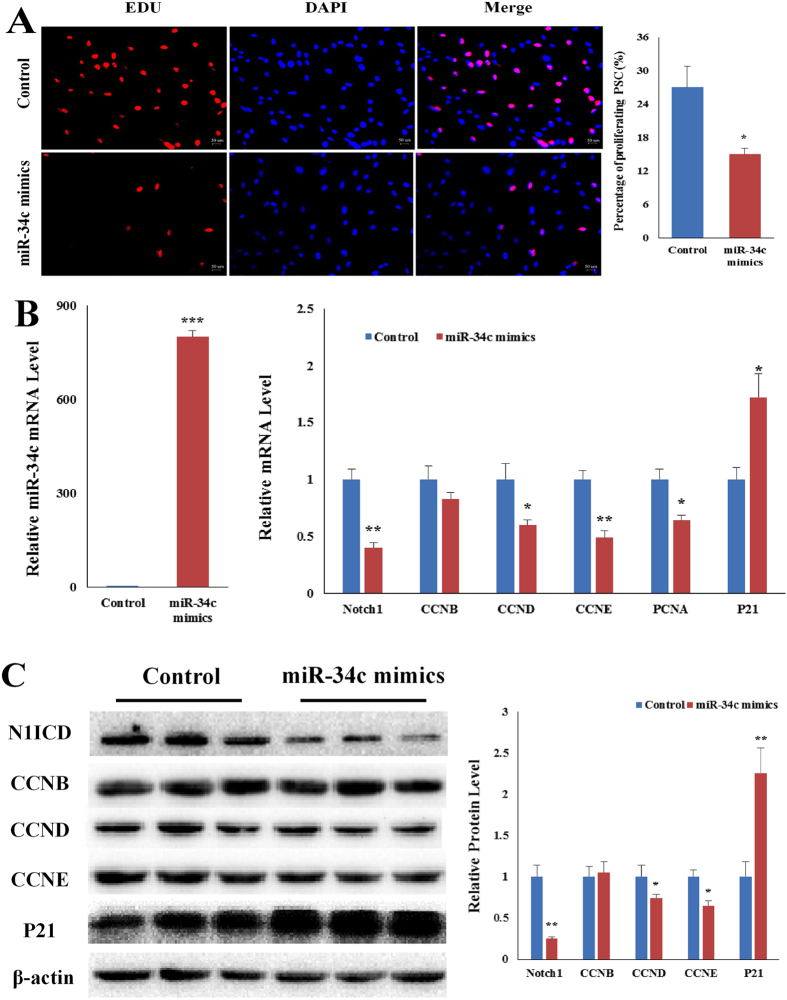
First, we investigated the role of miR-34 in PSCs proliferation. MiR-34c mimics was transfected into PSCs. Overexpressing miR-34c reduced the percentage of Edu positive cells (p < 0.05, Fig. 2A). qRT-PCR result showed miR-34c mimics increased (p < 0.001) cellular miR-34c level at 24 h after transfection (Fig. 2B). The miR-34c mimics decreased Notch1, CCND, CCNE and PCNA mRNA levels and increased p21 mRNA level (p < 0.05, Fig. 2B). Western blot confirmed that miR-34c mimics reduced (p < 0.05) Notch1, CCND and CCNE protein level, and increased (p < 0.01) p21 protein level (Fig. 2C). However, miR-34c mimics had no effect on CCNB mRNA and protein levels (Fig. 2B and C).
Figure 2.
Overexpressing miR-34c mimics inhibits PSCs proliferation. (A) Overexpressing miR-34c in PSCs lowed PSCs proliferation. Representative images of the immunofluorescent staining for proliferating PSCs are shown. Proliferating PSCs were labeled with Edu fluorescent dye (red). (B) qRT-PCR confirmed miR-34c is negatively correlated with proliferation-related genes in PSCs transfected with miR-34c mimics. (C) Western blot result showed the protein level is corresponding to the qRT-PCR result. *p < 0.05; **p < 0.01. The results are presented as Mean ± S.E.M. of three replicates for each group. Magnification 200×. The scale bar on the photomicrographs represents 50 μm. The full-length blot images are presented in Supplementary Figure 2.
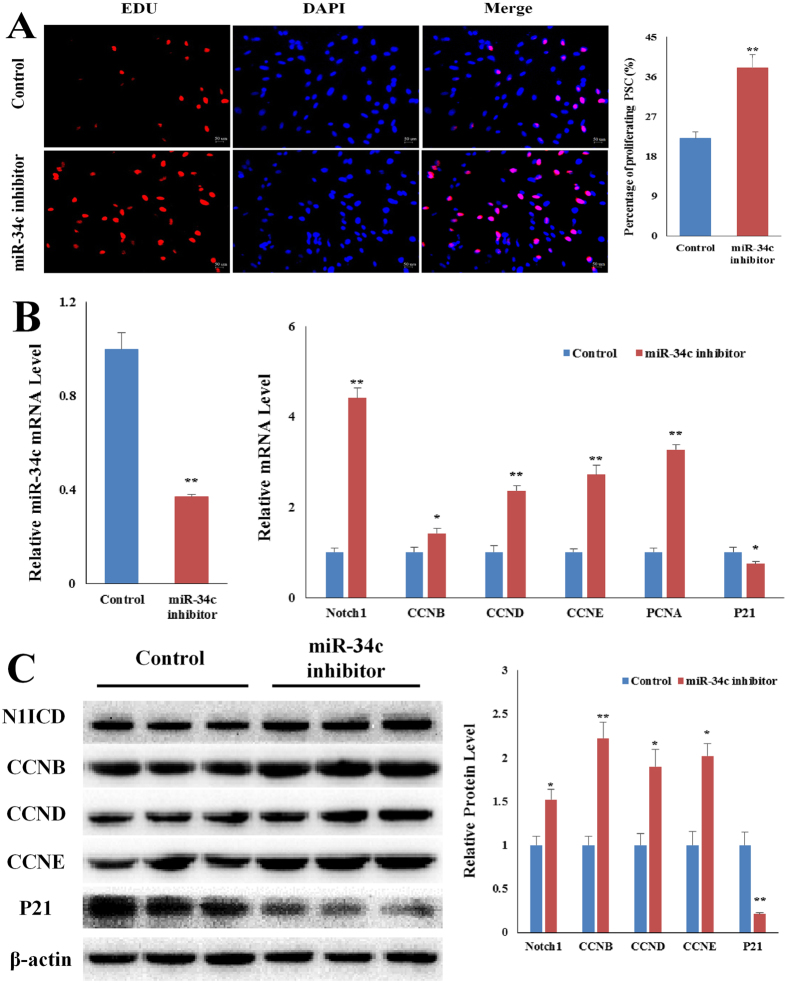
Next, we used miR-34c inhibitor to further ascertain the role of miR-34c on PSCs proliferation. MiR-34c inhibitor increased the percentage of Edu positive cells (p < 0.01, Fig. 3A). MiR-34c inhibitor reduced the endogenous miR-34c level (Fig. 3B). After transfected with miR-34c inhibitor, the mRNA level of Notch1, CCNB, CCND and CCNE and PCNA were increased and p21 was decreased (p < 0.05, Fig. 3B). And the western blot result confirmed the qRT-PCR result, the protein level of Notch1, CCNB, CCND and CCNE were increased and p21 was decreased (p < 0.05, Fig. 3C). Taken together, these results demonstrate miR-34c inhibits PSCs proliferation in vitro.
Figure 3.
Overexpressing miR-34c inhibitor increase PSCs proliferation. (A) miR-34c inhibitor increased PSCs proliferation. Representative images of immunofluorescent staining of PSCs transfected with miR-34c inhibitor. (B) PSCs treated with either miR-34c inhibitor or Control confirmed miR-34c is negatively correlated with proliferation-related genes. (C) Western blot result showed protein level is corresponding to the qRT-PCR result. *p < 0.05; **p < 0.01. The results are presented as Mean ± S.E.M. of three replicates for each group. Magnification 200×. The scale bar on the photomicrographs represents 50 μm. The full-length blot images are presented in Supplementary Figure 2.
Overexpressing miR-34c promotes PSCs differentiation
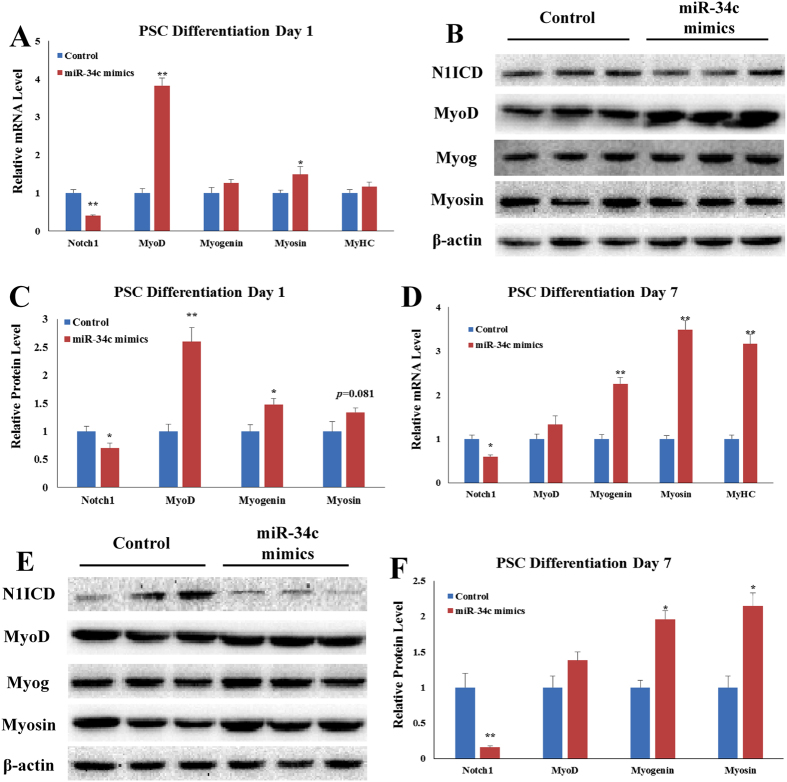
The medium was changed to differentiation medium for 1 and 7 days to study the function of miR-34c on PSCs differentiation. The expression of Notch1, MyoD, myogenin, myosin, and MyHC were measured. Overexpression of miR-34c reduced Notch1 mRNA, and elevated MyoD mRNA and myosin mRNA on differentiation day 1 (p < 0.05, Fig. 4A). As for protein level of these genes, western blot result showed overexpression of miR-34c reduced Notch1, and elevated MyoD and myogenin on differentiation day 1 (p < 0.05, Fig. 4B). However, on differentiation day 7, overexpression of miR-34c showed no significant influence on MyoD mRNA and protein levels (Fig. 4C and D). The mRNA level of myogenin, myosin, and MyHC was elevated by the miR-34c overexpression on differentiation day 7 (p < 0.01, Fig. 4C), and protein level of myogenin and myosin follows the same pattern as the mRNA (p < 0.05, Fig. 4D).
Figure 4.
Overexpressing miR-34c mimics promotes PSCs differentiation. We measured the expression of Notch1 and the myogenic marker genes after PSCs were transfected with miR-34c mimics. (A) qRT-PCR confirmed miR-34c increased mRNA level of myogenic marker genes on day 1 after differentiation induction. (B) We used western blot to measure protein level of Notch1 and myogenic marker genes on day 1 after differentiation induction. (C) Western blot showed the protein level result is corresponding to the qRT-PCR result on day 1 after differentiation induction. (D) qRT-PCR confirmed miR-34c increased mRNA level of myogenic marker genes on day 7 after differentiation induction. (E) We used western blot to measure protein level of Notch1 and myogenic marker genes on day 7 after differentiation induction. (F) Western blot showed the protein level result is corresponding to the qRT-PCR result on day 7 after differentiation induction. The results are presented as Mean ± S.E.M. of three replicates for each group. *p < 0.05; **p < 0.01. *Indicates a difference between Control and miR-34c mimics. The full-length blot images are presented in Supplementary Figure 3.
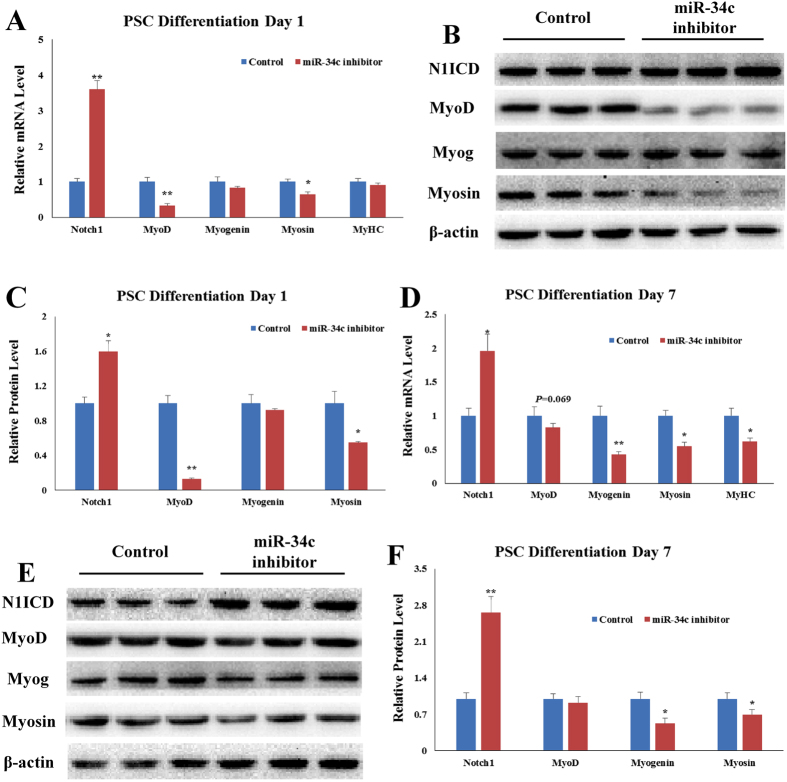
Next, miR-34c inhibitor or Control (miR-34c inhibitor and Control are both synthetic oligonucleotide sequences; see Table S1 for details) was transfected into PSCs, and the cells were induced to differentiation for 1 day and 7 days. PSCs transfected with miR-34c inhibitor increased the mRNA and protein levels of N1ICD on both differentiation day 1 and day 7 (Fig. 5). MiR-34c inhibitor decreased MyoD and myosin mRNA level on differentiation day 1 (p < 0.05, Fig. 5A). But miR-34c inhibitor had no effect (p > 0.05) on myogenin and MyHC mRNA level on differentiation day 1 (Fig. 5A). On differentiation day 1, western blot result shown miR-34c inhibitor decreased (p < 0.05) MyoD and myosin protein levels, but had no effect on myogenin (Fig. 5B). MiR-34c inhibitor decreased (p < 0.05) myogenin, myosin and MyHC genes mRNA level on differentiation day 7 (Fig. 5D). On differentiation day 7, western blot result shown miR-34c inhibitor decreased (p < 0.05) myogenin and myosin protein levels. But miR-34c inhibitor had no effect on Myod gene expression on differentiation day 7 (Fig. 5D and E). These results indicate elevated miR-34c promotes PSCs differentiation.
Figure 5.
Overexpressing miR-34c inhibitor reduces PSCs differentiation. We measured the expression of Notch1 and the myogenic marker genes after PSCs were transfected with miR-34c inhibitor. (A) qRT-PCR confirmed miR-34c inhibitor decreased mRNA level of myogenic marker genes on day 1 after differentiation induction. (B) We used western blot to measure protein level of Notch1 and myogenic marker genes on day 1 after differentiation induction. (C) Western blot showed the protein level result is corresponding to the qRT-PCR result on day 1 after differentiation induction. (D) qRT-PCR confirmed miR-34c inhibitor decreased mRNA level of myogenic marker genes on day 7 after differentiation induction. (E) We used western blot to measure protein level of Notch1 and myogenic marker genes on day 7 after differentiation induction. (F) Western blot showed the protein level result is corresponding to the qRT-PCR result on day 7 after differentiation induction. The results are presented as Mean ± S.E.M. of three replicates for each group. *p < 0.05; **p < 0.01. *Indicates a difference between Control and miR-34c mimics. MiR-34c mimics and Control are both synthetic oligonucleotide sequences; see Table S1 for details. The full-length blot images are presented in Supplementary Figure 3.
Reciprocal regulation between Notch1 and miR-34c
As overexpressing N1ICD decreases miR-34c expression during PSCs development (Fig. 1) and miR-34c mimics decreases Notch1 gene expression in the proliferation period (Fig. 2), differentiation day 1and differentiation day 7 (Fig. 4), these results suggest a regulatory feedback may exist between Notch1 and miR-34c.
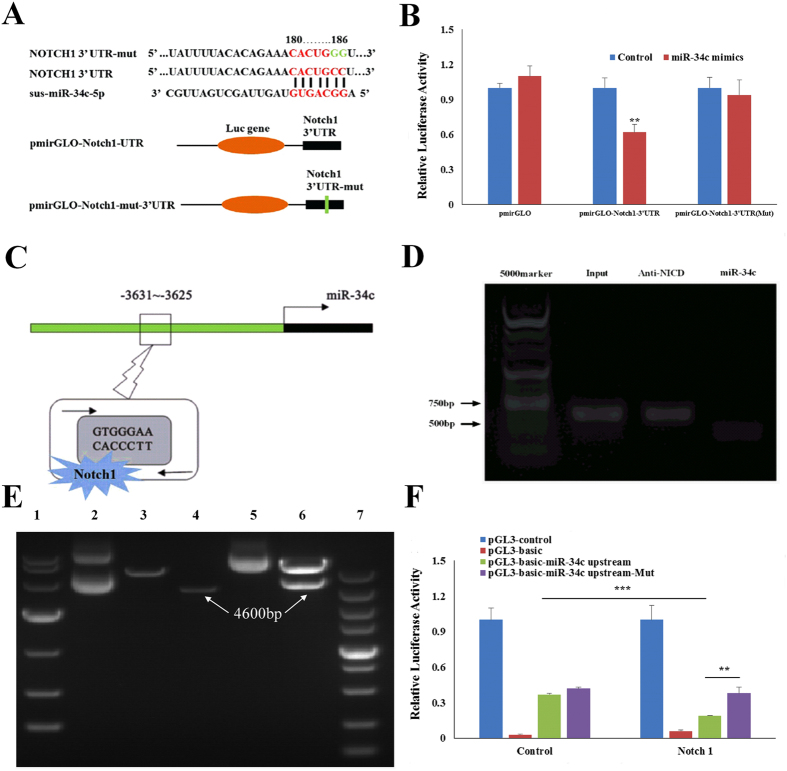
Since N1ICD expression was decreased after PSCs transfected with miR-34c mimics (Figs 2 C and 4B), Notch1 may be a potential target gene of miR-34c. A dual-luciferase reporter assay was used to identify the miR-34c binding sites on Notch1-3′UTR (Fig. 6A). We found relative luciferase activity was decreased (p < 0.01; Fig. 6B) when HEK-293T cells were co-transfected with miR-34c mimics and pmirGLO-Notch1-3′UTR. However, mutated pmirGLO-Notch1-3′UTR did not change relative luciferase activity (Fig. 6B). These results suggest that Notch1 is the direct target gene of miR-34c.
Figure 6.
Negative feedback between miR-34c and Notch1. (A) Schematic representation of the sequence of Notch1 3′UTR, Notch1 3′UTR-mut, miR-34c, and luciferase reporter constructs. (B) Luciferase reporters shown in (A) were transfected into HEK-293T cells with either miR-34c mimics or Control respectively. Luciferase activity was determined 24 h after transfection. (C) Schematic representation of Notch1 binds at the upstream of miR-34c in the genome. (D) PSCs transfected with pEGFP-N1ICD and were cultured in growth medium for 24 h. These PSCs were immune precipitated with the Notch1 antibody. The precipitated DNA was used to amplify the fragments of -3631 upstream of miR-34c gene by primers (arrows). (E) Cloned miR-34 upstream of its genomic site (about 4600 bp, the fourth lane), and constructed pGL3-basic-miR-34 upstream recombinant vector (the fifth and sixth lanes). Lanes 1–7 represent DL 10000, pGL3-basic, double digested pGL3-basic, double digested miR-34 upstream of its genomic site, pGL3-basic-miR-34 upstream recombinant vector, double digested pGL3-basic-miR-34 upstream recombinant vector, DL5000, respectively. (F) N1ICD decreased the pGL3-basic-miR-34 upstream recombinant vector relative luciferase activity. N1ICD with pGL3-basic-miR-34 upstream recombinant vector relative or pGL3-basic-miR-34 upstream (mut) recombinant vector were transfected into HEK-293T cells respectively. Luciferase activity was determined 24 h after transfection. *p < 0.05; **p < 0.01; ***p < 0.001; Student’s t-test. The results are presented as Mean ± S.E.M. of three replicates for each group.
As the miR-34c level is reduced when N1ICD was overexpressed during PSCs development (Fig. 1A) and a CSL-N1ICD complex binding site (GTGGGAA) exists at upstream of the miR-34c genomic site (Fig. 6C), we measured the DNA fragment binding with CSL-N1ICD complex by ChIP. A fragment about 400 bp long located at -3631 upstream of miR-34c was amplified (Fig. 6D). The CSL-N1ICD complex binds directly to the -3631~-3625 sites of miR-34c. This result indicates CSL-N1ICD complex may regulate miR-34c transcription. So we constructed the pGL3-basic-miR-34 upstream recombinant vector (pGL3-basic-miR-34 upstream). As shown in Fig. 6E the miR-34 upstream of its genomic site (about 4600 bp) is inserted into the pGL3-basic vector. Through the dual-luciferase reporter assay, we found N1ICD decreased the pGL3-basic-miR-34 upstream recombinant vector relative luciferase activity, but this inhibition was abolished by the mutated CSL-N1ICD complex binding site (GTGGGAA) (Fig. 6F). These findings establish there exists a regulatory loop between Notch1 and miR-34c.
MiR-34c represses muscle development in vivo
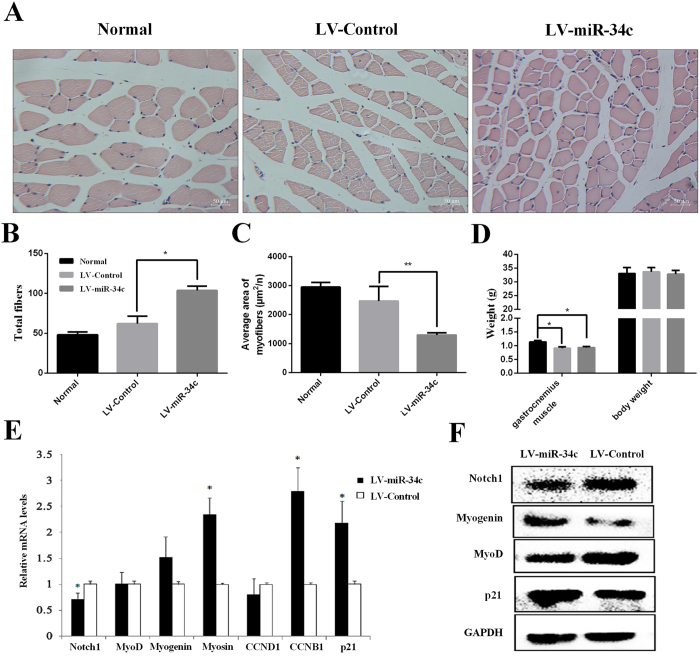
To evaluate the function of miR-34c in vivo, we injected lentivirus expressing miR-34c mimics or Control into mice gastrocnemius muscle. Both miR-34c mimics and Control are synthetic oligonucleotide sequences delivered by the lentiviral vector. Body weight and characteristics of gastrocnemius muscle were recorded, and total RNA and protein were extracted a week later. Total myofibers in a single field were increased (p < 0.01, Fig. 7A,B) in the LV-miR-34c injected group compared with the Normal group and LV-Control (p < 0.05, Fig. 7A,B). Consistent with this result is the average area of myofibers decreased (p < 0.01, Fig. 7C), which means miR-34c repressed muscle development in vivo. Body weights were no difference among all groups, but the weights of gastrocnemius muscle were decreased in the LV-miR-34c treatment group and Normal groups (p < 0.05, Fig. 7D). After injecting LV-miR-34c, MyoD protein level was decreased (Fig. 7F) and myogenin protein level was increased (Fig. 7F), but their mRNA levels were not different (Fig. 7E). LV-miR-34c injection also increased myosin mRNA level (p < 0.05, Fig. 7E). CCND1 mRNA level was decreased after LV-miR-34c injection, but mRNA levels of CCNB1 and p21 were increased (p < 0.05, Fig. 7E). Meanwhile, p21 protein level was also increased (Fig. 7F). Notch1 mRNA level was decreased in the LV-miR-34c injection group (p < 0.05, Fig. 7F). Notch1 protein level was also decreased (Fig. 7F). These results demonstrate that injecting LV-miR-34c miR-34c represses muscle development in vivo.
Figure 7.
MiR-34c injection represses muscle development. (A) H&E stained cross-section of the gastrocnemius muscle (GM) from Normal (physiological saline), LV-Control and LV-miR-34c. MiR-34c or microRNA-control was delivered by a lentiviral vector (LV). (B). Total fibers in one field were increased after injection with LV-miR-34c. (C) The average area of muscle fiber cross-sections was decreased after injection with LV-miR-34c. (D) Body weight and gastrocnemius muscle weight seven days after LV-Control or LV-miR-34c injection, respectively. (E) qRT-PCR analysis of Notch1, myogenic markers, cell cycle proteins, and p21 in the gastrocnemius muscle after LV-Control or LV-miR-34c injection for 7 days. (F) Western blots of proteins isolated from gastrocnemius muscles, which were injected either LV-Control or LV-miR-34c after day 7, respectively. The results are presented as Mean ± S.E.M. of three replicates for each group. *p < 0.05, **p < 0.01; *Indicates differences between LV-Control and LV-miR-34c in mice gastrocnemius muscle except the Fig. 6D. In Fig. 6D *Indicates differences between Normal and LV-Control or Normal and LV-miR-34c in mice gastrocnemius muscle. Magnification 200×. The scale bar on the photomicrographs represents 50 μm.
Discussion
Skeletal muscle is the major component of lean body mass in livestock. Postnatal muscle growth mainly depends on the hypertrophy of existing muscle fibers, and muscle satellite cells have been shown to be responsible for this process30. The development of muscle satellite cells is affected by many factors. The microenvironment or niche where muscle satellite cells locate is relative stable. The niche contains extracellular matrix proteins, vasculature system, muscle residence cells and muscle fibers31. However, the homeostasis of the niche is broken under stress or injury. Heat stress is well established to affect porcine growth rate negatively. Heat stress produces cellular stress and changes the rates of PSCs proliferation, protein turnover and abundance of heat shock proteins (Hsp) mRNA and protein in PSCs cultures32. Furthermore, the activity of PSCs changes with age. Mesires et al.33 isolated PSCs from the hind limb muscles of pigs at 1, 7, 14, and 21 weeks of age. The number of PSCs in these muscles decreased from 30% at one week to 14% at seven weeks of age and remained at a constant level after week 14. That may reflect a decrease in the number of differentiating PSCs or accelerated fusion into myofibers33.
Notch signaling has been shown to stimulate the proliferation and inhibit the differentiation of muscle satellite cells34. Wen et al.6 used mice skeletal muscle satellite cells as a model and constructed a myogenic cell lineage in which N1ICD was constitutively activated. The CSL-N1ICD complex binds to the DNA binding site (GTGGGAA) upstream of PAX7 and promotes the “self-renewal” of muscle satellite cells6. Consequently, we would like to know whether CSL-N1ICD complex binds to the binding site upstream of any miRNAs to affect its transcription. The miRNAs are critical regulators of many biological processes by modulating expression of genes at the post-transcriptional level. Proliferation and differentiation are mutually exclusive during myogenesis, and miRNAs are critically involved in balancing these two processes35. Many miRNAs, such as miR-1, miR-133, miR-29, miR-214, miR-206, miR-486, miR-208b, and miR-499 were involved in the regulation of skeletal myogenesis by binding to its target genes36, 37. To establish N1ICD regulates miR-34c expression, N1ICD was transfected into PSCs in primary culture. N1ICD was constitutively activated in these transfected PSCs. These PSCs were used to conduct our in vitro studies. Using ChIP assay, we showed that CSL-N1ICD complex binds directly to the −3631~−3625 sites of miR-34c. We confirmed the N1ICD reduces the transcriptional activity of the miR-34c promoter through dual-luciferase reporter assay. We have also established Notch1 is a direct target gene of miR-34c using a dual-luciferase reporter assay. These results demonstrate there exists a regulatory loop between Notch1 and miR-34c in PSCs development.
Postnatal muscle development involves muscle satellite cells activation, re-entering the cell cycle for replication, and subsequently differentiation into myoblasts38, 39. MiR-34c has been shown to be a new modulator of VSMC proliferation through targeting SCF 27. But, smooth and skeletal muscle tissues are composed of distinct cell types40. These two muscle cell types are also believed to have distinct embryonic origins. Smooth and skeletal muscle cells express distinct isoforms of the structural genes used for contraction41. To our knowledge, there is no report about the miR-34c function on skeletal muscle development. Our study ascertains that miR-34c inhibits PSCs proliferation by inhibiting Notch1 expression. PSCs proliferation is dependent on an active cell cycle. In the cell cycle, CCND is the first cyclin produced; thus, we measured CCND expression as the PSCs proliferation marker. CCND binds to existing CDK4, forming the active CCND-CDK4 complex. Then, CCND-CDK4, in turn, phosphorylates the retinoblastoma susceptibility protein (Rb). Rb dissociates from the E2F/DP1/Rb complex, activating E2F. E2F promotes various genes such as CCNE and DNA polymerase transcription. Therefore, we also measured CCNE expression as the PSCs proliferation marker. CCNE binds to CDK2, forming the CCNE-CDK2 complex, which pushes the cell from G1 to S phase. CCNB-CDK1 complex activation causes breakdown and prophase initiation, subsequently, CCNB-CDK1 deactivation causes the cell to exit mitosis42–44. The p21 protein is a cell cycle regulator, and it binds to cyclin-CDKs complexes and inhibits their activities45, 46. Thus, CCNB and P21 were also used as the PSCs proliferation markers in our study. In our study, we used Edu assay to confirm miR-34c inhibits PSCs proliferation, and our qRT-PCR and western blot results show miR-34c is positively correlated with p21, and negatively correlated with CCNB, CCND, and CCNE.
For PSCs differentiation, MyoD is an early myogenic marker47, and myogenin and MyHC are terminal differentiation markers48, 49. Thus, in our study, MyoD and myogenin/MyHC showed different expression patterns on differentiation day 1 and day 7.
The in vitro study is commonly used as an experimental model for the animals. However, results obtained using in vitro studies need to be verified by the in vivo studies to confirm the validity of the in vitro data. Thus, to ascertain the miR-34c function observed in our cell culture studies, we conducted the in vivo study in mice. We injected miR-34c into the gastrocnemius muscle of the mice to establish the changes in muscle growth and gene expressions. Since the mice used in this study were 4-week-old, they were in rapid growing stage. Thus the presence of virus affected muscle development, which explains the gastrocnemius muscle weight of the injected virus groups (LV-Control and LV-miR-34c) was lower than that of the Normal group. After birth, the skeletal muscle myofiber number remains constant, and muscle growth depends on the satellite cell proliferation and fusion to the existing myofibers50–52. But miR-34c inhibited the skeletal muscle satellite cell proliferation by targeting Notch1, and miR-34c reduced the number of satellite cells to be fused to existing myofibers, resulting in smaller muscle fiber diameter after miR-34c injection.
This study would be better to have the miR-34c inhibitor group in the in vivo study. However, after considerable deliberations, we decided to forgo the miR-34c inhibitor group for the following reason: the mice used in this study are 4-week-old. Mice at this age most likely are in the rapid muscle-growing stage. Therefore, injection of LV-miR-34c inhibitor may not obtain additional muscle growth; thus, the value of using more mice may be questionable following the principle of a minimal number of animals to be used in any experiments. Also, the experimental cost was considerably lower with fewer animals used. Since our in vitro study has shown that overexpressing miR-34 inhibits muscle development, we believe the miR-34c overexpression experiment alone would be sufficient to demonstrate the role of miR-34c in vivo. Furthermore, the objective of this particular experiment was to ascertain whether the miR-34c inhibits muscle development in vivo. The postnatal muscle development involves muscle satellite cells activation, re-entering the cell cycle for replication, and subsequent differentiation into myoblasts. In this study, the gastrocnemius muscles were dissected one week after injection; the activated PSCs would have already differentiated into myoblasts. Thus, muscle fiber diameter is the most intuitive phenotypic data for muscle development. Therefore, we did not conduct additional immunostaining of any myogenic markers in the mice gastrocnemius muscle.
In conclusion, our research demonstrates that miR-34c inhibits PSCs proliferation but promotes PSCs differentiation in vitro, and miR-34c represses pig muscle development in vivo. These results expand our understanding of muscle development mechanism in which miRNAs and genes participate in a regulatory loop that controls skeletal muscle development. Furthermore, this finding has implications for both human health and animal breeding.
Materials
All animals used in this study were with the approval of the College of Animal Science, South China Agricultural University. All experiments were conducted following ‘the instructive notions with respect to caring for laboratory animals’ issued by the Ministry of Science and Technology of the People’s Republic of China.
Isolation and culture of primary porcine satellite cells (PSCs)
Three one-day old male Landrace piglets (approximately 1.5 kg) were anesthetized using sodium pentobarbital, and muscles from legs were used for PSCs isolation. The PSCs isolation method used has been described previously28, 53. When the PSCs reached approximately 80–90% confluent, cells were allowed to differentiate in DMEM/F12 with 2% horse serum, 100 U/mL penicillin, and 100 μg/ml streptomycin (GIBCO BRL) for 1 day or 7 days. After differentiation, the cells were washed twice with cold phosphate-buffered saline (PBS) before protein extraction or total RNA extraction.
PSCs transfection
PSCs were seeded in 6-well plates (2 × 105 cells per well). PSCs transfected with pCDNA3.1-N1ICD, miR-34c inhibitor, miR-34c mimics or Control by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. After 6 hours, the medium was replaced with new growth medium and cells were maintained in growth medium for an additional 12 h before myogenic differentiation induction. MiR-34c mimics and inhibitor were (see Table S1) purchased from GENEWIZ (Suzhou, China).
RNA extraction and PCR analysis
Total RNAs were extracted from muscles or porcine satellite cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After digesting with DNase I (Takara Bio Inc., Japan), total RNAs (0.5 μg) were reverse transcribed to cDNA using PrimeScriptTM RT Master Mix (TaKaRa, Otsu, Shiga, Japan) according to the manufacturer’s instructions. SYBR Green Real-time PCR Master Mix reagents (Toyobo Co., Ltd., Osaka, Japan) were used for real-time quantitative polymerase chain reaction (PCR), and PCR reactions carried out on a CFX96™ Optical Reaction Module (BIO-RAD, USA). The relative expression of mRNAs and microRNA were normalized with GAPDH or U6 levels using the ΔΔCt method54. ΔΔCt is defined as the ratio of the relative mRNA level of the target gene between the experimental group and the control group. Primers were designed using Primer Premier 5 according to the pig genes sequence obtained from NCBI. Primers used for PCR are shown in Table S1.
Edu labeling
PSCs transfected with miR-34c mimics, miR-34c inhibitor or Control and maintained in growth medium. Twenty-four hours later, these PSCs were used for Edu labeling by Cell-Light™ Edu Apollo®488 In Vitro Imaging Kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. The Edu labeled PSCs were washed with PBS, then observed and recorded using a Nikon TE2000-U inverted microscope (Nikon Instruments, Tokyo, Japan). The Edu positive PSCs were counted using Image Pro Plus (Media Cybernetics, Inc., Silver Spring, MD, USA).
Western blot analysis
PSCs and mice gastrocnemius muscle were lysed in RIPA buffer containing 1 μM PMSF. About 20 μg protein lysates were separated with SDS-PAGE and then electroblotted onto polyvinylidene fluoride membranes (BIO-RAD, USA). The membranes were blocked at room temperature for 2 hours and incubated with different diluted antibodies at 4 °C overnight. Finally, the polyvinylidene fluoride membranes were incubated with horseradish peroxidase-conjugated secondary antibodies at room temperature for 40 minutes. The antibodies used in this study are listed in Table S2.
Luciferase Reporter Assay
Notch1 3′ UTR sequence was amplified and inserted into pmirGLO Vector (Ambion, Carlsbad, CA, USA). For the luciferase reporter assay, HEK 293 T cells were co-transfected with pmirGLO-Notch1-3′UTR plus either miR-34c mimics or Control for 48 hours. Both pmirGLO and pmirGLO-Notch1-3′UTR-mut were used as the controls for pmirGLO-Notch1-3′UTR. The activities of firefly and Renilla luciferases were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA), and firefly luciferase activity was normalized to that of Renilla luciferase.
A ~4600 bp sequence upstream of miR-34c in genomic DNA was amplified and inserted into pGL3-basic Vector (Ambion, Carlsbad, CA, USA). For the luciferase reporter assay, HEK 293 T cells were co-transfected with pGL3-basic-miR-34c upstream, pRL-TK plus either pCDNA3.1-N1ICD or pCDNA3.1. Either pGL3-basic-miR-34c upstream-mut or pGL3-control was used as a control for pGL3-basic-miR-34c upstream. The activities of firefly and Renilla luciferases were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA), and firefly luciferase activity was normalized to that of Renilla luciferase.
ChIP assay
The ChIP-IT® Express Magnetic Chromatin Immunoprecipitation Kit & Sonication Shearing Kit (Catalog Nos 53008) was purchased from Active Motif (Carlsbad, CA). For ChIP assay, the DNA was immunoprecipitated with the anti-N1ICD antibody (1:1000 dilution, Cell Signaling Technology, Danvers, MA, United States), and ChIP analysis was performed according to the manufacturer’s protocol. Primers used for ChIP assay are shown in Table S1. DNA samples before immunoprecipitation were used as a template for input control.
Tissue sampling and H&E
Mice were purchased from Guangdong Medical Lab Animal Center, and lentivirus containing miR-34c mimics or Control were purchased from Shanghai JiKai Gene Chemical Technology Co., LTD. Nine mice were divided into three groups, and each group contained three mice. Mice were injected with physiological saline, LV-Control or LV-miR-34c. Gastrocnemius muscles were dissected from each mouse one week after injection for extracting total RNA and proteins. Mice gastrocnemius muscles were stained with Hematoxylin and Eosin (H&E), and the cross section area of individual myofibers was photographed using Nikon TE2000-U inverted microscope (Nikon Corporation, Tokyo, Japan) and measured using Image-Pro Plus (IPP) 6.0 software (Media Cybernetics, Inc., Silver Spring, MD, USA).
Statistical analysis
All data are expressed as the mean ± standard error of the mean (S.E.M.). For all the experimental data in this study, including miR-34c inhibiting PSCs proliferation, miR-34c promoting PSCs differentiation and miR-34c repressing mice muscle development, we are comparing only the control group to the treatment group; thus we used the student’s t-test for the statistical analyses (SPSS 18.0, Chicago, IL, USA).
Data availability statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
This work was supported by the National High Technology Research and Development Program 863 (#2013AA102502); the National Natural Science Foundation of China (#31372283); The Team Project of Guangdong Agricultural Bureau (#2016LM2148) and the Natural Fund Key Projects of Guangdong Province (#2015A030311006). C.H. is supported by the USDA National Institute of Food and Agriculture, Hatch project HAW-H2037, managed by the College of Tropical Agriculture and Human Resources, University of Hawaii at Manoa.
Author Contributions
All authors made contributions to this study. C.W., C.H. and J.X. designed the study. L.H., Y.J. and J.O. performed the experiments and developed the methods. L.H. and H.L. analyzed the data. L.H., C.W. and C.H. wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Lianjie Hou and Jian Xu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09688-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Montarras D, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 2.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen X, Klionsky DJ. Autophagy is a key factor in maintaining the regenerative capacity of muscle stem cells by promoting quiescence and preventing senescence. Autophagy. 2016;12:617–618. doi: 10.1080/15548627.2016.1158373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dayanidhi S, Lieber RL. Skeletal muscle satellite cells: mediators of muscle growth during development and implications for developmental disorders. Muscle & nerve. 2014;50:723–732. doi: 10.1002/mus.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pallafacchina G, et al. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Wen Y, et al. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol Cell Biol. 2012;32:2300–2311. doi: 10.1128/MCB.06753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 8.Iso T, Chung G, Hamamori Y, Kedes L. HERP1 is a cell type-specific primary target of Notch. The Journal of biological chemistry. 2002;277:6598–6607. doi: 10.1074/jbc.M110495200. [DOI] [PubMed] [Google Scholar]
- 9.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Science’s STKE: signal transduction knowledge environment. 2006;2006 doi: 10.1126/stke.3642006cm7. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura T, et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. The Journal of clinical investigation. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo D, Renault VM, Rando TA. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Seminars in cell & developmental biology. 2005;16:612–622. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Mourikis P, et al. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem cells. 2012;30:243–252. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]
- 13.Bjornson CR, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. The EMBO journal. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 17.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 19.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299 doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 21.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. Rna. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/S1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 23.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 24.Chen T, et al. Exploration of microRNAs in porcine milk exosomes. BMC genomics. 2014;15 doi: 10.1186/1471-2164-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, et al. MicroRNA identity and abundance in developing swine adipose tissue as determined by Solexa sequencing. J Cell Biochem. 2011;112:1318–1328. doi: 10.1002/jcb.23045. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q, et al. miRNA-34a reduces neointima formation through inhibiting smooth muscle cell proliferation and migration. Journal of molecular and cellular cardiology. 2015;89:75–86. doi: 10.1016/j.yjmcc.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Choe N, et al. The microRNA miR-34c inhibits vascular smooth muscle cell proliferation and neointimal hyperplasia by targeting stem cell factor. Cell Signal. 2015;27:1056–1065. doi: 10.1016/j.cellsig.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Qin L, et al. Notch1-mediated signaling regulates proliferation of porcine satellite cells (PSCs) Cell Signal. 2013;25:561–569. doi: 10.1016/j.cellsig.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Xu J. The Interaction between Notch Signaling Pathway and MicroRNAs in the Regulation of Porcine Skeletal Muslce Development. Thesis of South China Agricultural University. 2015;1 [Google Scholar]
- 30.Rhoads RP, et al. Extrinsic regulation of domestic animal-derived myogenic satellite cells II. Domestic animal endocrinology. 2009;36:111–126. doi: 10.1016/j.domaniend.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Wilschut KJ, Haagsman HP, Roelen BA. Extracellular matrix components direct porcine muscle stem cell behavior. Experimental cell research. 2010;316:341–352. doi: 10.1016/j.yexcr.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Kamanga-Sollo E, Pampusch MS, White ME, Hathaway MR. & Dayton, W. R. Effects of heat stress on proliferation, protein turnover, and abundance of heat shock protein messenger ribonucleic acid in cultured porcine muscle satellite cells. Journal of animal science. 2011;89:3473–3480. doi: 10.2527/jas.2011-4123. [DOI] [PubMed] [Google Scholar]
- 33.Mesires NT, Doumit ME. Satellite cell proliferation and differentiation during postnatal growth of porcine skeletal muscle. American journal of physiology. Cell physiology. 2002;282:C899–906. doi: 10.1152/ajpcell.00341.2001. [DOI] [PubMed] [Google Scholar]
- 34.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Developmental cell. 2002;3:397–409. doi: 10.1016/S1534-5807(02)00254-X. [DOI] [PubMed] [Google Scholar]
- 35.Kovanda A, Rezen T, Rogelj B. MicroRNA in skeletal muscle development, growth, atrophy, and disease. Wiley interdisciplinary reviews. RNA. 2014;5:509–525. doi: 10.1002/wrna.1227. [DOI] [PubMed] [Google Scholar]
- 36.Ge Y, Chen J. MicroRNAs in skeletal myogenesis. Cell Cycle. 2011;10:441–448. doi: 10.4161/cc.10.3.14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou X, et al. Discovery of MicroRNAs associated with myogenesis by deep sequencing of serial developmental skeletal muscles in pigs. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nature reviews. Molecular cell biology. 2011;13:127–133. doi: 10.1038/nrm3265. [DOI] [PubMed] [Google Scholar]
- 39.Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clinical genetics. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 40.Wobus AM, Guan K, Yang HT, Boheler KR. Embryonic stem cells as a model to study cardiac, skeletal muscle, and vascular smooth muscle cell differentiation. Methods in molecular biology. 2002;185:127–156. doi: 10.1385/1-59259-241-4:127. [DOI] [PubMed] [Google Scholar]
- 41.Dean DA. Nonviral gene transfer to skeletal, smooth, and cardiac muscle in living animals. American journal of physiology. Cell physiology. 2005;289:C233–245. doi: 10.1152/ajpcell.00613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nature reviews. Molecular cell biology. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 43.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nature reviews. Molecular cell biology. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 44.Gopinathan L, Ratnacaram CK, Kaldis P. Established and novel Cdk/cyclin complexes regulating the cell cycle and development. Results and problems in cell differentiation. 2011;53:365–389. doi: 10.1007/978-3-642-19065-0_16. [DOI] [PubMed] [Google Scholar]
- 45.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Srivastava RK, Chen Q, Siddiqui I, Sarva K, Shankar S. Linkage of curcumin-induced cell cycle arrest and apoptosis by cyclin-dependent kinase inhibitor p21(/WAF1/CIP1) Cell Cycle. 2007;6:2953–2961. doi: 10.4161/cc.6.23.4951. [DOI] [PubMed] [Google Scholar]
- 47.Cao Y, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faralli H, Dilworth FJ. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comparative and functional genomics. 2012;2012 doi: 10.1155/2012/836374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bentzinger, C. F., Wang, Y. X. & Rudnicki, M. A. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol4, doi:10.1101/cshperspect.a008342 (2012). [DOI] [PMC free article] [PubMed]
- 50.Lee SJ. Regulation of muscle mass by myostatin. Annual review of cell and developmental biology. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- 51.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 52.Peault B, et al. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Molecular therapy: the journal of the American Society of Gene Therapy. 2007;15:867–877. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 53.Qin LL, et al. Mechano growth factor (MGF) promotes proliferation and inhibits differentiation of porcine satellite cells (PSCs) by down-regulation of key myogenic transcriptional factors. Mol Cell Biochem. 2012;370:221–230. doi: 10.1007/s11010-012-1413-9. [DOI] [PubMed] [Google Scholar]
- 54.Shu G, et al. Phloretin promotes adipocyte differentiation in vitro and improves glucose homeostasis in vivo. The Journal of nutritional biochemistry. 2014;25:1296–1308. doi: 10.1016/j.jnutbio.2014.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).