ABSTRACT
West Nile virus (WNV) is a mosquito-borne flavivirus that causes epidemics of encephalitis and viscerotropic disease worldwide. This virus has spread rapidly and has posed a significant public health threat since the outbreak in New York City in 1999. The interferon (IFN)-mediated antiviral response represents an important component of virus-host interactions and plays an essential role in regulating viral replication. Previous studies have suggested that multifunctional nonstructural proteins encoded by flaviviruses antagonize the host IFN response via various means in order to establish efficient viral replication. In this study, we demonstrated that the nonstructural protein 1 (NS1) of WNV antagonizes IFN-β production, most likely through suppression of retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) activation. In a dual-luciferase reporter assay, WNV NS1 significantly inhibited the activation of the IFN-β promoter after Sendai virus infection or poly(I·C) treatment. NS1 also suppressed the activation of the IFN-β promoter when it was stimulated by interferon regulatory factor 3 (IRF3)/5D or its upstream molecules in the RLR signaling pathway. Furthermore, NS1 blocked the phosphorylation and nuclear translocation of IRF3 upon stimulation by various inducers. Mechanistically, WNV NS1 targets RIG-I and melanoma differentiation-associated gene 5 (MDA5) by interacting with them and subsequently causing their degradation by the proteasome. Furthermore, WNV NS1 inhibits the K63-linked polyubiquitination of RIG-I, thereby inhibiting the activation of downstream sensors in the RLR signaling pathway. Taken together, our results reveal a novel mechanism by which WNV NS1 interferes with the host antiviral response.
IMPORTANCE WNV Nile virus (WNV) has received increased attention since its introduction to the United States. However, the pathogenesis of this virus is poorly understood. This study demonstrated that the nonstructural protein 1 (NS1) of WNV antagonizes the induction of interferon beta (IFN-β) by interacting with and degrading retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which are crucial viral sensors in the host innate immune system. Further experiments suggested that NS1-mediated inhibition of the RIG-I-like receptor (RLR) signaling pathway involves inhibition of RIG-I K63-linked polyubiquitination and that the proteasome plays a role in RIG-I degradation. This study provides new insights into the regulation of WNV NS1 in the RLR signaling pathway and reveals a novel mechanism by which WNV evades the host innate immune response. The novel findings may guide us to discover new therapeutic targets and develop effective vaccines for WNV infections.
KEYWORDS: flavivirus, IFN-β, MDA5, NS1, RIG-I, West Nile virus
INTRODUCTION
West Nile virus (WNV) belongs to the genus Flavivirus in the family Flaviviridae and causes annual epidemics of encephalitis and viscerotropic disease worldwide (1). WNV was originally isolated in the West Nile province of Uganda in 1937 and then introduced to many other parts of the world (2). Since its introduction into North America in 1999, WNV has posed a significant public health risk in the United States (3, 4).
The flavivirus genome is a single-stranded, positive-sense RNA, comprising approximately 11,000 nucleotides. The genomic RNA consists of a 5′ untranslated region (UTR), a single open reading frame (ORF), and a 3′ UTR. The single ORF encodes three structural proteins (capsid, membrane, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (5–7). The structural proteins play essential roles in virus entry, fusion, and assembly. The nonstructural proteins are the main components of viral replication complexes; they also play a role in the evasion of the host immune response. A number of studies have previously reported that flavivirus NS2A, NS4A, NS4B, and NS5 play a crucial role in blocking the expression of interferon (IFN) and downstream antiviral signaling (8–13).
The innate immune response is the first line of host defense against viral infections (14). During a viral infection, host pattern recognition receptors (PRRs) recognize viral components, known as pathogen-associated molecular patterns, and then trigger the antiviral responses by producing type I IFNs (15). Viral RNAs are mainly recognized by two classes of PRRs: Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs). Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which are viral sensors in the innate immune system, can recognize the invading viruses and trigger the immune response (1). After sensing the cytoplasmic viral RNAs, RIG-I or MDA5 interacts with mitochondrial antiviral signaling protein (MAVS; also known as VISA, IPS1, or Cardif) and then activates the downstream inhibitor of κB kinase ε (IKKε), which then leads to the phosphorylation of a critical transcription factor, interferon regulatory factor 3 (IRF3). The phosphorylated IRF3 is translocated to the nucleus and induces type I IFN production and inflammatory responses (16–21).
To combat the IFN antiviral responses, different viruses have evolved various strategies to antagonize IFN production. Many viruses can interact with IRF3 directly or inhibit the activation of IRF3 to combat the IFN response (21–25). For example, bovine herpesvirus 1 and varicella-zoster virus suppress the antiviral response by degrading IRF3 (26, 27). The V proteins of paramyxoviruses interact with MDA5 to block the activation of the IFN-β promoter (28). The US11 protein of herpes simplex virus 1 (HSV-1) prevents the production of IFN-β via direct interaction with RIG-I and MDA5 (29). The flavivirus has been shown to affect both innate and adaptive immune responses (30–33). WNV NS1 may inhibit the activation of the IFN-β promoter and nuclear factor κB (NF-κB) promoter by inhibiting IRF3 and NF-κB nuclear translocation by influencing the Toll-like receptor 3 (TLR3) signaling pathway (31).
However, there are conflicting results showing that the NS1 proteins from WNV, yellow fever virus, and dengue virus 2 (DENV-2 [DV2]) do not inhibit TLR3 signaling (34). Recently, the crystal structures of WNV and DENV NS1 were shown to have three domains: a “β-roll,” a “wing,” and a “β-ladder” (35). Interestingly, the NS1 wing domain is similar to the helicase domain of RIG-I and MDA5 (16, 18, 35). The structural similarity between the host receptors and the viral protein implies that the flavivirus NS1 may interfere with the RLR signaling pathway to assist the flaviviruses in the evasion of the host immune response.
Since the mechanism underlying the role of the flavivirus NS1 in the evasion of the host immune response is unclear, we systematically investigated the functions of the WNV NS1 related to the IFN response. We demonstrated that NS1 antagonizes the production of IFN-β by interacting with and promoting the degradation of RIG-I and MDA5. In addition, our results showed that NS1 suppresses the RLR signaling pathway by inhibiting RIG-I K63-linked polyubiquitination (but not K48-linked polyubiquitination) and that the proteasome may play a role in the degradation of RIG-I. Our study indicates a novel mechanism by which WNV NS1 can evade the host immune response.
RESULTS
WNV NS1 blocks the production of IFN-β.
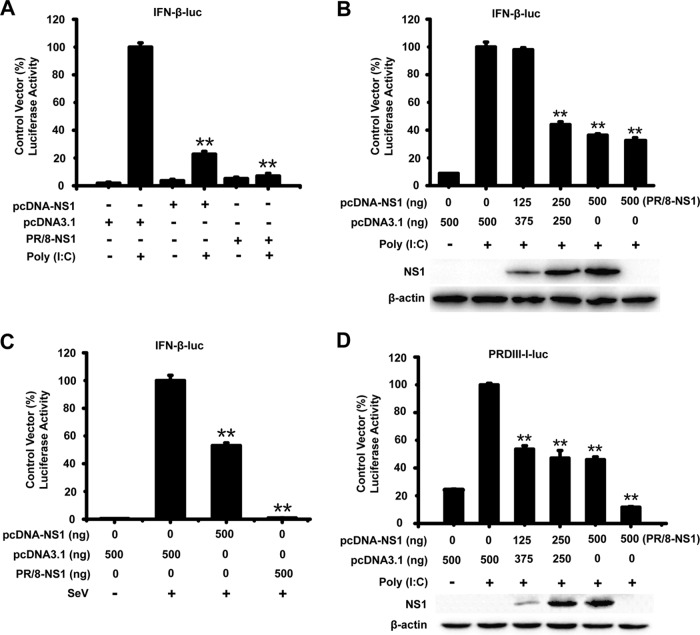
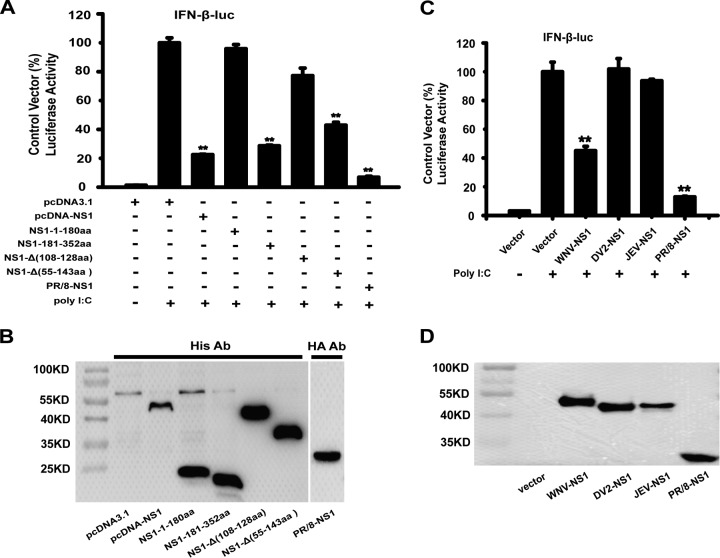
To examine whether WNV NS1 antagonizes the production of IFN-β, HEK-293T cells were transfected with a reporter plasmid [pIFN-β-Luc or PRD(III-I)-Luc], the internal control plasmid (pRL-TK), and an expression plasmid encoding the WNV NS1 protein or an empty vector, pcDNA3.1. At 24 h after transfection, the cells were infected or mock infected with Sendai virus (SeV) and treated or mock treated with poly(I·C) for 16 h. A plasmid encoding the NS1 protein of the influenza virus PR/8 (an inhibitor of IFN-β) was used as the positive control (36). As shown in Fig. 1A and B, the expression of WNV NS1 significantly inhibited the activation of the IFN-β promoter [induced by poly(I·C)] in a dose-responsive manner. WNV NS1 also suppressed the activation of the IFN-β promoter after the cells were infected with SeV (Fig. 1C). The reporter plasmid pIFN-β-Luc contains the full length of the transcription factor-responsive regions, whereas the PRD(III-I)-Luc plasmid contains only the IRF3-responsive region of the IFN-β promoter. As shown in Fig. 1D, upon poly(I·C) treatment, WNV NS1 inhibited the activation of the PRD(III-I)-Luc promoter in a dose-dependent manner. These results indicate that WNV NS1 can inhibit the activation of the IFN-β promoter after the promoter is induced by various stimulators.
FIG 1.
WNV NS1 blocks the production of IFN-β. HEK-293T cells were cotransfected with pIFN-β-Luc, pRL-TK, and the indicated amount of NS1 or PR/8-NS1 expression plasmid for 24 h. The cells were then treated or mock treated with poly(I·C) for 24 h or infected or mock infected with SeV for 16 h (C). Reporter activity was detected by dual-luciferase reporter assays. HEK-293T cells were cotransfected with PRD(III-I)-Luc, pRL-TK, and the indicated amount of NS1 expression plasmid for 24 h, followed by treatment or mock treatment with poly(I·C) for 24 h (D). Reporter activity was determined by dual-luciferase reporter assays. The resultant ratios for the samples were normalized using the renilla luciferase values, and they were expressed as a percentage of the value induced in cells transfected with an empty vector with poly(I·C) treatment or SeV infection. The data are representative of three independent experiments, with each determination performed in triplicate (mean ± standard deviation of the fold change). Two asterisks indicate significant differences between groups (**, P < 0.01, as determined by Student's t test). The cell lysates were analyzed by immunoblotting with anti-His Ab (lower panels) to detect the expression of pcDNA3.1-His-NS1, and immunoblotting of β-actin was used as the loading control.
NS1 affects the IRF3 signaling pathway.
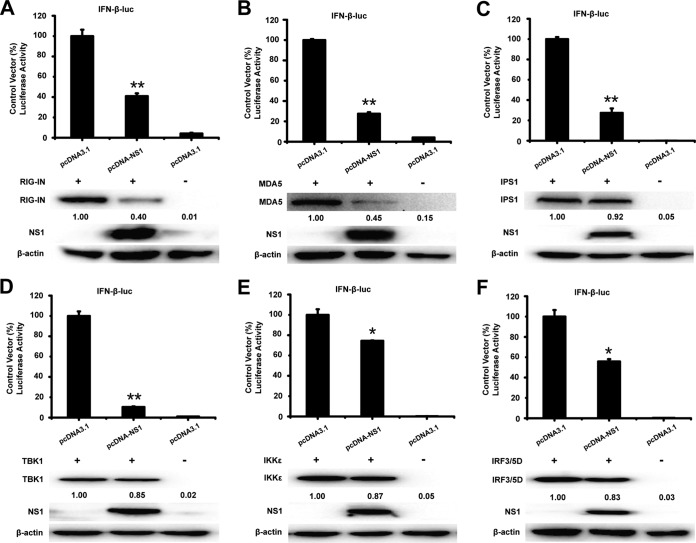
To analyze the potential mechanism underlying the NS1-mediated antagonism of IFN-β promoter activation, HEK-293T cells were cotransfected with pcDNA3.1-His-NS1 and a series of expression plasmids encoding molecules from the IRF3 signaling pathway (including RIG-IN, MDA5, IPS1, TBK1, IKKε, and IRF3/5D), together with the luciferase reporter plasmid p-IFN-β-Luc and pRL-TK. The luciferase activity was assessed at 36 h after cotransfection. As shown in Fig. 2A to F, overexpression of the signaling molecules activated the IFN-β promoter. However, the activation of the IFN-β promoter induced by IRF3/5D and its upstream molecules was inhibited by WNV NS1 (Fig. 2A to F). The results indicate that NS1 blocks the production of IFN-β by targeting the signaling molecules of the IRF3 pathway.
FIG 2.
NS1 affects the IRF3 signaling pathway. HEK-293T cells were cotransfected with pIFN-β-Luc, the pRL-TK plasmid, and pcDNA3.1-His-NS1 together with constructs expressing RIG-IN (A), MDA5 (B), IPS1 (C), TBK1 (D), IKKε (E), or IRF3/5D (F). Luciferase assays were performed 36 h after transfection. The resultant ratios were normalized to the value for the cells cotransfected with pIFN-β-Luc, pRL-TK, and an empty vector together with the relevant constructs. The results represent the means and standard deviations of data from three independent experiments in duplicate. The asterisks indicate significant differences between groups (*, P < 0.05; **, P < 0.01, as determined by Student's t test). The cell lysates were detected by immunoblotting (lower panels) with anti-FLAG Ab to detect the expression of RIG-IN, MDA5, IPS1, TBK1, IKKε, or IRF3/5D. The expression of NS1 was detected with anti-His Ab, and immunoblotting of β-actin was used as the loading control. The expression level of RIG-IN, MDA5, IPS1, TBK1, IKKε, and IRF3/5D was quantified by carrying out a densitometric analysis using Image Lab software, and the data were normalized to the band densities of the first lane.
NS1 blocks IRF3 phosphorylation and nuclear translocation.
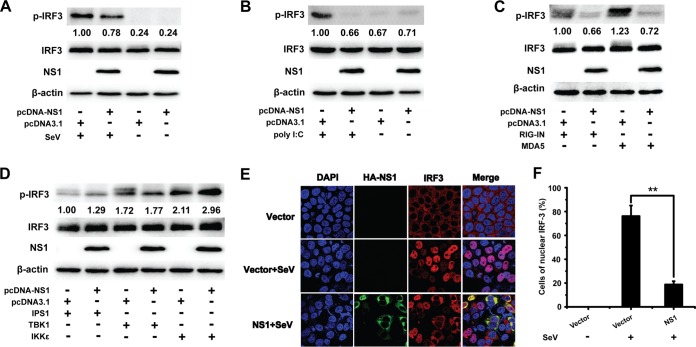
IRF3 is a key transcription factor in the type I IFN signaling pathway. The IFN-β promoter is activated after IRF3 is phosphorylated, translocated to the nucleus, and bound to the promoter sequence (18, 37, 38). As WNV NS1 inhibited the IRF3 signaling pathway, we investigated the effect of NS1 on the phosphorylation and nuclear translocation of IRF3. The phosphorylation of IRF3 was analyzed in 293T cells transfected with pcDNA3.1-His-NS1 or an empty vector followed by SeV infection or poly(I·C) treatment. As expected, SeV infection and poly(I·C) treatment increased IRF3 phosphorylation. However, the phosphorylation of IRF3 induced by SeV and poly(I·C) was markedly inhibited by WNV NS1. In contrast, the expression level of IRF3 was not altered by NS1 expression (Fig. 3A and B). This result suggests that NS1 inhibits the phosphorylation of IRF3 induced by SeV infection or poly(I·C) treatment.
FIG 3.
NS1 blocks IRF3 phosphorylation and nuclear translocation. (A and B) Immunoblot analysis of total and phosphorylated IRF3 in HEK-293T cells transfected with empty vector or NS1 expression plasmid and treated or not treated with SeV (A) or poly(I·C) (B) for 8 h after transfection. The expression of NS1 and β-actin in the same cell lysates was analyzed by immunoblotting. (C and D) Immunoblot analysis of total and phosphorylated IRF3 in HEK-293T cells transfected with various combinations of plasmids for FLAG-tagged RIG-IN, MDA5, IPS1, TBK1, or IKKε plus an empty vector or expression vector encoding NS1. The expression of NS1 in the same cell lysates was analyzed by immunoblotting. The expression level of phosphorylated IRF3 was quantified by carrying out a densitometric analysis using Image Lab software, and the data were normalized to the band densities of the first lane. (E) Fluorescence microscopy of IRF3 in HeLa cells transfected with an empty vector or expression vector encoding HA-tagged NS1 and then infected or not infected with SeV for 12 h. The cells were incubated with both mouse anti-HA Ab and rabbit anti-IRF3 PAb, followed by Cy3-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse IgG secondary Abs. Then cells were stained with DAPI and observed under a confocal microscope (Nikon A1 MP Storm). (F) The percentage of IRF3-positive nuclei was quantified in a number of fields. Two asterisks indicate significant differences between groups (**, P < 0.01, as determined by Student's t test).
To investigate the effect of NS1 on the phosphorylation of IRF3 induced by signaling molecules in the IRF3 pathway (including RIG-IN, MDA5, IPS1, TBK1, and IKKε), HEK-293T cells were cotransfected with an NS1-expressing plasmid or an empty vector and expression plasmids encoding molecules from the IRF3 signaling pathway. IRF3 phosphorylation was detected at 36 h after cotransfection. As shown in Fig. 3C and D, WNV NS1 significantly inhibited the phosphorylation of IRF3 induced by RIG-I and MDA5 but not that induced by IPS1, TBK1, or IKKε. Consistent with the inhibition results, the nuclear translocation of IRF3 was also blocked by NS1, whereas the vector had a negligible effect on the nuclear translocation of IRF3 induced by SeV (Fig. 3E and F). These results suggest that WNV NS1 blocks the phosphorylation and nuclear translocation of IRF3, leading to decreased expression of IFN-β.
NS1 inhibits RIG-I and MDA5 expression.
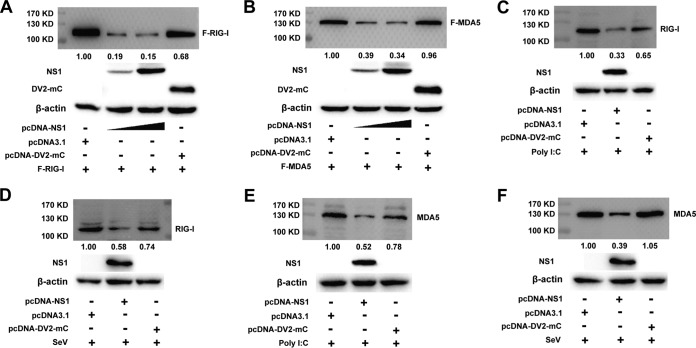
We next examined how WNV NS1 blocks the activation of IFN-β and IRF3. HEK-293T cells were cotransfected with an increasing dose of plasmid pcDNA3.1-His-NS1 and a fixed dose of plasmid expressing RIG-I or MDA5. At 36 h after cotransfection, the cell lysates were probed for RIG-I or MDA5 expression using Western blotting. As shown in Fig. 4A and B, increasing the level of NS1 expression decreased the level of RIG-I and MDA5 expression. As a negative control, cotransfection of pcDNA3.1-DV2-mC (a plasmid expressing DV2 capsid protein) did not inhibit the expression level of RIG-I or MDA5.
FIG 4.
NS1 inhibits RIG-I and MDA5 expression. (A and B) Immunoblot analysis of exogenous RIG-I and MDA5 expression in HEK-293T cells transfected with a plasmid for FLAG-RIG-I (A) or FLAG-MDA5 (B) plus an empty vector or pcDNA3.1-DV2-mC or increasing doses of plasmid for NS1 (wedge) (0.5 and 2 μg). The negative-control plasmid pcDNA3.1-DV2-capsid was cotransfected with RIG-I or MDA5 into the cells. The expression of NS1, DV2-mC, and β-actin in the same cell lysates was analyzed by immunoblotting. (C to F) Immunoblot analysis of RIG-I (C and D) and MDA5 (E and F) in HEK-293T cells transfected with an empty vector or pcDNA3.1-DV2-mC or 2 μg plasmid for NS1, followed by treatment or no treatment with SeV (D and F) or poly(I·C) (C and E). The negative-control plasmid pcDNA3.1-DV2-mC was transfected into the cells. The expression of NS1 and β-actin in the same cell lysates was analyzed by immunoblotting. The expression level of RIG-I and MDA5 was quantified by carrying out a densitometric analysis using Image Lab software, and the data were normalized to the band densities of the first lane. One representative experiment out of two is shown.
To clarify whether NS1 inhibits endogenous expression of RIG-I and MDA5, HEK-293T cells were cotransfected with pcDNA3.1-His-NS1, an empty vector, or the negative-control vector pcDNA3.1-DV2-mC followed by SeV infection or poly(I·C) treatment. At 24 h after transfection, the level of endogenous RIG-I and MDA5 was lowered by NS1 in the poly(I·C)-treated cells, whereas the expression of DV2-mC had a weak effect on the endogenous expression of RIG-I and MDA5 (Fig. 4C and E). In the SeV-infected cells, NS1 also blocked the endogenous expression of RIG-I and MDA5 protein, whereas DV2-mC had a negligible effect on the endogenous expression of RIG-I and MDA5 (Fig. 4D and F). These results suggest that overexpression of WNV NS1 inhibits the exogenous and endogenous levels of RIG-I and MDA5 protein.
The amino acid (aa) 181 to 352 (181–352aa) domain of NS1 inhibits IFN-β production.
To find the essential domain of WNV NS1 involved in the inhibition of IFN-β production, we constructed four truncation mutants: NS1-1–180aa, NS1-181–352aa, NS1-Δ(108–128aa), and NS1-Δ(55–143aa). These four mutants were designed based on the NS1 crystal structure. HEK-293T cells were cotransfected with the reporter plasmid pIFN-β-Luc, the internal control plasmid pRL-TK, and either the NS1 expression plasmid, an NS1 truncation mutant expression plasmid, or an empty vector for 24 h followed by stimulation with poly(I·C) or no poly(I·C) treatment for 16 h. The influenza virus PR/8-NS1 was used as the positive control. As shown in Fig. 5A, the results showed that the N-terminal domain of NS1, that is, the NS1-1–180aa truncation mutant, barely inhibited the IFN-β promoter activation induced by poly(I·C), whereas the C-terminal domain of NS1, that is, the NS1-181–352aa truncation mutant, significantly inhibited the activation of the IFN-β promoter. A previous study showed that NS1 55–143aa, an α/β subdomain of the NS1 wing domain, is similar to the superfamily 2 (SF2) helicase domain found in both RIG-I and MDA5 (35). However, deletion of this domain of NS1, that is, the NS1-Δ(55–143aa) truncation mutant, still inhibited the IFN-β promoter activation to some degree (Fig. 5A). The expression of the WNV NS1 and NS1 truncation mutant expression plasmids was confirmed using immunoblotting as shown in Fig. 5B.
FIG 5.
The 181–352aa domain of NS1 inhibits IFN-β production. (A and C) HEK-293T cells in 24-well plates were cotransfected with pIFN-β-Luc and pRL-TK along with an NS1 or truncated NS1 expression plasmid (A) or a PR/8-NS1, JEV-NS1, or DV2-NS1 expression plasmid (C) for 24 h. The cells were then treated or not treated with poly(I·C) for 16 h. Reporter activity was detected by dual-luciferase reporter assays. The resultant ratios were normalized to the value for the cells cotransfected with pIFN-β-Luc, pRL-TK, and an empty vector and treated with poly(I·C). The results represent the means and standard deviations of data from three independent experiments. Two asterisks indicate significant differences between groups (**, P < 0.01 as determined by Student's t test). (B and D) The cell lysates were detected by immunoblotting with anti-His Ab or anti-HA Ab to detect the expression of pcDNA3.1-His-NS1 (B) or the truncated NS1 expression plasmids or the JEV-NS1, DV2-NS1, or PR/8-NS1 proteins (D).
To further identify the mechanism underlying the effect of the NS1 protein of other flaviviruses on IFN-β activation, we also analyzed the function of Japanese encephalitis virus (JEV) and DENV-2 NS1 on the activation of IFN-β using dual-luciferase reporter assays. However, the data showed that the NS1 proteins encoded by JEV and DENV-2, which have a high degree of amino acid sequence homology and similar effects on flavivirus replication, did not have the same inhibitory effect as WNV NS1 (Fig. 5C). The expression of JEV NS1, DENV-2 NS1, and the influenza virus PR/8-NS1 was detected as expected by immunoblotting using antihemagglutinin (anti-HA) antibody (Ab) (Fig. 5D).
NS1 and RIG-I/MDA5 interactions based on BiFC system and colocalization assay.
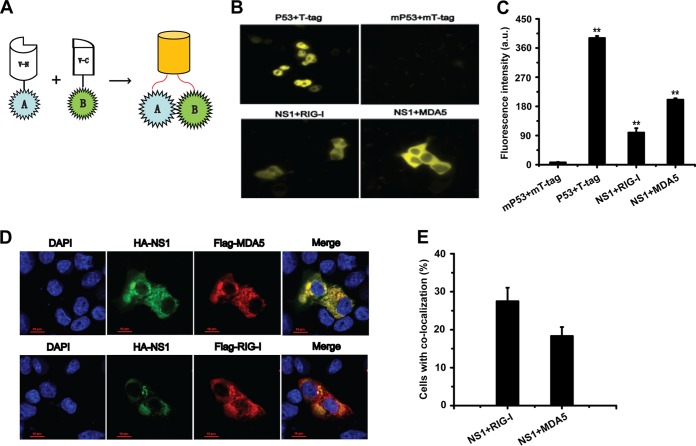
To investigate the possible interaction between NS1 and RIG-I/MDA5, we first evaluated the potential interaction between NS1 and RIG-I/MDA5 using a Venus-based bimolecular fluorescence complementation (BiFC) system. A schematic diagram of this system is shown in Fig. 6A. In this system, two proteins (A and B) that potentially interact with each other are fused to V-N and V-C fragments, respectively. When the two proteins interact, the two fragments of the fluorescent protein are brought together, which allows visualization of the protein interactions. According to a previous report (39), the Venus fluorescent protein can be split at the point between amino acids (aa) 173 and 174, generating the V-N and V-C fragments. In our study, these two fragments were fused to NS1 and RIG-I/MDA5, respectively.
FIG 6.
NS1 and RIG-I/MDA5 interactions based on a BiFC system and colocalization assay. (A) Schematic of the basic principles of the BiFC system. N- and C-terminal fragments of Venus fluorescent protein are fused to proteins A and B, respectively. The interaction between A and B brings the N- and C-terminal fragments of Venus in proximity to reconstitute the intact fluorescent protein. (B) 293T cells were cotransfected with V-N-NS1 and V-C-RIG-I/MDA5 plasmids and incubated at 37°C for 12 h. Plasmids V-N-p53 and V-C-T-tag were used as the positive controls and cotransfected into the cells, while V-N-mp53 and V-C-mT-tag were used as the negative controls and cotransfected into the cells. Fluorescent images generated by BiFC in the cells were visualized using an inverted Delta Vision Restoration microscopy system (Applied Precision, WA, USA) with excitation and emission wavelengths of 500/24 nm and 542/27 nm, respectively. (C) Quantitative analysis of the BiFC efficiency using Volocity software; a.u., arbitrary units. (D) Colocalization of NS1 and RIG-I/MDA5. HA-NS1 and FLAG-RIG-I/MDA5 plasmids were cotransfected into Vero cells. The cells were then incubated with both mouse anti-HA Ab and rabbit anti-FLAG Ab, followed by Cy3-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse IgG secondary Abs. Then, cells were stained with DAPI and observed under a confocal microscope (Nikon A1 MP Storm). Colocalization of HA-NS1 (green) and FLAG-RIG-I/MDA5 (red) appears as yellow. (E) The percentage of cells with colocalization of NS1 and RIG-I/MDA5 was quantified in a number of fields.
As shown in Fig. 6B and C, yellow fluorescence was detected in the HEK-293T cells after cotransfection with V-N-NS1 and V-C-RIG-I/MDA5 at 12 h. To assess the reliability of the Venus-based BiFC system, p53 and T-tag were selected for use in a positive-control assay due to their well-known strong interaction. Strong yellow fluorescence signals were detected after 293T cells were cotransfected with the fusion proteins V-N-p53 and V-C-T-tag. In addition, mp53 and mT-tag were used in a negative-control assay (40), and there were no fluorescence signals detected after V-N-mp53 and V-C-mT-tag coexpression in the 293T cells.
Immunofluorescence confocal microscopy was then used to detect the colocalization of NS1 and RIG-I/MDA5. HA-NS1 and FLAG-RIG-I/MDA5 expression plasmids were cotransfected into Vero cells, and the colocalization of the two proteins was then observed under a confocal microscopy. As shown in Fig. 6D, both NS1 and RIG-I/MDA5 were predominantly located in the cytoplasm. The colocalization between NS1 and RIG-I/MDA5 suggested a potential association between these proteins.
Analysis of the interaction between NS1 and RIG-I using a co-IP assay.
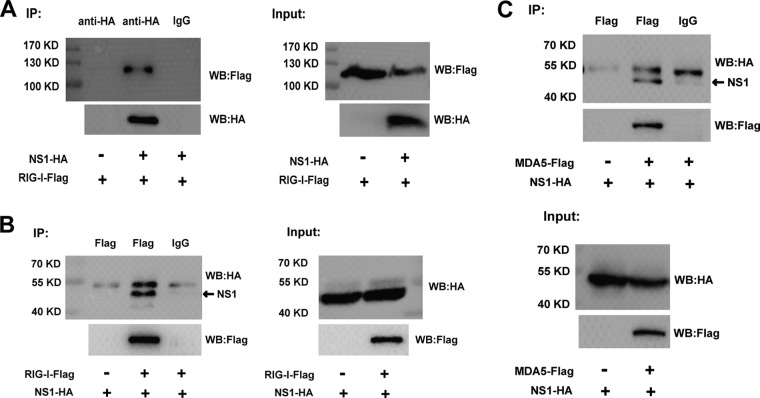
To identify the physical interaction between WNV NS1 and RIG-I/MDA5, we then used a coimmunoprecipitation (co-IP) assay. HEK-293T cells were cotransfected with the expression plasmids pCAGGS-HA-NS1 and FLAG-tagged RIG-I. The cell lysates were then immunoprecipitated with rabbit anti-HA or mouse anti-FLAG monoclonal antibody (MAb). As shown in Fig. 7A, RIG-I coprecipitated with NS1 when the anti-HA Ab was used. In addition, NS1 coprecipitated with RIG-I when the anti-FLAG MAb was used (Fig. 7B). These results indicate that there was a direct interaction between NS1 and RIG-I. To identify the physical interaction between WNV NS1 and MDA5, HEK-293T cells were cotransfected with the expression plasmids pCAGGS-HA-NS1 and FLAG-tagged MDA5 expression plasmid. As shown in Fig. 7C, NS1 coprecipitated with MDA5 when the anti-FLAG MAb was used. These results further confirm the potential physical interaction between NS1 and RIG-I/MDA5.
FIG 7.
Analysis of the interaction between NS1 and RIG-I/MDA5 by co-IP assay. HEK-293T cells were cotransfected with plasmid expressing HA-tagged NS1 and a FLAG-tagged RIG-I/MDA5 expression plasmid or an empty vector for 36 h. The cells were lysed and subjected to immunoprecipitation with rabbit anti-HA or mouse anti-FLAG Ab. Rabbit or mouse normal IgG was used as the negative control. The IP products and input samples were analyzed by immunoblotting using mouse anti-HA and mouse anti-FLAG Abs. One representative experiment out of three is shown. WB, Western blot.
NS1 decreases K63-linked polyubiquitination of RIG-I.
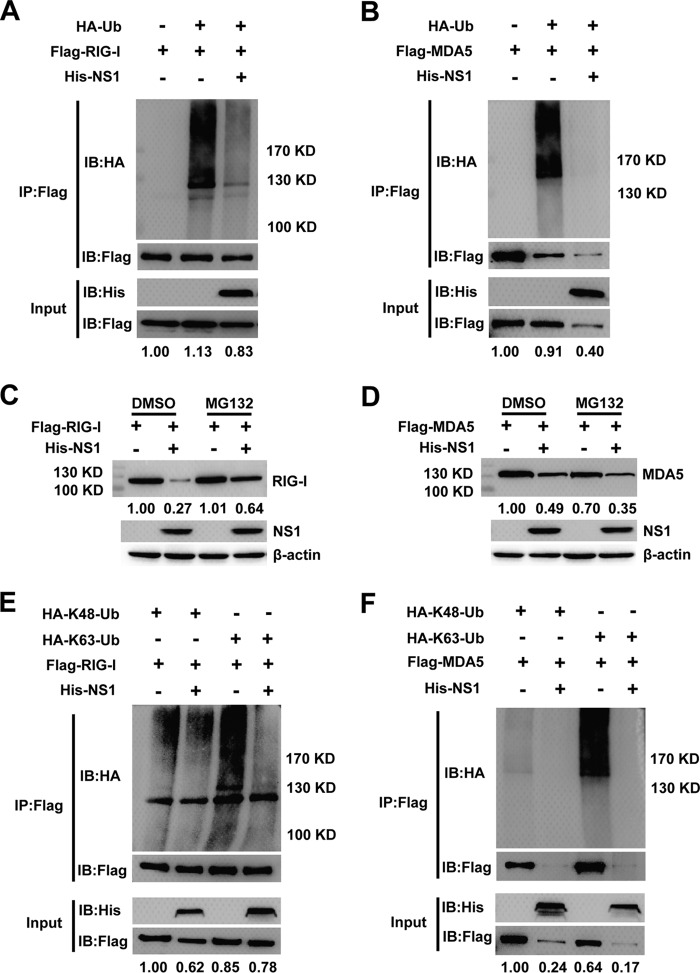
Ubiquitination is an important posttranslational modification in the regulation of innate immune signaling pathways. To clarify the role of NS1 in RIG-I and MDA5 ubiquitination, RIG-I and MDA5 plasmids were coexpressed with hemagglutinin (HA)-ubiquitin (Ub) and His-NS1 or an empty vector. RIG-I ubiquitination was easily detected when the cells were cotransfected with HA-tagged ubiquitin, and the ubiquitination was remarkably decreased when the cells were cotransfected with the NS1 plasmid (Fig. 8A). Meanwhile, the expression of RIG-I with cotransfected NS1 did not change when the cells were treated with the proteasome inhibitor MG132 rather than an empty vector (Fig. 8A and C). Degradation of RIG-I induced by NS1 was blocked by MG132, which indicated that NS1 may cause the degradation of RIG-I via the proteasome pathway (Fig. 8C). Although MDA5 ubiquitination was significantly decreased when the cells were cotransfected with NS1, the level of MDA5 protein was remarkably reduced when the cells were treated with MG132 (Fig. 8B and D).
FIG 8.
NS1 decreases K63-linked polyubiquitination of RIG-I. (A and B) Lysates from HEK-293T cells transiently cotransfected with FLAG-RIG-I/MDA5, His-NS1, and HA-Ub plasmids and treated with MG132 were subjected to immunoprecipitation with anti-FLAG Ab followed by an immunoblotting analysis. Anti-HA Ab was used to detect polyubiquitinated RIG-I/MDA5. (C and D) Immunoblot analysis of extracts of HEK-293T cells transfected with plasmid for FLAG-RIG-I/MDA5 and His-NS1 and treated with dimethyl sulfoxide (DMSO) or MG132. (E and F) Lysates from HEK-293T cells transiently cotransfected with FLAG-RIG-I/MDA5, His-NS1, and HA-K48-Ub or HA-K63-Ub plasmids and treated with MG132 were subjected to immunoprecipitation with anti-FLAG Ab followed by an immunoblotting analysis. Anti-HA Ab was used to detect polyubiquitinated RIG-I/MDA5. The expression level of RIG-I and MDA5 was quantified by carrying out a densitometric analysis using Image Lab software, and the data were normalized to the band densities of the first lane. One representative experiment out of two is shown.
Next, we examined whether NS1 specifically affects K48 and K63 polyubiquitination of RIG-I and MDA5. As shown in Fig. 8E, transfection of the ubiquitin mutant vectors HA-K48-Ub and HA-K63-Ub (two ubiquitin mutants retaining only a single lysine residue, K48 and K63, respectively) indicated that polyubiquitination of RIG-I involves both types of polyubiquitin chain and that NS1 greatly decreased the K63-linked polyubiquitination but not the K48-linked polyubiquitination of RIG-I. This indicates that NS1 specifically deubiquitinates K63-linked polyubiquitin chains from RIG-I. Similarly, polyubiquitination of MDA5 involves both the K48 and K63 polyubiquitin chains, and NS1 significantly decreased both polyubiquitin chains, but the MDA5 expression was also remarkably inhibited when the cells were treated with MG132 (Fig. 8F). In summary, these results suggest that NS1-mediated inhibition of the RLR signaling pathway involves inhibition of RIG-I K63-linked polyubiquitination and that proteasomes may play a role in RIG-I degradation, but the inhibition does not involve K48-linked polyubiquitination of RIG-I.
DISCUSSION
The IFN system is the first line of the host's defense against viral infection. Viruses have developed various strategies to disrupt the IFN signaling pathway. The viral IFN antagonists are usually multifunctional proteins, and their different properties may play different roles in the antagonization of IFN production at different stages of the virus replication cycle. Previous studies have shown that several nonstructural proteins encoded by flaviviruses antagonize the IFN response (9, 41–44). In the present study, we demonstrated that WNV NS1 inhibits the production of IFN-β by preventing downstream activation of the RLR signaling pathway. Our results showed that the WNV NS1 protein targets RIG-I and MDA5 by interacting directly with them and causing them to be degraded, leading, in turn, to the inhibition of both the activation of IRF3 and the expression of IFN-β. These results provide novel insights into the underlying mechanism by which WNV NS1 antagonizes the host antiviral response.
Regarding the interaction between a flavivirus infection and the host antiviral response, flaviviruses have developed various strategies to antagonize the host antiviral response. Previous studies have showed that flaviviruses can be recognized by TLRs and RLRs, which leads to the activation of type I IFN immune signaling pathways (45, 46). However, nonstructural flavivirus proteins can regulate the host innate immune response to improve the viral replication efficiency. For example, the flavivirus NS2A protein inhibits IFN-β promoter-driven transcription (9). The protease NS2B3 of DENV targets the human mediator of IRF3 activation (MITA) to subvert MITA-triggered antiviral signaling (41). The flavivirus NS4B protein blocks IFN-α/β signaling by preventing signal transducer and activator of transcription 1 and 2 (STAT1 and STAT2) phosphorylation and nuclear translocation (8, 10). The flavivirus NS5 of protein also plays an essential role in antagonizing type I IFN signaling by various means (7). WNV NS5 antagonizes host IFN-β antiviral responses by inhibiting the IFN-mediated Janus kinase (JAK)-STAT signaling pathway, including by preventing STAT1 phosphorylation and nuclear translocation (13). Furthermore, the NS4B protein of hepatitis C virus (which is closely genetically related to other flaviviruses in the family Flaviviridae) has been reported to suppress IFN-β production by directly interacting with stimulator of interferon genes (STING), thereby abrogating RIG-I-mediated type I IFN-dependent innate immunity (43). These findings suggest that flavivirus nonstructural proteins play crucial roles in the suppression of host antiviral responses. However, the precise underlying mechanisms are yet to be fully elucidated.
In the present study, we first analyzed the role of the WNV NS1 protein on IFN-β expression using luciferase reporter assays. Our results showed that WNV NS1 inhibits the production of IFN-β by targeting the RLR-mediated signaling pathway (Fig. 1A to C). The activation of the IFN-β promoter depends on IRF3 phosphorylation and nuclear translocation to allow it to bind to its target sequences (18, 37). Our data demonstrate that WNV NS1 inhibited the activity of the PRD(III-I)-Luc promoter [induced by transfected poly(I·C)] in a dose-dependent manner (Fig. 1D). In addition, WNV NS1 also blocked RIG-IN, MDA5, IPS1, TBK1, IKKε, and IRF3/5D-induced activation of IFN-β promoter, suggesting that NS1 inhibits the production of IFN-β by targeting upstream signaling molecules in the IRF3 signaling pathway (Fig. 2A to F). More importantly, IRF3 phosphorylation induced by RIG-I and MDA5, but not that induced by IPS1, TBK1, and IKKε, was inhibited in the presence of NS1 (Fig. 3C and D). These data suggest that WNV NS1 targets RIG-I and MDA5, leading to inhibition of IRF3 activation and IFN-β production.
Furthermore, the phosphorylation and nuclear translocation of IRF3 induced by SeV or poly(I·C) treatment were also blocked by WNV NS1 (Fig. 3A, B, and E). Comparisons of different types of truncated mutants indicated that the 181–352aa domain of NS1 is crucial for preventing the induction of the IFN-β promoter (Fig. 5A). Further studies should be conducted to identify the functional sites of NS1 involved in inhibition of IFN-β production. In addition, our data showed that the NS1 proteins encoded by JEV and DENV-2 failed to prevent the induction of the IFN-β promoter as they lacked the inhibitory effect of WNV NS1, indicating that the function of the enigmatic NS1 protein may not be universal in the genus Flavivirus (Fig. 5C).
The RLRs RIG-I and MDA5 (which are both members of the DExD/H box RNA helicase family) play major roles in sensing RNA virus infections and triggering antiviral responses (18). To drive type I IFN expression, the RLRs activate downstream transcription factors, which allow the host to control the virus infection using an intracellular immune response.
Meanwhile, viruses and their multifunctional viral proteins have developed various strategies to antagonize RLR-mediated immune responses. A number of studies have suggested that many viral proteins target RIG-I or MDA5 in order to antagonize the host IFN signaling pathway using various mechanisms. For example, one vital mechanism of influenza A virus NS1 for inhibiting the transcriptional activation of IFN-β is directly interacting with RIG-I (47). In addition, the 3C protein of enterovirus 71 suppresses RIG-I-mediated IRF3 activation and IFN-β induction associating with RIG-I (48). In addition, human respiratory syncytial virus encodes nonstructural protein NS2, which also antagonizes the transcriptional activation of IFN-β by directly interacting with RIG-I (49). Previous reports have also shown that the Z protein of New World arenaviruses can bind to RIG-I, leading to inhibition of the downstream activation of the RIG-I-mediated signaling pathway and interfering with IFN-β transcriptional induction (50). Furthermore, the V proteins of paramyxoviruses inhibit IFN-β promoter activation by targeting and binding to MDA5 directly (28). Interestingly, HSV-1 US11 acts as an antagonist of IFN-β induction, binding to RIG-I and MDA5 and inhibiting downstream activation of the RLR-mediated signaling pathway, leading to the prevention of IFN-β production (29). These reports suggested that these viruses have developed independent mechanisms to inhibit the activation of IFN-β by targeting RIG-I or MDA5.
In this study, the results suggest that overexpression of WNV NS1 was able to inhibit exogenous and endogenous RIG-I and MDA5 expression (Fig. 4A to F). In support of this, further analyses based on a BiFC assay and colocalization detection revealed that there is a potential interaction between WNV NS1 and the RIG-I and MDA5 proteins (Fig. 6B to E). The result of the co-IP assay further demonstrated that there is a direct association between NS1 and RIG-I/MDA5 (Fig. 7A to C). We speculate that the direct interactions of NS1 and RIG-I/MDA5 and the subsequent NS1-mediated inhibition of RIG-I/MDA5 are responsible for the inhibition of IRF3 activation and IFN-β induction. Moreover, our data suggest that NS1 greatly decreased the K63-linked polyubiquitination of RIG-I (Fig. 8E). Additionally, the degradation of RIG-I caused by cotransfected NS1 was clearly prevented when the cells were treated with MG132, indicating that proteasome may play a role in RIG-I degradation (Fig. 8C and E).
Ubiquitination is an important posttranslational modification in the regulation of host innate immune signaling pathways (51). RIG-I activation requires both RNA binding and ubiquitination involving K63-linked polyubiquitin chains, mediated by E3 ubiquitin ligases such as tripartite motif-containing protein 25 (TRIM25) and Riplet (52). The activated RIG-I interacts with MAVS to trigger the downstream signaling pathway. The unanchored K63-linked polyubiquitin chains can also activate MDA5 to trigger type I IFN signaling (53). Previous studies have shown that many viral proteins antagonize the IFN response by regulating RIG-I ubiquitination. The NS3-4A proteases of hepatitis C virus have been reported to target Riplet and inhibit RIG-I K63-linked polyubiquitination in order to suppress type I IFN production (54). There is evidence suggesting that the glycoprotein NS1 of WNV, including the secreted form, may block the activation of the IFN-β promoter by preventing IRF3 nuclear translocation via the TLR3 signaling pathway (31, 55). In the present study, our data suggest that WNV NS1 antagonizes the production of IFN-β by targeting RIG-I and MDA5 via the RLR signaling pathway. This indicates that, in order to suppress IFN-β activation, the enigmatic NS1 protein may target two receptor signaling pathways.
The NS1 protein of WNV, as an antagonist of IFN-β induction, may use multiple strategies to interrupt the host immune response. However, further studies should be carried out to develop a fuller understanding of the NS1 functions regarding the inhibition of the host antiviral response. In conclusion, we have demonstrated that the NS1 protein targets RIG-I and MDA5 by interacting with them and causing their degradation. Additionally, NS1 decreases the K63-linked polyubiquitination of RIG-I and subsequently blocks the downstream activation of the RLR signaling pathway. To our knowledge, this is the first time that the nonstructural flavivirus protein NS1 has been shown to interfere with the host immune system by targeting RIG-I and MDA5. These results provide insight into a novel mechanism by which WNV NS1 antagonizes the host antiviral response.
MATERIALS AND METHODS
Cells, viruses, and reagents.
HEK-293T, BHK-21, Vero, and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Gibco, USA) at 37°C in a 5% CO2 incubator. Sendai virus (SeV) was propagated in 12-day-old embryonated eggs and titrated using a hemagglutination assay.
Anti-RIG-I (3743), anti-MDA5 (5321), anti-MAVS (3993), anti-hemagglutinin (HA) tag (3724), anti-His tag (2366), and anti-p-IRF3 at Ser396 (4947s) antibodies (Abs) were purchased from Cell Signaling Technology (CST; Boston, MA); anti-IRF3 Ab was purchased from Proteintech (11312-1-AP) (China); anti-FLAG tag Ab was purchased from Sigma (F1804) (Sigma-Aldrich, MO, USA); anti-β-actin Ab was purchased from Santa Cruz Biotechnology (sc-81178) (Santa Cruz, CA, USA); and Cy3-conjugated goat anti-rabbit IgG (BA1032) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (BA1101) were purchased from Boster (China). Poly(I·C) was purchased from InvivoGen (tlrl-pic) (USA). The proteasome inhibitor MG132 (C2211) and N-ethylmaleimide (NEM) (E3876) were purchased from Sigma (Sigma-Aldrich, MO, USA).
Plasmids.
All the primers used for plasmid construction are listed in Table 1. The pcDNA3.1(+) expression plasmid encoding the WNV NS1 and expression plasmids involving the following four truncation mutants, which lacked specific amino acids, were constructed according to NS1 crystal structures (35): NS1-1–180aa, NS1-181–352aa, NS1-Δ(108–128aa) (lacking aa 108 to 128), and NS1-Δ(55–143aa) (lacking aa 55 to 143). The last 24 aa of the envelope protein was also included as a signal peptide sequence for NS1 expression. In addition, the NS1 expression plasmid and the four truncation mutant expression plasmids each contained a C-terminal His6 tag to facilitate their detection. The NS1 mutations were generated by overlap PCR using standard procedures.
TABLE 1.
Primers used in PCR for plasmid construction
| Gene | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| WNV | ||
| NS1 | GATAGGTCCATAGCTCTCACG | AGCATTCACTTGTGACTGCAC |
| NS1-1–180aa | GATAGGTCCATAGCTCTCACG | GTCACATTCAGTTGTGTTGCTC |
| Overlap PCR | ||
| NS1-181–352aa | GATAGGTCCATAGCTCTCACG | ATGATCTTCGAAGCGTGCACGTTCACGGAGAGG |
| ACGTGCACGCTTCGAAGATCATTGGAACGGCTG | AGCATTCACTTGTGACTGCACG | |
| NS1-Δ(108–128aa) | GATAGGTCCATAGCTCTCACG | GTGTTGTTGGCGGTGGCGGTGAGGCGTTTAGGT |
| TCACCGCCACCGCCAACAACACCTTTGTGGTTG | AGCATTCACTTGTGACTGCACG | |
| NS1-Δ(55–143aa) | GATAGGTCCATAGCTCTCACG | TTCTGAGTCGGCACTCCTTCCTTATGAGCTTTC |
| AGGAAGGAGTGCCGACTCAGAATCGCGCTTGGA | AGCATTCACTTGTGACTGCACG | |
| JEV and DV2 | ||
| JEV NS1 | GACCGATCAATTGCTTTGGCC | AGCATCAACCTGTGATCTGAC |
| DV2 NS1 | GCTAGCACCTCACTGTCTGTGTCA | GGCTGTGACCAAAGAGTTGAC |
| DV2 mC | GCTAATAACCAACGGAAAAAGGCG | TCTGCGTCTCCTGTTCAAG |
| BiFC assay constructs | ||
| V-N-NS1 | ACCATGGATAGGTCCATAGCT | ACCGGTGGAGCATTCACTTGTGACT |
| V-C-RIG-I | ACCATGACCACCGAGCAGCGACGC | ACCGGTGGTTTGGACATTTCTGCTG |
| V-C-MDA5 | ACCATGTCGAATGGGTATTCCACA | ACCGGTGGATCCTCATCACTAAAT |
| V-N-P53 | ACCATGGAGGAGCCGCAGTCA | ACCGGTGGGTCTGAGTCAGGCCCT |
| V-C-T-tag | ACCATGACTGATGAATGGGAGCAG | ACCGGTGGTGTTTCAGGTTCAGGGG |
The open reading frame encoding DENV-2 (DV2) mC was amplified by PCR and cloned into pcDNA3.1(+) expression vector with a His tag at the C terminus. For other constructs, the open reading frames encoding WNV NS1, DV2 NS1, and Japanese encephalitis virus (JEV) NS1 were amplified by PCR. These genes were each associated with the signal peptide sequence, and they were cloned into a pCAGGS expression vector with an HA tag at the C terminus. The WNV infectious cDNA clone pFLWNV (2) (GenBank no. AF404756) served as the template for the cloning the WNV NS1 gene. The infectious clones of pACYC-NGC (56) and pACYC-JEV-SA14 (57) served as the templates for the cloning of the DV2 and JEV NS1 genes, respectively. All the constructs were verified by DNA sequencing.
The yellow fluorescent protein Venus, which was obtained via multiple-site mutagenesis of enhanced yellow fluorescent protein, as previously reported (58), was used as a reporter in bimolecular fluorescence complementation (BiFC) experiments. Venus can be expressed as two separate proteins: the N-terminal amino acids (V-N, 1 to 173) and the C-terminal amino acids (V-C, 174 to 239) of the full length as reported previously (39). Expression plasmids V-N-NS1 and V-C-RIG-I/MDA5 were generated by inserting the coding sequences for WNV NS1 and RIG-I/MDA5 with the coding sequences for VN and VC, respectively. To assess the reliability of the Venus-based BiFC system, the positive-control plasmids V-N-p53 and V-C-T-tag were generated by inserting the coding sequences for p53 and T-tag to V-N and V-C (due to the well-known strong interaction of p53 and T-tag). Mutant p53 (mp53) and mutant T-tag (mT-tag) were used as negative controls (40). The negative-control plasmids V-N-mp53 and V-C-mT-tag were generated as described previously (59).
The expression plasmids for wild-type IRF3 (pIRES-hrGFP/IRF-3-FLAG) and its constitutively active mutant IRF3/5D (pIRES-hrGFP/IRF-3/5D-FLAG) were provided by Yi-Ling Lin (45). The PRD(III-I)-Luc reporter plasmid was provided by Stephan Ludwig (University of Muenster, Muenster, Germany) (60). The expression plasmid for the constitutively active RIG-I mutant (pEF-FLAG-RIG-IN) was provided by Takashi Fujita (Kyoto University, Kyoto, Japan) (61). The pcDNA3.1-TBK1-FLAG and pcDNA3.1-IKKε-FLAG expression plasmids were kindly provided by Katherine Fitzgerald (University of Massachusetts Medical School, Worcester, MA, USA) (37). The reporter plasmid p-IFN-β-Luc and internal control plasmid phRL-TK have been described previously (25).
Transfection and luciferase reporter assays.
HEK-293T cells plated in 24-well plates were cotransfected with reporter plasmid pIFN-β-luc or PRD(III-I)-Luc (125 ng), renilla luciferase plasmid pRL-TK (25 ng), and the relevant expression plasmids [pcDNA3.1-NS1, NS1-1–180aa, NS1-181–352aa, NS1-Δ(108–128aa), and NS1-Δ(55–143aa)] or an empty control plasmid. The transfections were performed by using calcium phosphate reagents. The cells were stimulated with 100 HA U ml−1 SeV for 16 h or transfected with 2 μg poly(I·C) using Lipofectamine 2000 (Invitrogen) for 24 h after transfection. The cells were lysed after stimulation, and the luciferase activity was determined using a the dual-luciferase reporter assay system (Promega) with a multifunction microplate reader (Varioskan Flash; Thermo Scientific) according to the manufacturer's instructions. The data were determined by normalization of the firefly luciferase activities to the renilla luciferase activities.
For overexpression of RIG-IN, MDA5, IPS1, TBK1, IKKε, or IRF3/5D, HEK-293T cells were transfected with 25 ng of the plasmids encoding the inducers, 125 ng of the reporter plasmids [pIFN-β-Luc or PRD(III-I)-Luc], 25 ng of the pRL-TK plasmid, and 500 ng of the relevant WNV NS1 expression plasmids or an empty vector. At 36 h transfection, the cells were lysed and luciferase activities were measured.
Immunoblotting analysis.
For the transient-transfection experiments, HEK-293T cells were transfected with the plasmids in a 3.5-cm dish. The whole-cell protein extracts were prepared after transfection or stimulation with appropriate stimulators with 200 μl lysis buffer containing 1% Triton X-100, 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF) for 10 min on ice. The lysates were boiled at 95°C for 10 min and then subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto polyvinylidene difluoride (PVDF) membranes (0.2 μm; Millipore), followed by blocking with 5% nonfat milk in Tris-buffered saline with Tween 20 (TBST; 50 mM Tris-HCl, 150 mM NaCl, and 0.1% Tween 20, at pH 7.4) for 1 h at room temperature. The blots were then probed with appropriate primary Abs at room temperature (RT) for 1.5 h. After being washed three times with TBST, the blots were incubated with goat anti-mouse IgG or goat anti-rabbit IgG conjugated with horseradish peroxidase (Bio-Rad) at RT for 1 h. Protein bands were visualized with a chemiluminescence system (ChemiDoc; Bio-Rad) after the chemiluminescent substrate was added (Millipore).
Immunofluorescence microscopy.
For the colocalization assay, Vero cells grown on 35-mm glass-bottom plates were cotransfected with plasmids pCAGGS-HA-WNV NS1 and FLAG-RIG-I/MDA5. At 36 h after transfection, the cells were washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100 for 15 min at RT. For the immunofluorescence assay to detect the effect of WNV NS1 on IRF3 nuclear translocation, HeLa cells grown on glass-bottom plates were transfected with a pCAGGS-NS1-HA plasmid or an empty vector. At 24 h after transfection, the cells were treated with or without SeV (100 HA U ml−1) for 12 h and then fixed and permeabilized as described above. After being washed three times in PBS, cells were incubated in PBS containing 5% bovine serum albumin at 4°C overnight and then incubated with both mouse anti-HA Ab and rabbit anti-FLAG Ab or rabbit anti-IRF3 polyclonal Ab (PAb) at a dilution of 1:100 for 2 h at RT. After three washes with PBS, the cells were incubated with Cy3-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse IgG at a dilution of 1:100 for 1 h at RT. The cells were then washed with PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min at RT. After three washes with PBS, the cells were observed under a fluorescence microscope (Nikon A1 MP Storm; Japan).
BiFC imaging.
For the BiFC experiments, HEK-293T cells were cotransfected with V-N-NS1 and V-C-RIG-I/MDA5 plasmids and incubated at 37°C for 12 h as described above. To assess the reliability of the Venus-based BiFC system, p53 and T-tag were selected as positive controls due to their well-known strong interactions. In addition, mutant p53 (mp53) and mutant T-tag (mT-tag) were used as negative controls (40). The positive controls, V-N-p53 and V-C-T-tag, were cotransfected into HEK-293T cells and incubated as described above. In addition, HEK-293T cells were cotransfected with V-N-mp53 and V-C-mT-tag plasmids as negative controls and incubated at 37°C for 12 h as described above. The fluorescent images generated by BiFC in the cells were visualized using an inverted Delta Vision Restoration microscopy system (Applied Precision, WA, USA) with excitation and emission wavelengths of 500/24 nm and 542/27 nm, respectively.
Co-IP assay.
HEK-293T cells in 10-cm culture dishes were cotransfected with a plasmid expressing FLAG-tagged RIG-I/MDA5 and an HA-tagged NS1 expression plasmid or an empty vector for 36 h. The cells were lysed on ice for 20 min in 600 μl lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, and 1% Triton X-100) containing a protease inhibitor mixture plus the protease inhibitor phenylmethylsulfonyl fluoride (PMSF). The cell lysates were then immunoprecipitated overnight at 4°C with mouse anti-FLAG Ab or rabbit anti-HA Ab and protein G agarose (Millipore, USA) or protein A/G agarose beads (Beyotime, China). Mouse or rabbit normal IgG (Beyotime) was used as a negative control. The immunoprecipitates were washed four times with phosphate-buffered saline with Tween 20 (PBST) and then subjected to an immunoblotting analysis.
Ubiquitination assays.
To analyze the ubiquitination of RIG-I/MDA5, HEK-293T cells were cotransfected with FLAG-RIG-I/MDA5, HA-Ub, or HA-Ub mutants and His-NS1 and then treated with the proteasome inhibitor MG132 for 4 h. The cells were washed twice in PBS supplemented with 10 mM NEM and lysed with 1% SDS lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 1% SDS) containing the protease inhibitor PMSF and 10 mM NEM. The cell lysates were then boiled for 5 min, diluted to 0.1% SDS with immunoprecipitation buffer (20 mM Tris [pH 7.5], 150 mM NaCl, and 1% Triton X-100), and incubated overnight with mouse anti-FLAG Ab and then incubated with protein G agarose (Millipore) for 3 h at 4°C. After washing three times with lysis buffer, the immunoprecipitates were boiled at 95°C for 10 min and subjected to an immunoblotting analysis.
ACKNOWLEDGMENTS
We thank the Core Facility and Technical Support staff at Wuhan Institute of Virology and Wuhan Key Laboratory on Emerging Infectious Diseases and Biosafety for their helpful support during the course of the work.
This work was supported by the National Key Research and Development Program of China (2016YFD0500400), and the Key Program of the Chinese Academy of Sciences (ZDRW-ZS-2016-4). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Suthar MS, Aguirre S, Fernandez-Sesma A. 2013. Innate immune sensing of flaviviruses. PLoS Pathog 9:e1003541. doi: 10.1371/journal.ppat.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol 76:5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suthar MS, Diamond MS, Gale M Jr. 2013. West Nile virus infection and immunity. Nat Rev Microbiol 11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 4.Lazear HM, Diamond MS. 2014. New insights into innate immune restriction of West Nile virus infection. Curr Opin Virol 11C:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Dong H, Zhou Y, Shi PY. 2008. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5′ stem-loop of genomic RNA. J Virol 82:7047–7058. doi: 10.1128/JVI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Dong H, Ye H, Tilgner M, Shi PY. 2010. Genetic analysis of West Nile virus containing a complete 3′CSI RNA deletion. Virology 408:138–145. doi: 10.1016/j.virol.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Shi PY. 2014. Flavivirus NS5 prevents the inSTATement of IFN. Cell Host Microbe 16:269–271. doi: 10.1016/j.chom.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A 100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J Virol 78:12225–12235. doi: 10.1128/JVI.78.22.12225-12235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol 79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol 80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol Rev 227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 17.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Loo YM, Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. 2012. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo H, Zhang Z, Zheng Z, Ke X, Zhang X, Li Q, Liu Y, Bai B, Mao P, Hu Q, Wang H. 2013. Human bocavirus VP2 upregulates IFN-beta pathway by inhibiting ring finger protein 125-mediated ubiquitination of retinoic acid-inducible gene-I. J Immunol 191:660–669. doi: 10.4049/jimmunol.1202933. [DOI] [PubMed] [Google Scholar]
- 21.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. 2014. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. 2007. Binding of Kaposi's sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol 81:10950–10960. doi: 10.1128/JVI.00183-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cloutier N, Flamand L. 2010. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J Biol Chem 285:7208–7221. doi: 10.1074/jbc.M109.018838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Zheng Z, Luo H, Meng J, Li H, Li Q, Zhang X, Ke X, Bai B, Mao P, Hu Q, Wang H. 2012. Human bocavirus NP1 inhibits IFN-beta production by blocking association of IFN regulatory factor 3 with IFNB promoter. J Immunol 189:1144–1153. doi: 10.4049/jimmunol.1200096. [DOI] [PubMed] [Google Scholar]
- 26.Saira K, Zhou Y, Jones C. 2007. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol 81:3077–3086. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. 2011. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 85:11079–11089. doi: 10.1128/JVI.05098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A 101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J Virol 86:3528–3540. doi: 10.1128/JVI.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A 103:19111–19116. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson JR, de Sessions PF, Leon MA, Scholle F. 2008. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol 82:8262–8271. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youn S, Cho H, Fremont DH, Diamond MS. 2010. A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition. J Virol 84:9516–9532. doi: 10.1128/JVI.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison CR, Scholle F. 2014. Abrogation of TLR3 inhibition by discrete amino acid changes in the C-terminal half of the West Nile virus NS1 protein. Virology 456-457:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baronti C, Sire J, de Lamballerie X, Querat G. 2010. Nonstructural NS1 proteins of several mosquito-borne flavivirus do not inhibit TLR3 signaling. Virology 404:319–330. doi: 10.1016/j.virol.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 35.Akey DL, Brown WC, Dutta S, Konwerski J, Jose J, Jurkiw TJ, DelProposto J, Ogata CM, Skiniotis G, Kuhn RJ, Smith JL. 2014. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343:881–885. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 37.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 38.Ramos HJ, Gale M Jr. 2011. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shyu J, Liu H, Deng X, Hu C-D. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 40.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. 2006. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev 20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, Shu HB, Zhong J. 2013. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol 59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S, Tasaka-Fujita M, Asahina Y, Yoneyama M, Fujita T, Watanabe M. 2013. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 44.Dalrymple NA, Cimica V, Mackow ER. 2015. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. mBio 6:e00553-15. doi: 10.1128/mBio.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang TH, Liao CL, Lin YL. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect 8:157–171. doi: 10.1016/j.micinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M Jr. 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol 82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei X, Liu X, Ma Y, Sun Z, Yang Y, Jin Q, He B, Wang J. 2010. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J Virol 84:8051–8061. doi: 10.1128/JVI.02491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling Z, Tran KC, Teng MN. 2009. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol 83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan L, Briese T, Lipkin WI. 2010. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol 84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhoj VG, Chen ZJ. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 52.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oshiumi H, Miyashita M, Matsumoto M, Seya T. 2013. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog 9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crook KR, Miller-Kittrell M, Morrison CR, Scholle F. 2014. Modulation of innate immune signaling by the secreted form of the West Nile virus NS1 glycoprotein. Virology 458-459:172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. 2013. Membrane topology and function of dengue virus NS2A protein. J Virol 87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li XD, Li XF, Ye HQ, Deng CL, Ye Q, Shan C, Shang BD, Xu LL, Li SH, Cao SB, Yuan ZM, Shi PY, Qin CF, Zhang B. 2014. Recovery of a chemically synthesized Japanese encephalitis virus reveals two critical adaptive mutations in NS2B and NS4A. J Gen Virol 95:806–815. doi: 10.1099/vir.0.061838-0. [DOI] [PubMed] [Google Scholar]
- 58.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 59.Fan JY, Cui ZQ, Wei HP, Zhang ZP, Zhou YF, Wang YP, Zhang XE. 2008. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem Biophys Res Commun 367:47–53. doi: 10.1016/j.bbrc.2007.12.101. [DOI] [PubMed] [Google Scholar]
- 60.Ehrhardt C, Kardinal C, Wurzer WJ, Wolff T, von Eichel-Streiber C, Pleschka S, Planz O, Ludwig S. 2004. Rac1 and PAK1 are upstream of IKK-epsilon and TBK-1 in the viral activation of interferon regulatory factor-3. FEBS Lett 567:230–238. doi: 10.1016/j.febslet.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 61.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]