ABSTRACT
Syncytin genes are envelope genes of retroviral origin that have been exapted for a role in placentation. They are involved in the formation of a syncytial structure (the syncytiotrophoblast) at the fetomaternal interface via their fusogenic activity. The mouse placenta is unique among placental mammals since the fetomaternal interface comprises two syncytiotrophoblast layers (ST-I and ST-II) instead of one, as observed in humans and all other hemochorial placentae. Each layer specifically expresses a distinct mouse syncytin, namely, syncytin-A (SynA) for ST-I and syncytin-B (SynB) for ST-II, which have been shown to be essential to placentogenesis and embryo survival. Their cognate cellular receptors, which are necessary to mediate cell-cell fusion and syncytiotrophoblast formation, are still unknown. By devising a sensitive method that combines a cell-cell fusion assay with the screening of a mouse cDNA library, we succeeded in identifying the glycosylphosphatidylinositol (GPI)-anchored membrane protein lymphocyte antigen 6E (Ly6e) as a candidate receptor for SynA. Transfection of cells with the cloned receptor led to their fusion to cells expressing SynA, with no cross-reactive fusion activity with SynB. Knocking down Ly6e greatly reduced SynA-induced cell fusion, thus suggesting that Ly6e is the sole receptor for SynA in vivo. Interaction of SynA with Ly6e was further demonstrated by a competition assay using the soluble ectodomain of Ly6e. Finally, reverse transcription-quantitative PCR (RT-qPCR) analysis of Ly6e expression on a representative panel of mouse tissues shows that it is significantly expressed in the mouse placenta together with SynA.
IMPORTANCE Syncytin genes are envelope genes of endogenous retroviruses, co-opted for a physiological function in placentation. Syncytins are fusogenic proteins that mediate cell-cell fusion by interacting with receptors present on the partner cells. Here, by devising a sensitive in vitro fusion assay that enables the high-throughput screening of normalized cDNA libraries, we identified the long-sought receptor for syncytin-A (SynA), a mouse syncytin responsible for syncytiotrophoblast formation at the maternofetal interface of the mouse placenta. This protein, Ly6e (lymphocyte antigen 6E), is a GPI-anchored membrane protein, and small interfering RNA (siRNA) experiments targeting its deletion as well as a decoy assay using a recombinant soluble receptor show that Ly6e is the necessary and sufficient partner of SynA. Its profile of expression is consistent with a role in both ancestral endogenization of a SynA founder retrovirus and present-day placenta formation. This study provides a powerful general method to identify genes involved in cell-cell fusion processes.
KEYWORDS: endogenous retrovirus, envelope protein, mouse, placenta, receptor, syncytin
INTRODUCTION
The sequencing of mammalian genomes has shown that sequences from retroviral origins occupy a significant portion: about 8% of the human genome and 10% of the mouse genome (1, 2). Most of these endogenous retroviral sequences are not functional as the result of the accumulation of mutations and genome rearrangements. However, among the functional ones, syncytin genes are envelope (env) genes from ancestrally endogenized retroviruses that have been co-opted for a role in placenta formation (reviewed in reference 3). Their expression at the cell surface induces cell-cell fusion, and they are involved in the formation of a syncytial structure at the interface between fetal and maternal blood, the syncytiotrophoblast. The syncytin genes have been found in all major clades of placental mammals and result from independent captures of env genes in each lineage where they have been identified (4–11). The syncytin genes display conserved characteristics from env genes but also lineage-specific differences which correlate with placental structure differences.
The structure of the mouse placenta is unique among placental mammals: at variance with all other described placentae, the fetomaternal interface comprises two syncytiotrophoblast layers (ST-I and ST-II) instead of one, as observed in humans and all other hemochorial placentae (12). Each of these syncytiotrophoblast layers expresses a different syncytin: syncytin-A (SynA) for ST-I and syncytin-B (SynB) for ST-II (13). Both syncytins have been demonstrated to be required for placenta development, with altered structures of the fetomaternal interface in knockout (KO) mice (14, 15). SynA KO mice display the most severe phenotype, resulting in death of embryos at midgestation.
By analogy with other retroviral Envs, syncytin-induced cell-cell fusion is thought to be mediated by the interaction of the syncytin with a specific membrane receptor expressed on neighboring cells (16). In this model, syncytin-mediated cell-cell fusion is initiated when the surface subunit (SU) of the syncytin glycoprotein binds to a specific receptor expressed on the surface of a neighboring cell. Attachment induces a series of conformational changes of the SU and transmembrane (TM) subunits of the syncytin, which results in the fusion of the plasma membranes.
It was previously demonstrated that SynA and -B overexpression induces cell-cell fusion ex vivo (10). As SynA and -B overexpression does not induce fusion in the same cell lines, it was hypothesized that their cellular receptors had to be distinct (10). Among the 11 mammalian syncytins characterized so far, the receptors for human syncytin-1 and -2 (human ASCT2/SLC1A5/RDR and human MFSD2/NLS1, respectively) and rabbit syncytin-Ory1 (rabbit ASCT2) have been identified by using pseudotyping assays (5, 17, 18). Although SynA and -B were first identified more than 10 years ago, the corresponding receptors have not been identified yet. This was mainly due to the difficulty in assessing interactions between membrane proteins, the impossibility of generating functional pseudotypes with SynA or -B (data not shown), and the lack of appropriate assays to screen cDNA libraries for cell-cell fusion per se. To address these challenges, we report here the development of a new technique to screen a cDNA expression library using a cell-cell fusion assay based on α-complementation of β-galactosidase. The β-galactosidase can be split into two fragments, an N-terminal α-peptide (Galα) and a C-terminal ω-peptide (Galω). These fragments are enzymatically inactive separately but complement each other and recover enzymatic activity when mixed together (19, 20). We applied this α-complementation property to perform a large-scale screen of cDNAs in order to identify the specific receptor that is required for SynA-dependent cell-cell fusion. In this study, we identify the membrane protein Ly6e as the receptor for SynA. Its role in cell-cell fusion in cellulo is then investigated, as well as its ability to interact with SynA and its tissue expression profile.
RESULTS
Development of a cell fusion-based screening method.
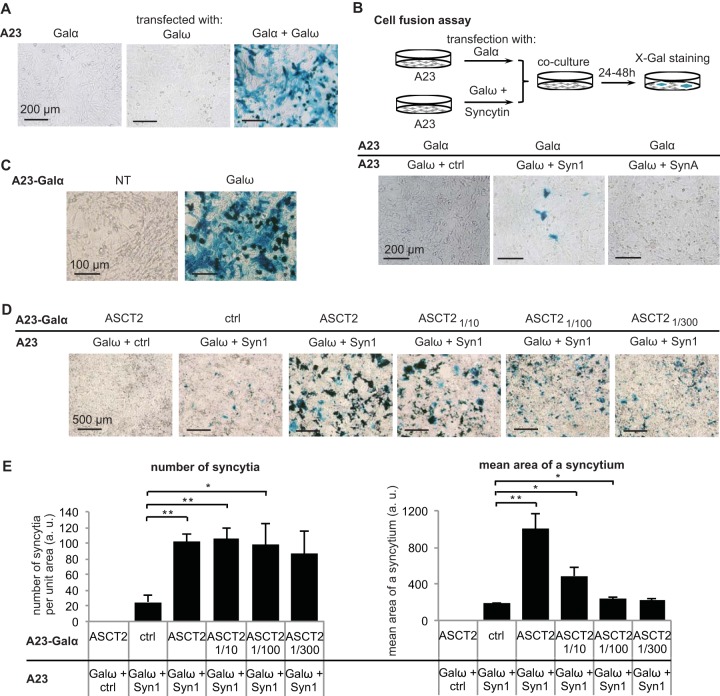
Given that SynA does not generate functional pseudotypes (probably due to improper incorporation into viral particles), we searched for a cell-cell fusion assay which would be simple and sensitive enough to screen a cDNA library. α-Complementation of β-galactosidase was previously used to detect HIV envelope glycoprotein-mediated cell fusion (19). We first checked that α-complementation was possible in our model cell lines. To do so, hamster A23 or human 293T cells were transiently transfected with expression vectors which expressed the β-galactosidase fragment Galα or Galω or both fragments (Fig. 1A and data not shown). Cells transfected with the Galα or Galω expression vector remained unstained after 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining, whereas 60 to 95% of the cells (depending on cell transfection efficiency) displayed blue staining when cotransfected with both the Galα and Galω expression vectors.
FIG 1.
Syncytin-mediated cell-cell fusion assay: detection, quantification, and sensitivity. (A) α-Complementation of β-galactosidase was tested in A23 cells by independently transfecting or cotransfecting Galα and Galω expression vectors and analyzing β-galactosidase activity after fixation and X-Gal staining at 24 h posttransfection. (B) Syn1 and SynA fusogenicity was tested in A23 cells by a cell-cell fusion assay as shown by the scheme. (C) Stable expression of Galα in an A23-Galα cell clone was assessed by X-Gal staining at 24 h after transfection of Galω expression vector. NT, not transfected. (D and E) ASCT2/Syn1 partnership was tested with various ratios of ASCT2 expression vector mixed with a control empty vector by a cell-cell fusion assay as described for panel B using A23 and A23-Galα cells. Representative images of at least three independent experiments are shown in panels A to D. (E) The number of syncytia and the mean area of a syncytium were assessed using ImageJ software on microscope images. Data are the means ± SEM (four independent experiments; *, P < 0.05; **, P < 0.01, Student's t test). a.u., arbitrary units.
We then checked that the α-complementation assay was sensitive enough to detect syncytin-induced cell-cell fusion. We used human syncytin-1 (Syn1), which induces fusion when overexpressed in A23 cells that naturally express the Syn1 receptor, i.e., ASCT2. We transfected a first group of A23 cells with the Galω expression vector together with a vector expressing Syn1 or an empty vector and a second group of A23 cells with Galα. At 1 day posttransfection, the two groups of cells were mixed. After 2 days of coculture, the cells were stained with X-Gal to detect blue syncytia formed following fusion of cells from both groups. Blue syncytia could indeed be detected under the condition where Syn1 was present, whereas none were detected with a control empty vector (Ctrl) (Fig. 1B).
We have shown previously that SynA ectopic expression induces cell-cell fusion in various cell lines from human, mouse, or monkey (including 293T and HeLa cells), suggesting that its receptor is expressed in these particular cells (10). To identify SynA receptor(s), we searched for a cell line where ectopic expression of SynA is not fusogenic, suggesting that its receptor is not significantly expressed. Using the α-complementation fusion assay, we found that SynA, in contrast to human Syn1, was not fusogenic per se in the A23 cell line (Fig. 1B).
In order to screen a cDNA library, we constructed a subclone of A23 cells stably expressing Galα (A23-Galα) in >95% of cells to avoid having to cotransfect Galα when transfecting plasmids from the library. We checked that, when transiently transfected with Galω, this A23-Galα subclone displayed blue staining after X-Gal treatment (Fig. 1C). We then tested whether the cell-cell fusion assay using the A23-Galα cells was sensitive enough to detect the fusogenic effect of a known syncytin/receptor pair even when the receptor was ectopically expressed at low levels. The sodium-dependent neutral amino acid transporter type 2, ASCT2, was previously identified as the receptor for syncytin-1 (5). A23-Galα cells transfected with an ASCT2 expression vector mixed with a control empty vector (Ctrl) at various ratios (ASCT21/10 [1:10 dilution of ASCT2], ASCT21/100, and ASCT21/300) were cocultured with A23 cells transfected with Galω and Syn1 expression vectors. After X-Gal staining, we analyzed two different parameters to measure the amount of cell-cell fusion induced by the Syn1/ASCT2 pair under each condition: the number of syncytia per surface unit and the mean area of a syncytium (Fig. 1D and E). We found that even with a 100-fold dilution of ASCT2, the number of syncytia per surface unit and the mean area of a syncytium were both significantly higher than the values obtained with a control empty vector. For the rest of this study, we chose to use mainly the number of blue syncytia per surface area as this parameter was the most simply measured to assess the amount of cell-cell fusion. However, we noted that when the percentage of syncytial area was high (Fig. 1E, ASCT2/Syn1 and ASCT21/10/Syn1 conditions), the number of syncytia per surface reached a plateau and was less representative of the amount of cell-cell fusion in this case than the mean area of a syncytium.
Screening of a mouse cDNA library using the cell-cell fusion assay.
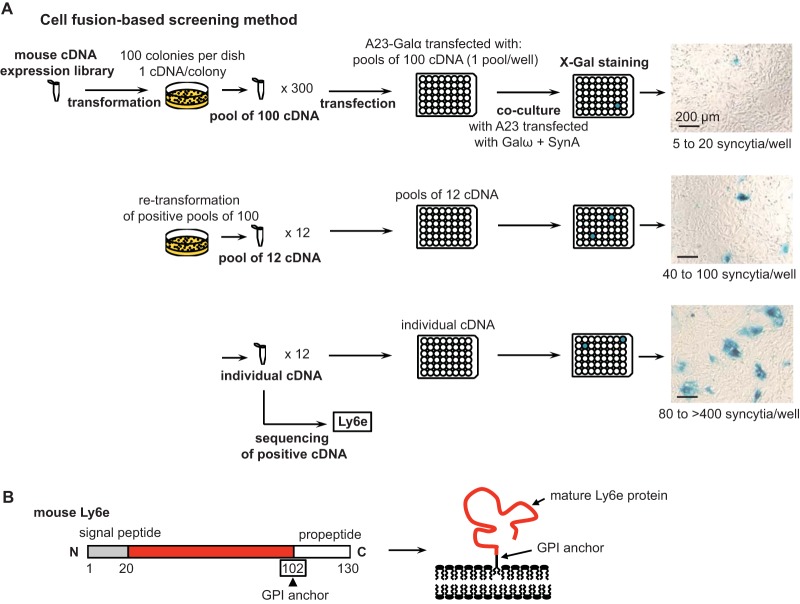
To search for the receptor for SynA, we used a normalized cDNA library prepared from mouse brain tissues, which are known among all tissues to express a wide range of genes (21). Given the results of the dilution testing, we reasoned that we would have a good signal-to-noise ratio even if the cDNA of the SynA receptor was diluted 100 times among other cDNAs. We therefore prepared pools of 100 cDNAs from the mouse library and tested their ability to induce fusion in the presence of SynA using the cell-fusion assay. A23-Galα cells transfected with a pool of 100 cDNAs were cocultured with an equal number of A23 cells transfected with SynA and Galω. After 2 days of coculture, the number of blue spots was analyzed. A total of 300 different pools of 100 cDNAs each were tested (Fig. 2A). Among them, only three pools displayed a number of blue spots per surface unit that was reproducibly higher than that of the control condition (Fig. 2A, top right photo). We then subcloned each positive pool of 100 cDNAs into pools of 12 cDNAs and tested them using our cell-cell fusion assay. Positive pools (Fig. 2A, middle right photo) were then subcloned into individual cDNA clones. As expected, the number of syncytia per surface unit increased at each subcloning step for the positive pools (Fig. 2A, right panels). After sequencing, all positive clones contained the same open reading frame (ORF). This ORF corresponds to a previously characterized gene encoding the lymphocyte antigen 6E (Ly6e), also known as thymic shared antigen 1 (Tsa-1) or stem cell antigen 2 (Sca-2). As expected for partners of SynA, Ly6e is a membrane protein. It belongs to a category of cell surface proteins which interact with the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor. Here, the GPI anchor is attached to alanine 102 of mature Ly6e (Fig. 2B) after cleavage of the C-terminal propeptide (amino acids 103 to 130).
FIG 2.
cDNA expression library screening to identify the syncytin-A partner of fusion. (A) Scheme of the screening of a mouse cDNA expression library by cell-cell fusion, which resulted in the identification of Ly6e as the potential receptor of SynA. Representative cell images of each step of the screening are shown with the range of syncytium numbers per well. (B) Scheme of mouse immature and mature Ly6e protein. In the mature form, the extracellular domain (in red) is attached to the plasma membrane by a GPI anchor on the amino acid residue boxed in the immature form after cleavage of the signal peptide (in gray) and the propeptide sequences (in white).
Ly6e is the specific partner of SynA in cellulo.
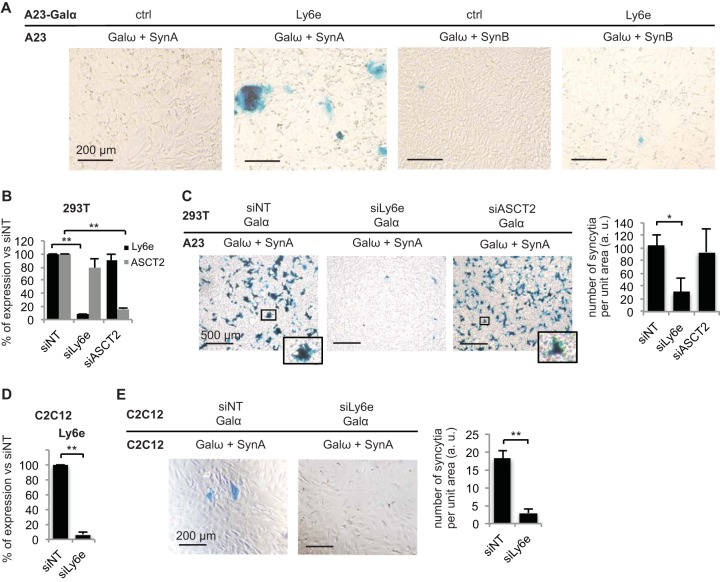
As the two mouse syncytins SynA and SynB are structurally related, with 67% identity between the two proteins, we tested whether the identified candidate receptor for SynA was also able to induce cell-cell fusion in the presence of SynB. Using the cell-cell fusion assay, we showed that, in contrast to results with cells expressing SynA, mixing A23 cells expressing SynB and Galω with A23-Galα cells expressing Ly6e does not increase the number of syncytia compared to that under the control condition without the candidate receptor (Fig. 3A). Thus, Ly6e is specific to SynA and not SynB, consistent with the fact that their overexpression does not induce fusion in the same cell lines (10).
FIG 3.
Ly6e is necessary and sufficient for syncytin-A mediated cell-cell fusion and is not a syncytin-B receptor. The cell-cell fusion assay was used to assess the role of Ly6e in cellulo and the specificity of the SynA/Ly6e partnership. (A) The specificity of the Ly6e partnership with SynA and not SynB was assessed by transfecting the proteins as indicated in A23 and A23-Galα cells and analyzing the formation of syncytia (presence or absence) after coculture of the two groups of cells and subsequent X-Gal staining. (B to E) The role of endogenous Ly6e in SynA-induced cell fusion was tested in human 293T embryonic kidney cells and in mouse C2C12 undifferentiated myoblasts, as indicated, by knocking down Ly6e by specific siRNAs (siLy6e) and analyzing the number of SynA-induced syncytia in comparison to those formed with nontargeting (siNT) or ASCT2 (siASCT2)-specific siRNAs. siRNA efficiency was verified by RT-qPCR (B and D), and the number of syncytia per well was analyzed by ImageJ (C) or by visual counting (E). Representative images of at least three independent experiments are shown. Data are the means ± SEM (at least three independent experiments; *, P < 0.05; **, P < 0.01, Student's t test).
As we have previously shown that SynA ectopic expression is fusogenic per se in cell lines from different species, including the easily transfectable 293T cells where SynA induces massive cell-cell fusion (10), we tested whether Ly6e was involved in cellulo in SynA-induced cell-cell fusion. We first checked that Ly6e was endogenously expressed in 293T cells and knocked it down by specific small interfering RNAs (siRNAs) (Fig. 3B). Using the cell-cell fusion assay, we then analyzed the number of syncytia per surface unit after coculture with Chinese hamster A23 cells transfected with the SynA expression vector (Fig. 3C). Interestingly, we observed that SynA-induced cell-cell fusion greatly decreases when Ly6e is knocked down by siRNA in comparison to expression with a control nontargeted siRNA or with an siRNA targeting the human syncytin-1 receptor, namely, ASCT2 (Fig. 3B and C). This result demonstrates that human Ly6e (hLy6e) is the main receptor for SynA in cellulo, at least in 293T cells. This result was also true in other human cell lines, including HeLa and U-2 OS cells (data not shown). Using the same methodology, we knocked down Ly6e in a mouse cell line where it is endogenously expressed, i.e., the C2C12 myoblast cell line (Fig. 3D), and demonstrated that mouse Ly6e (mLy6e) is also the main receptor for SynA in mouse cells (Fig. 3E).
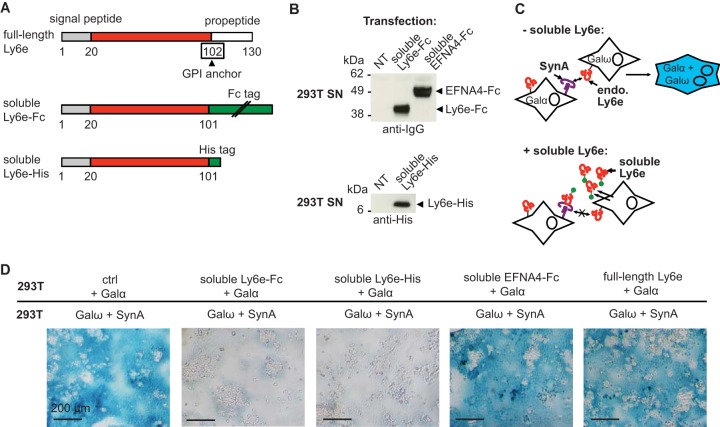
To further investigate whether the role of Ly6e in SynA-induced cell-cell fusion involves an interaction with SynA, we constructed an expression vector for an Fc-tagged soluble form of mouse Ly6e (Ly6e-Fc) (Fig. 4A). In this construct, Ly6e is truncated just before the alanine residue 102, where the GPI anchor is normally attached, and tagged with an Fc tag at the C terminus. We checked that this protein is secreted in the supernatant of transfected cells (Fig. 4B). We then tested whether this soluble form of Ly6e acts as a competitor for endogenous membrane-bound Ly6e, as follows. A first group of 293T cells was transfected with Ly6e-Fc and Galα vectors, whereas the second group of cells was transfected with SynA and Galω (Fig. 4C and D). We used as a negative control ephrin A4 (EFNA4-Fc), another Fc-tagged soluble protein. EFNA4 was previously identified as a GPI-anchored receptor for the envelope of the mouse IAPE (intracisternal A-type particles elements with an envelope) endogenous retrovirus (22). If the ectopically expressed soluble form of Ly6e is still able to bind to SynA, it will compete with the endogenous full-length Ly6e expressed at the membranes of 293T cells (Fig. 4C). This competition should result in a decrease in fusion of the two groups of cells, which would lead to a decrease in blue staining (Fig. 4C). Indeed, we observed that the soluble Ly6e-Fc but not the control EFNA4-Fc inhibits the fusion of the two groups of cells (Fig. 4D). This inhibitory effect is not due to the Fc tag because another soluble form of Ly6e with a polyhistidine tag (Ly6e-His) is also able to inhibit SynA-induced cell-cell fusion (Fig. 4D). Of note, we still observed unstained syncytial structures in the presence of Ly6e-Fc or Ly6e-His, which likely resulted from the fusion that took place inside the group of cells transfected with SynA and Galω before the mixing with the second group of cells expressing Galα and soluble Ly6e.
FIG 4.
Inhibition of syncytin-A mediated cell-cell fusion by soluble Ly6e recombinant proteins. (A) Scheme of mouse full-length immature Ly6e and soluble Fc- and His-tagged Ly6e. The signal peptide is in gray, the propeptide is in white, the extracellular domain is in red, the tags are in green, and the amino acid to which the GPI anchor is attached is boxed. (B) Supernatants of 293T cells untransfected or transfected with the indicated expression vectors were analyzed by Western blotting using anti-IgG and anti-His antibodies. NT, not transfected. (C) Scheme of the expected results of coculturing a first group of 293T cells overexpressing SynA and Galω and a second group overexpressing either Galα alone or Galα and a secreted soluble form of Ly6e. In the absence of soluble Ly6e, endogenous Ly6e interacts with SynA, which leads to cell-cell fusion and blue-stained syncytia; conversely, the presence of secreted soluble Ly6e will compete with endogenous Ly6e for SynA interaction, resulting in decreased formation of blue syncytia. (D) The decoy effect of soluble Ly6e (Ly6e-His and Ly6e-Fc) in comparison to an irrelevant Fc-tagged soluble receptor (EFNA4-Fc), the full-length Ly6e, or a control empty vector (ctrl) was investigated by a cell-cell fusion assay using 293T cells transfected with the indicated expression vectors. Representative images of X-Gal-stained cells are shown.
To summarize, the above data strongly suggest that Ly6e is the receptor for SynA involved in cell-cell fusion and that an interaction between the two membrane proteins is required to mediate fusion.
Ly6e is expressed in the mouse placenta.
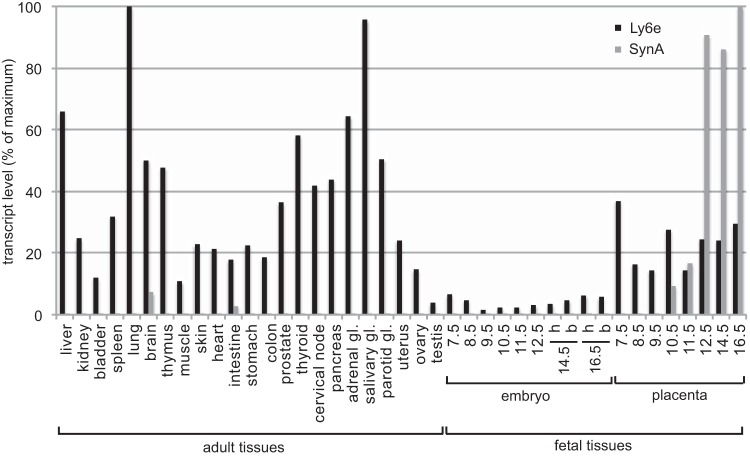
As a candidate receptor for SynA, Ly6e is expected to be expressed in the mouse placenta where SynA is involved in cytotrophoblast fusion into syncytiotrophoblasts. We thus investigated Ly6e expression by quantitative reverse transcription-PCR (RT-PCR) in a large panel of mouse adult tissues as well as fetal tissues, including the placenta. We observe that Ly6e is rather ubiquitously expressed in mouse adult tissues, with maximal expression in the lung and the salivary gland, in contrast with SynA levels, which are low in the same adult tissues (Fig. 5). The expression of Ly6e is strikingly lower in the fetal tissues tested, except for the placenta: at day 12.5 of embryonic development, the Ly6e level in embryo represents less than 4% of the lung expression level, whereas the placenta level reaches 24% (Fig. 5). More generally, we observed that Ly6e is expressed in the placenta at the earliest stage tested (day 7.5) of embryo development and remains highly expressed between days 12.5 and 16.5, when SynA becomes significantly expressed.
FIG 5.
Expression profile of Ly6e and syncytin-A in a panel of adult and fetal mouse tissues: coexpression in the placenta. Ly6e and SynA transcript levels were assessed by real-time quantitative RT-PCR on tissues of adult male mice (liver to parotid gland [gl] and testis), adult female mice (uterus and ovary), embryo head and body, and placenta at indicated days of embryonic development. The embryo head (h) and body (b) were analyzed separately whenever possible. Tissue samples were analyzed in duplicate, and transcript levels were normalized relative to the amount of the RPLP0 housekeeping gene transcript. Transcript levels are represented as the percentage of maximal levels, reached in adult lung for Ly6e and in the placenta at embryonic day 16.5 for SynA.
DISCUSSION
In this study, we managed to combine a sensitive cell-cell fusion assay with a specific cDNA library screening procedure in order to identify proteins involved in the formation of syncytial structures. We applied this technique to search for the receptor for mouse SynA and identified Ly6e as a candidate receptor. Among the retroviral Env receptors described so far, most of them are cellular proteins that contain several transmembrane domains; some of them display a single transmembrane domain, and a few receptors are attached to the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor (22). Ly6e belongs to the latter group of membrane proteins, together with HYAL2, TVA800, and EFNA1 to EFNA5, the receptors for jaagsiekte sheep retrovirus (JSRV) and enzootic nasal tumor virus (ENTV), avian sarcoma and leukosis virus subgroup A (ASLV-A), and IAPE retroviruses, respectively (22–25).
We show that Ly6e induces SynA-dependent syncytium formation in our cell-cell fusion assay. In every model cell line tested where ectopic expression of SynA is enough to induce cell-cell fusion (thus implying that its receptor is expressed), Ly6e depletion by siRNA drastically reduced the level of SynA-induced cell-cell fusion. These results prompted us to hypothesize that Ly6e is the sole receptor for SynA in cellulo.
The GPI-anchored Ly6e, also known as Tsa-1 for thymic shared antigen 1, was previously described as a lymphostromal membrane protein and was suspected to play a role in the development of thymus through interaction with an unknown partner at the surface of thymocytes (26, 27). Interestingly, the study of Ly6e knockout embryos previously revealed that Ly6e depletion is embryonically lethal (28). No abnormality was observed in the thymus of the Ly6e null embryos. Their death was instead attributed to cardiac defect although Ly6e was not significantly expressed in the heart of wild-type embryos. In the light of our results on the Ly6e and SynA partnership in cell-cell fusion and of the fact that we previously showed that SynA null mouse embryos also die in utero (14), we suspect that the embryonic death of Ly6e null embryos could instead be the result of placental defects due to deficient cell-cell fusion involved in formation of the syncytiotrophoblast ST-I layer. This hypothesis is strengthened by the fact that our results show that Ly6e is significantly expressed in the mouse placenta, with at least a 4-fold higher expression level than in the rest of the embryo at the same developmental stage. Furthermore, Ly6e mRNA had been previously demonstrated to be expressed in the placental ST-I layer, which is consistent with SynA-mediated formation of ST-I (29). Outside its partnership with SynA during placental development, we suspect from our reverse transcription-quantitative PCR (RT-qPCR) results in adult tissues that Ly6e is involved in a more ubiquitous physiological process in adult mice, which remains to be characterized.
The process of retroviral endogenization begins with germ line infection by an ancestral retrovirus. The SynA-encoding ancestral retrovirus is related to a group of endogenous gammaretroviruses, the human endogenous retrovirus F/H family (10; data not shown), and its entry into the genome of rodents dates back to about 40 million years ago (30). The expression of the cellular receptor for the retroviral envelope in germ line cells is a prerequisite for germ line infection and subsequent integration into the genome of infected organisms. Our RT-qPCR data show that Ly6e is expressed in the ovary and, to a lesser extent, in the testis of mice, thus supporting a possible role of Ly6e as the original receptor for SynA during early steps of retroviral endogenization.
In addition to its role in placental development, we recently showed, using syncytin-B KO mice, that SynB was involved in myoblast fusion during muscle development as an “add-on” contribution to myogenesis in male mice (31). We also observed that SynA depletion reduced the amount of cell-cell fusion in vitro, suggesting that both mouse syncytins could be involved in myoblast fusion in vivo (31). Here, we show that Ly6e depletion impairs SynA-induced cell-cell fusion in mouse undifferentiated C2C12 myoblasts, suggesting that the Ly6e/SynA partnership could be involved in myoblast fusion in mice.
More generally, cell-cell fusion is a critical process in several physiological conditions, including not only placental development and muscle formation but also fertilization and bone homeostasis (32). The molecular mechanisms involved in membrane fusion during these processes are the subject of numerous studies. Outside of the placenta, several membrane proteins involved in mammalian cell-cell fusion have been identified, such as myomaker, myomixer, BAI1, and SynB in the muscles (31, 33–35), Izumo and Juno in fertilization (36, 37), and dendritic cell-specific transmembrane protein (DC-STAMP) in bone homeostasis (38). However, the protein partners (including true fusogens such as the syncytins) which are both necessary and sufficient to induce cell-cell fusion in the above-mentioned processes are not completely characterized. As illustrated by the Ly6e/SynA partnership, we believe that the cell-cell fusion-based screening assay described here will be of great use to unravel the precise molecular determinants of cell fusion in any physiological process where it is involved.
MATERIALS AND METHODS
Cell lines and expression vectors.
Human 293T embryonic kidney cells, Chinese hamster A23 fibroblasts, and mouse C2C12 myoblasts were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 100 μg/ml streptomycin (Life Technologies), and 100 U/ml penicillin (Life Technologies). A23-Galα cells were grown in the same growth medium supplemented with 1 mg/ml of G418 (Life Technologies). The vectors for expression of ASCT2 and syncytin-1 (5), an empty control (derived from phCMV-G; GenBank accession number AJ318514 [gift from F.-L. Cosset]), the vectors for expression of syncytin-A and syncytin-B (10), EFNA4-Fc (39), the α fragment of β-galactosidase (Galα vector; formerly pSCTZ-α-N85), the ω fragment of β-galactosidase (Galω vector; formerly pSCTZ-omega) (gifts from N. R. Landau [20]), and the G418 resistance gene (pSVneo) (40) were previously described. Ly6e-Fc and Ly6e-His expression vectors were constructed as follows: mouse Ly6e cDNA was PCR amplified from the Ly6e expression vector found in the cDNA library, using a BamHI-containing forward primer with a Kozak sequence before the initiation codon of Ly6e and a PstI-containing reverse primer specific to the C-terminal Ly6e sequence just upstream of the codon encoding the GPI-anchored residue 102. Primer sequences were, respectively, 5′-ATACATGGATCCGCCACCATGAGAGTCTTCCTGCCTG-3′ and 5′-ATACATCTGCAGGGCTGAAGTTGCAGAAGGAG-3′. After restriction with BamHI and PstI, the PCR products were cloned into a vector derived from phCMV-G and containing the sequence encoding a hexahistidine or rabbit Fc tag just downstream of the PstI restriction site. Constructs were verified by sequencing.
Production of A23-Galα cells.
To obtain A23-Galα stable cells, A23 cells were cotransfected with the G418 resistance expression vector pSVneo and Galα vector at a mass ratio of 1:9, using Fugene 6 transfection reagent (Promega), according to the manufacturer's protocol. A23 cells stably expressing G418 resistance (and potentially Galα) were selected in growth medium supplemented with 1 mg/ml G418. After 12 days of selection, resistant cell clones were transferred into different wells of 24-well plates using cloning disks (Sigma-Aldrich) soaked with trypsin-EDTA (Life Technologies) and amplified in selection medium for 2 to 4 weeks. Galα expression was tested by Lipofectamine LTX-mediated transfection (Life Technologies) of Galω vector, followed by X-Gal staining 1 to 3 days after transfection. The best A23-Galα clones (with >95% Galα-expressing cells) were selected.
Cell-cell fusion assay by α-complementation.
A23, A23-Galα, 293T, or C2C12 cells were seeded at day 1 in 48-well plates at approximately 20% confluence (104 to 2 × 104 cells/well). When siRNAs were used, their transfection was done at day 2, using Lipofectamine RNAiMAX reagent (Life Technologies) according to the manufacturer's protocol. siRNAs against human and mouse Ly6e and against human ASCT2, as well as a nontargeting siRNA, were purchased from Dharmacon (siGENOME SMARTpool). The day after siRNA transfection (day 3) or at day 2 in the absence of siRNA transfection, a first group of cells was cotransfected using Lipofectamine LTX or Fugene 6 reagent with vectors encoding Galα and a candidate receptor (or a control empty vector) at a ratio of 1:1, except for A23-Galα cells, which were transfected with only the syncytin receptor (or the control vector). For the testing of ASCT2 at various ratios, an ASCT2 expression vector and a control empty vector were cotransfected in A23-Galα cells at a mass ratio of 1:9 (named ASCT21/10), 1:99 (ASCT21/100), or 1:299 (ASCT21/300). The second group of cells was cotransfected with vectors encoding Galω and a syncytin (or a control empty vector) at a ratio of 1:1. At 4 to 24 h after plasmid transfection, the two groups of cells were cocultured at a ratio of 1:1. At 16 to 48 h after cell mixing, cells were fixed and stained with X-Gal to identify syncytia by microscopy analysis.
cDNA library screening by cell-cell fusion.
Syncytin-A receptor was searched using a normalized mouse brain cDNA library (Express Genomics) with an average insert size of 1.65 kb under the control of a cytomegalovirus (CMV) promoter. The cDNA library was plated at a density of about 100 bacterial colonies per plate. A total of 300 pools of 100 colonies were harvested, and the plasmids were extracted using a miniprep kit (Macherey-Nagel). One day after A23-Galα plating into 48-well plates, 200 ng of each pool of 100 cDNAs was transfected into each well. The cell-cell fusion assay was then performed as described above. Positive pools were then retransformed into DH5α Escherichia coli (Life Technologies). Plasmids were purified from individual colonies grown in 96-well plates, pooled into groups of 12 cDNAs, and tested by cell-cell fusion assay. Individual plasmids of the positive pools of 12 were then finally tested by cell-cell fusion assay. The positive plasmids were sequenced using M13 forward (5′-GTAAAACGACGGCCAGT-3′) and reverse (5′-CAGGAAACAGCTATGAC-3′) primers, and the sequences were subjected to a blastn (NCBI) search against the Mus musculus nucleotide sequence collection. All the positive clones contained the same full-length ORF (GenBank accession number CT010244.1) encoding the mouse Ly6e protein.
Microscopy analysis by ImageJ software.
For automatic counting of syncytia, X-Gal-stained cells were imaged using a transmitted-light microscope and a digital camera (Nikon). Images were then analyzed by ImageJ software as follows. Images were converted to black and white (8 bit), and the detection threshold was adjusted above the background noise so that only blue syncytia were counted; the number and pixel area of syncytia were then automatically analyzed using the “analyze particles” function of ImageJ. Only syncytia above a size threshold of 670 pixels were counted to exclude cellular debris.
Western blotting on soluble proteins.
Ly6e-His, Ly6e-Fc, or EFNA4-Fc soluble protein was produced by 293T cells in a serum-free medium (Opti-MEM; Life Technologies) after transient transfection with the corresponding expression vectors, using Fugene 6 transfection reagent. The protein-containing supernatants were collected 2 days posttransfection, passed through 0.45-μm-pore-size filters, and 10 times concentrated in Vivaspin ultrafiltration spin columns (Sartorius). Samples were denatured for 10 min at 90°C after the addition of Laemmli buffer, separated by electrophoresis in 4 to 12% bis-Tris NuPAGE gels in morpholineethanesulfonic acid (MES) buffer (Life Technologies) according to the manufacturer's protocol, and analyzed by Western blotting. Antibodies used for Western blotting were purchased from Qiagen (horseradish peroxidase [HRP]-conjugated anti-His) and GE Healthcare Life Sciences (anti-mouse and rabbit HRP-conjugated IgG).
RNA extraction, RT-PCR, and real-time quantitative PCR.
Total RNAs from mouse tissues were extracted as previously described (10). Total RNAs from siRNA-transfected cells were extracted using an RNeasy extraction kit (Qiagen) and treated with DNase I (Ambion). One microgram was used for each reverse transcription reaction using murine leukemia virus (MLV) reverse transcriptase (Applied Biosystems). Quantitative PCR was done using 5 μl of a 1/25 dilution of the cDNAs in a final volume of 25 μl by using SYBR green PCR master mix (Applied Biosystems). PCR was carried out using an ABI Prism 7000 sequence detection system with primers 5′-TACTCCTGCCCGATAGATGA-3′ and 5′-CCGTTTTTCTTAACAGTGGGT-3′ for SynA, 5′-GGCTTGGTAGTGTTTGCCAT-3′ and 5′-GGGCAAAGAGTAAACCCACA-3′ for human ASCT2, 5′-CGGGCTTTGGGAATGTCAAC-3′ and 5′-GTGGGATACTGGCACGAAGT-3′ for mouse Ly6e, 5′-TCTGTACTGCCTGAAGCCGA-3′ and 5′-CCACACCAACATTGACGCCT-3′ for human Ly6e, and 5′-GAGGACCTCACTGAGATTCGG-3′ and 5′-TTCTGAGCTGGCACAGTGAC-3′ for the ribosomal protein lateral stalk subunit P0 (RPLP0). The transcript levels were normalized relative to the amount of RPLP0 transcripts using the ΔΔCT (where CT is threshold cycle) method. Samples were assayed in duplicate.
Statistical analysis.
For ImageJ analysis, the number and mean area of syncytia were analyzed in randomly chosen fields from at least three independent experiments. Visual counting of syncytia was performed, and the results were obtained by averaging the number of syncytia of two duplicate wells of 48-well plates; data are the means ± standard errors of the means (SEM) from at least three independent experiments. Student's t test was used to analyze the differences in mean syncytium number and area, which were considered significant when at P values of <0.05 and <0.01.
ACKNOWLEDGMENTS
We thank Sandro Rusconi and Nathaniel Landau for plasmids and advice, Christian Lavialle for critical reading of the manuscript, and members of the UMR9196 laboratory for technical assistance and helpful discussions.
This work was supported by the Centre National de la Recherche Scientifique (France) and the Agence Nationale de la Recherche (ANR Retro-Placenta, France, to T.H.).
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, et al. . 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, et al. . 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of “syncytins,” retroviral env genes exapted for a role in placentation. Philos Trans R Soc B Biol Sci 368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaise S, de Parseval N, Benit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A 100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 74:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Veron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci U S A 109:E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis G, Heidmann O, Degrelle SA, Vernochet C, Lavialle C, Letzelter C, Bernard-Stoecklin S, Hassanin A, Mulot B, Guillomot M, Hue I, Heidmann T, Dupressoir A. 2013. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc Natl Acad Sci U S A 110:E828–E837. doi: 10.1073/pnas.1215787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, Zeller U, Heidmann O, Dupressoir A, Heidmann T. 2015. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A 112:E487–E496. doi: 10.1073/pnas.1417000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis G, Vernochet C, Malicorne S, Souquere S, Tzika AC, Goodman SM, Catzeflis F, Robinson TJ, Milinkovitch MC, Pierron G, Heidmann O, Dupressoir A, Heidmann T. 2014. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc Natl Acad Sci U S A 111:E4332–E4341. doi: 10.1073/pnas.1412268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A 102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 12.Georgiades P, Ferguson-Smith AC, Burton GJ. 2002. Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 13.Simmons DG, Natale DR, Begay V, Hughes M, Leutz A, Cross JC. 2008. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 135:2083–2091. doi: 10.1242/dev.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A 106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, Pierron G, Heidmann T. 2011. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A 108:E1164–E1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosset FL, Lavillette D. 2011. Cell entry of enveloped viruses. Adv Genet 73:121–183. doi: 10.1016/B978-0-12-380860-8.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esnault C, Priet S, Ribet D, Vernochet C, Bruls T, Lavialle C, Weissenbach J, Heidmann T. 2008. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc Natl Acad Sci U S A 105:17532–17537. doi: 10.1073/pnas.0807413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. 2009. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland AU, Munk C, Lucero GR, Nguyen LD, Landau NR. 2004. Alpha-complementation assay for HIV envelope glycoprotein-mediated fusion. Virology 319:343–352. doi: 10.1016/j.virol.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Moosmann P, Rusconi S. 1996. Alpha complementation of LacZ in mammalian cells. Nucleic Acids Res 24:1171–1172. doi: 10.1093/nar/24.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. 2015. Proteomics tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 22.Dewannieux M, Vernochet C, Ribet D, Bartosch B, Cosset FL, Heidmann T. 2011. The mouse IAPE endogenous retrovirus can infect cells through any of the five GPI-anchored ephrin A proteins. PLoS Pathog 7:e1002309. doi: 10.1371/journal.ppat.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bates P, Young JA, Varmus HE. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 24.Dirks C, Duh FM, Rai SK, Lerman MI, Miller AD. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J Virol 76:2141–2149. doi: 10.1128/jvi.76.5.2141-2149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai SK, Duh FM, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, Miller AD. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci U S A 98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Classon BJ, Boyd RL. 1998. Thymic-shared antigen-1 (TSA-1). A lymphostromal cell membrane Ly-6 superfamily molecule with a putative role in cellular adhesion. Dev Immunol 6:149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randle ES, Waanders GA, Masciantonio M, Godfrey DI, Boyd RL. 1993. A lymphostromal molecule, thymic shared Ag-1, regulates early thymocyte development in fetal thymus organ culture. J Immunol 151:6027–6035. [PubMed] [Google Scholar]
- 28.Zammit DJ, Berzins SP, Gill JW, Randle-Barrett ES, Barnett L, Koentgen F, Lambert GW, Harvey RP, Boyd RL, Classon BJ. 2002. Essential role for the lymphostromal plasma membrane Ly-6 superfamily molecule thymic shared antigen 1 in development of the embryonic adrenal gland. Mol Cell Biol 22:946–952. doi: 10.1128/MCB.22.3.946-952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes M, Natale BV, Simmons DG, Natale DR. 2013. Ly6e expression is restricted to syncytiotrophoblast cells of the mouse placenta. Placenta 34:831–835. doi: 10.1016/j.placenta.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Vernochet C, Redelsperger F, Harper F, Souquere S, Catzeflis F, Pierron G, Nevo E, Heidmann T, Dupressoir A. 2014. The captured retroviral envelope syncytin-A and syncytin-B genes are conserved in the Spalacidae together with hemotrichorial placentation. Biol Reprod 91:148. doi: 10.1095/biolreprod.114.124818. [DOI] [PubMed] [Google Scholar]
- 31.Redelsperger F, Raddi N, Bacquin A, Vernochet C, Mariot V, Gache V, Blanchard-Gutton N, Charrin S, Tiret L, Dumonceaux J, Dupressoir A, Heidmann T. 2016. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet 12:e1006289. doi: 10.1371/journal.pgen.1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oren-Suissa M, Podbilewicz B. 2007. Cell fusion during development. Trends Cell Biol 17:537–546. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sanchez-Ortiz E, Bassel-Duby R, Olson EN. 2017. Control of muscle formation by the fusogenic micropeptide myomixer. Science 356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, Call JA, Klibanov AL, Yan Z, Mandell JW, Ravichandran KS. 2013. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497:263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millay DP, O'Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. 2013. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianchi E, Doe B, Goulding D, Wright GJ. 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue N, Ikawa M, Isotani A, Okabe M. 2005. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 38.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, Oike Y, Takeya M, Toyama Y, Suda T. 2005. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med 202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aasheim HC, Munthe E, Funderud S, Smeland EB, Beiske K, Logtenberg T. 2000. A splice variant of human ephrin-A4 encodes a soluble molecule that is secreted by activated human B lymphocytes. Blood 95:221–230. [PubMed] [Google Scholar]
- 40.Esnault C, Casella JF, Heidmann T. 2002. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res 30:e49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]