ABSTRACT
Viral replication in eukaryotes is a process inherently organized in both space and time. Viral components target subcellular organelles to access host machineries required for replication and spread. Diverse viruses are known to alter organelle shape, composition, function, and dynamics as part of their replication cycles. Here, we highlight recent advances in microscopy and proteomic methods that have helped and will continue to help define mechanisms used by viruses to exploit host proteome organization.
KEYWORDS: mass spectrometry, microscopy, organelle, proteomics, systems biology, virus-host interactions
INTRODUCTION
Spatial organization is an essential component of a viral infection process. Eukaryotic viruses replicate within highly organized host cells that compartmentalize their vast cellular proteomes within organelles. Each organelle performs a specialized set of functions that arise from its distinct morphology, composition, and dynamics, which are intimately linked to the local balance of protein, lipid, and inorganic ion contents. Regulating organelle function by modulating the abundance and structure of these components is crucial for cellular homeostasis. Viruses may simply take advantage of the specialized functions organelles offer or, like intrinsic host organelle regulation, repurpose organelles by modifying their composition. Regardless, use of organelle functions for the temporally ordered steps of the viral life cycle requires viral components to both be active at the proper time and targeted to specific subcellular locations.
VIRAL INFECTIONS DEPEND ON ORGANELLE FUNCTION, MORPHOLOGY, AND DYNAMICS
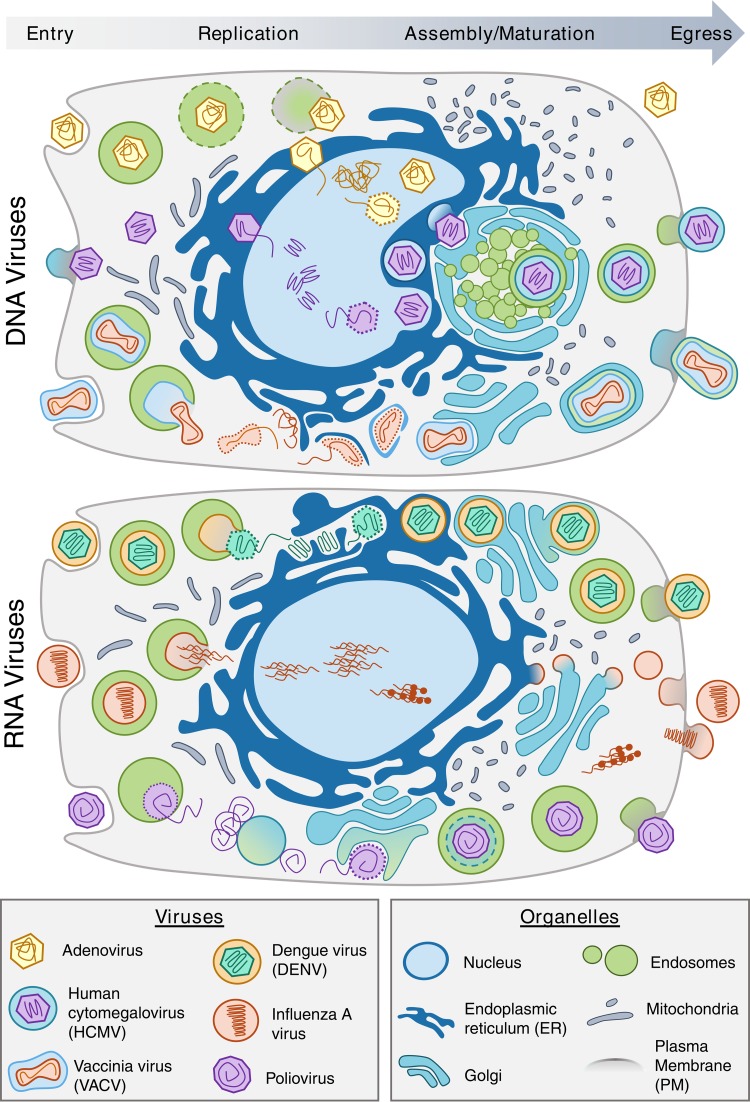
Viruses rely on organelle functions throughout their cellular life cycles: entry, replication, assembly, and egress (Fig. 1). Despite the large diversity in viral species, viruses meet and must overcome similar challenges, resulting in striking commonalities in the spatial-temporal organization of an infection. The interaction with the plasma membrane (PM) seems mostly nonnegotiable for mammalian viruses, and viral entry often requires the use of secretory organelles (1–4) (Fig. 1, entry). Enveloped viruses can enter the cell by fusing directly with the PM or, similar to most nonenveloped viruses, by receptor-mediated endocytosis. Endocytosed viruses face the problem of being sent to lysosomes for degradation. To avoid this, viruses often use pH-dependent escape mechanisms related to endosomal maturation. For example, flavivirus membranes (e.g., dengue virus [DENV]) fuse with early endosomes because of pH-induced conformational changes in its glycoproteins (4), while nonenveloped viruses (e.g., adenovirus and poliovirus) undergo partial uncoating, exposing capsid proteins that disrupt endosomal membranes and eject viral particles into the cytoplasm (3).
FIG 1.
Viruses exhibit both shared and specific spatial organization strategies during infection. Depicted is organelle regulation during infection with DNA and RNA viruses. Shown at the top are the infection cycles of the DNA viruses adenovirus, HCMV, and VACV. Shown at the bottom are the infection cycles of the RNA viruses DENV, influenza A virus, and poliovirus. The difference between longer (gray, left) and smaller (gray, right) mitochondria depicts mitochondrial fragmentation induced by diverse types of viral infections.
Following entry, viral replication and assembly in human cells can require further modulation of host organelles. The nucleus, often a site of viral genome replication (e.g., adenovirus, human cytomegalovirus [HCMV], and influenza A virus), provides proximity to polymerases and replication factors (Fig. 1, replication). Additionally, regardless of their site of replication, numerous viruses alter host transcription to manipulate cellular pathways, such as host defense, metabolism, and the cell cycle. Viral entry and exit from the nucleus rely on interactions with host machineries, including the use or disruption of the nuclear pore complex, lamina, or membranes, as described for herpesviruses (5). During maturation, several enveloped viruses acquire their membranes from cytoplasmic organelles, such as the endoplasmic reticulum (ER), Golgi apparatus, and endosomes. A common process in the herpesvirus family is that viruses undergo secondary envelopment at restructured organelles of the endomembrane system (6). For example, HCMV, a betaherpesvirus, forms a vesicular viral assembly complex by inducing structural remodeling (1) and proteome composition changes (7) in the aforementioned organelles (Fig. 1, replication, top panel). Other DNA viruses, as well as RNA viruses, also use the ER as a site for viral maturation and envelopment. For example, DENV (an RNA virus) replicates at ER membrane invaginations and buds into the ER to receive its primary envelope, while vaccinia virus (VACV; a DNA virus) generates ER-derived “crescent membranes” that mature to form the viral envelope (2, 3) (Fig. 1, replication and assembly). Even some enveloped viruses that do not directly interact with secretory organelles, such as influenza A virus, utilize the ER and Golgi apparatus to traffic viral components toward the PM to undergo final assembly (Fig. 1, assembly, bottom panel). While the use of organelles may seem obvious for enveloped viruses, some nonenveloped viruses also form crucial interactions with secretory organelles for maturation and assembly. Picornaviruses, single-stranded RNA viruses, generate replication sites from the trans-Golgi network (TGN) and tend to associate with autophagosomes late in infection (8) (Fig. 1, assembly, bottom panel). Rotaviruses, double-stranded RNA viruses, repurpose lipid droplet components to create dynamic inclusion bodies known as viroplasms, which serve as the initial site of viral genome replication and assembly before release into the ER (3).
Despite this established dependence of viruses on organelle function, morphology, and dynamics, many of the mechanisms underlying these extensive organelle-remodeling events remain unknown. Here, we highlight advances in microscopy and proteomic methods that have allowed virologists to examine virus-host interactions in unprecedented temporal and spatial detail, as well as emerging techniques that have yet to be applied to viral infection studies.
DETAILED MAPPING OF HOST AND VIRAL PROTEINS BY MICROSCOPY
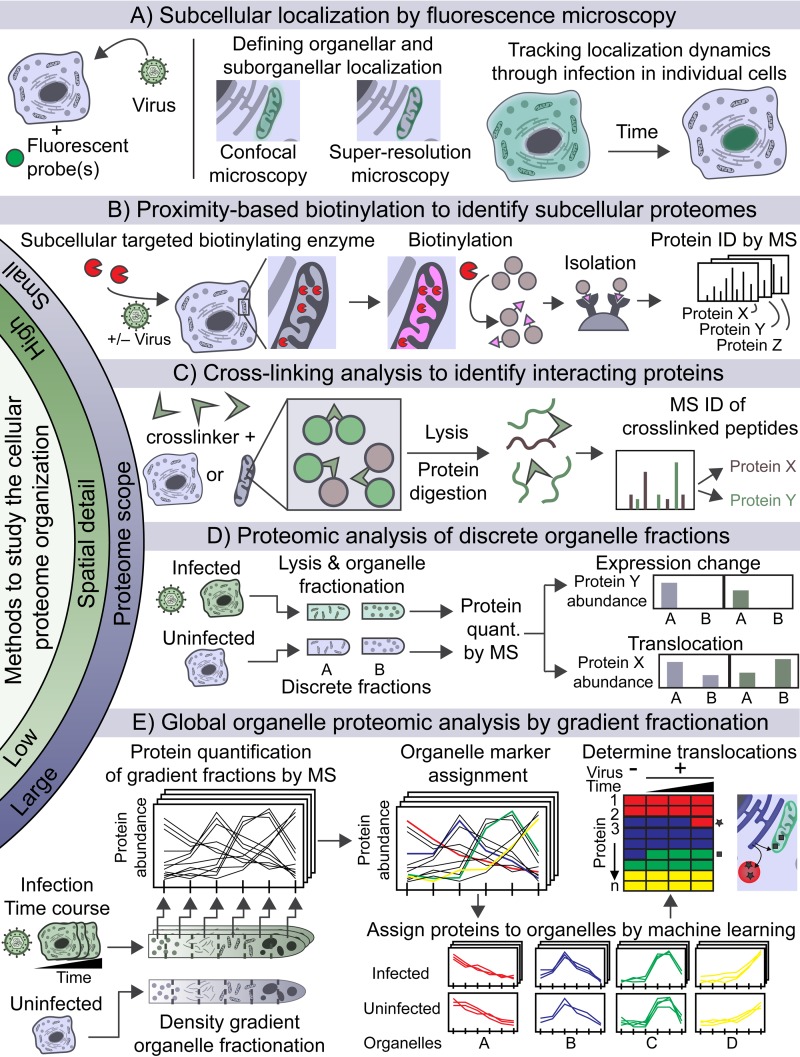
An increasing number of experimental tools are available with which to explore the dynamics of cellular proteome organization (Fig. 2). These technologies are tailored to specific biological questions based on the level of spatial detail provided (e.g., organelle versus suborganelle resolution) and the required depth of analysis (e.g., individual proteins versus whole organelle proteome). Confocal fluorescence microscopy is one of the most robust and frequently implemented tools for low-throughput assessment of viral and host protein localization to specific organelles (Fig. 2A). The colocalization of viral assembly proteins with organelle markers (i.e., proteins acting as markers or organelle-specific fluorescent dyes) has helped discover host organelles associated with viral assembly compartments. Examples of such findings include flavivirus replication at ER-derived membranes (2), picornavirus replication at repurposed TGN membranes (8), and HCMV maturation and envelopment at repurposed Golgi apparatus, ER, and endosomal membranes (1). The use of multicolor confocal imaging during time-lapse live microscopy has elevated the ability to investigate dynamic organelle processes during infection. The discovery that pseudorabies virus hijacks the constitutive secretion machinery for exocytosis was made possible by the imaging of fluorescently labeled Rab proteins, involved in vesicle trafficking, and virus labeling with pH-sensitive fluorescent protein (9).
FIG 2.
Methods used to study cellular proteome organization. (A) Confocal fluorescence microscopy can be used to determine the subcellular localization of a protein by colocalization with organelle markers. This includes translocation caused by viral infection. (B) Proximity-based biotinylation to identify subcellular proteomes. A promiscuous biotinylating enzyme (APEX) is genetically targeted to a subcellular compartment, which biotinylates adjacent molecules that are subsequently purified and identified by MS. ID, identification. (C) Cross-linking analysis to identify protein interactions and infer localization. Cross-linkers are added to whole cells or organelles to covalently bind adjacent proteins. Following lysis and digestion, the cross-linked peptides are enriched (e.g., by affinity purification or size exclusion chromatography) and analyzed by MS. (D) Proteomic analysis of discrete organelle fractions. Organelles from infected or uninfected cells are isolated into discrete fractions. The proteins within these fractions are identified and quantified by MS. A change in protein expression occurs if the abundance of a protein changes in an organelle, whereas a translocation is defined by an increase in abundance in an organelle accompanied by a decrease in abundance in another organelle. quant., quantification. (E) Global organelle proteomic analysis by gradient fractionation. Cells are lysed, and organelles are separated throughout a density gradient by centrifugation. Fractions from the gradient are analyzed by MS, which results in distinct distribution profiles of proteins found within the same organelle. A set of proteins are used as organelle markers to identify the distribution profile of specific organelles. The remaining proteins are then computationally assigned to organelles on the basis of their similarity to marker profiles. Virus-induced translocation events are determined when a protein is assigned to a different organelle in infected versus uninfected samples.
Superresolution microscopy has further expanded localization capabilities, reaching nanometer resolution and allowing spatial definition of viral and host components within suborganelles, such as within submitochondrial regions (10) (Fig. 2A). Viral proteins that localize to the mitochondria have been shown to induce mitochondrial fragmentation and changes in mitochondrial functions to inhibit innate immunity (e.g., influenza A virus protein PB1-F2) or apoptosis (e.g., viral mitochondrion-localized inhibitor of apoptosis [vMIA] in HCMV) or to upregulate oxidative phosphorylation (e.g., rabies virus P). Confocal microscopy was used to identify or confirm the location of these proteins to the mitochondria, yet submitochondrial localization could not be assessed. In the case of PB1-F2, its localization to the inner mitochondrial membrane was determined by a tour de force of biochemical fractionation of mitochondrial membranes (11). More recently, multicolor structured illumination microscopy (MSIM), a type of superresolution microscopy, revealed with high confidence that vMIA forms 100-nm clusters at the outer (versus the inner) mitochondrial membrane (10). Together with the knowledge of vMIA's association with the host proteins Bax, mitochondrial antiviral signaling (MAVS) protein, and viperin, these findings suggest the formation of vMIA protein complexes at the outer mitochondrial membrane to evade innate immunity and facilitate viral replication (10, 12). Another example of the value of superresolution microscopy comes from the study of viral replication organelles; the Herker lab implemented three-color, three-dimensional, direct stochastic optical reconstruction microscopy (dSTORM) to define the spatial organization of HCV replication structures at the periphery of lipid droplets (reviewed in reference 10).
To further expand protein localization and structural analysis capabilities, hybrid approaches that correlate microscopy methods have been and continue to be developed. Ultrastructural images from electron microscopy were correlated with fluorescent signals from light microscopy to elucidate the structure of herpesviruses budding during nuclear egress (5). The creative effort of several groups has been expanding this technology to correlate superresolution light and electron microscopy to provide detailed multiprotein information of virus-host protein interactions within suborganelle nanometer structures (e.g., see reference 13). More recently, Valm et al. coupled a powerful multispectral imaging technique with rigorous computational analysis to simultaneously image six fluorescently tagged organelles (ER, Golgi apparatus, lysosome, peroxisome, mitochondria, and lipid droplet) within live cells (14). This approach allowed the authors to analyze spatial-temporal interactions between organelles in three dimensions, providing a depiction of cellular organization and dynamics in both normal and perturbed states (i.e., starvation) at a previously unattainable level. Such multicolor fluorescent approaches, in conjunction with advanced quantitative techniques for analysis of large microscopy data sets, can be applied to a multitude of future studies to visualize virus-host interactions in live cells and understand the subcellular organization of an infection process.
DISCOVERING PROTEOME ORGANIZATION BY USING PROTEOMICS
Microscopy analyses, while providing high spatial resolution, are limited in the number of proteins that can be studied at one time. Mass spectrometry (MS)-based proteomics has become established as an important tool for sensitive detection and quantification of thousands of proteins in biological samples and has been successfully applied to diverse viral infection studies (15). Relevant to the spatial organization of the cell, subcellular fractionation, followed by MS analysis for protein identification, termed “spatial proteomics,” can inform about protein localization within specific organelles or other subcellular compartments (16–18). The integration of classic fractionation methods, such as density gradient centrifugation and differential detergent fractionation, with MS was proven valuable for assigning novel localizations to viral proteins. Examples include the above-mentioned discovery of rabies virus protein P localization to the mitochondria (19) and the temporal localization of HCMV protein pUL13 to the PM and mitochondria (7). Methods used to enrich subcellular compartments were also developed by taking advantage of protein biotinylation. Proximity-based biotinylation (bioID) was shown to identify proteins in subcellular spaces by targeting a promiscuous biotin ligase (BirA) to an organelle by using genetic tags and by purifying and identifying neighboring biotinylated proteins by MS (20). BioID can also identify protein interactions within a subcellular niche, as demonstrated for the HCMV tegument protein pUL103 (21). Further expanding the capabilities and applications of protein biotinylation, an engineered ascorbate peroxidase (APEX) was developed to provide faster biotinylation reactions and the ability to reach tight intracellular spaces via short-lived biotin-phenoxy radicals, as shown for mitochondrial compartments (18) (Fig. 2B). This approach has not yet been applied to infection but is a promising tool that can be used to identify the composition of subcellular spaces exclusively found in viral infections, such as cytoplasmic compartments for herpesviruses maturation and secondary envelopment (6).
Another proteomic method for capturing proteins in their cellular milieu that continues to gain attention is cross-linking MS. A cross-linker, generally amine reactive, is used to covalently link physically adjacent proteins, and the cross-linked peptides are identified by MS (Fig. 2C). This approach was applied to viral infection to study protein-protein interaction topologies of the capsid of potato leafroll virus (22). The development of cross-linkers that diffuse through membranes has provided the opportunity to identify interactions in live intact cells or within organelles, as demonstrated for mitochondria (23). This technology is further being extended to quantify alterations in interactions across multiple conditions by using isotopically labeled cross-linkers and MS. Future incorporation of cross-linking strategies promises to help address many unanswered questions, for example, by providing further detail about virus-induced dysregulation of respiratory supercomplexes within mitochondria (19) or mechanisms of viral maturation and assembly (1, 3, 6, 24).
THE DYNAMICS OF PROTEOME ORGANIZATION: PROTEIN ABUNDANCES AND TRANSLOCATIONS
In addition to cataloguing organelle components, the understanding of organelle functions during infection requires knowledge of the dynamic alterations in organelle composition that may occur via temporal changes in protein abundance or translocations of proteins from one compartment to another. Proteins and pathways regulated within organelles can be monitored by using quantitative MS-based proteomic and bioinformatic tools (Fig. 2D) (see reference 15 for a review of quantitative proteomic methods). Gudleski-O'Regan et al. characterized cell surface proteome changes during the progression of HCMV infection. Cell surface proteins were biotinylated, isolated by affinity purification, and quantified by MS at different time points of infection. Among the upregulated proteins, LRP1, a regulator of lipid metabolism, was shown to contribute to a host defense mechanism that decreases intracellular cholesterol and virion infectivity (25). Varjak et al. identified proteome alterations in endolysosomal replication complexes of Semliki Forest virus (26). Endolysosomes were isolated from infected or mock-infected cells by using endocytosed magnetic nanoparticles for relative protein quantification by MS. Functional network analysis revealed an enrichment of RNA-binding proteins in these organelles during infection, of which PCBP1 was found to be required for efficient replication.
Different mechanisms can contribute to infection-induced alterations in organelle protein abundances, including transcriptional regulation, posttranslational modification, and protein translocations from one compartment to another. A translocation can alter a protein's function or lifetime by bringing it into contact with new binding partners, substrates, or effector molecules. During DENV infection, for example, the host protein ERI3, a putative RNA exonuclease, was found to associate with DENV RNAs by RNA chromatography, followed by protein identification by MS (24). By using Airyscan superresolution microscopy, this study further demonstrated that ERI3 translocates from the Golgi apparatus to ER-derived viral replication organelles to facilitate viral RNA synthesis. HCMV was also shown to induce the translocation of many host proteins to the viral assembly complex, including the recently characterized unconventional myosin MYO18A, SNARE components, the ER-resident chaperone BiP, and mTOR kinase (1, 7, 12). Similarly, enterovirus induces the translocation of Golgi apparatus components to the TGN to form replication organelles (8). Translocation is also a mechanism underlying viral evasion of the host immune response; two HCMV proteins, US18 and US20, have been shown to translocate MICA, a host immune ligand, from the PM to lysosomes for immune evasion (27), and vMIA induces translocation of the antiviral protein viperin from the ER to the assembly compartment and mitochondria to avoid its antiviral effects and enhance viral production (12).
Protein translocations have been studied with a variety of tools, including microscopy, biochemical fractionation, and proteomic approaches. Proteomic workflows provide the ability to sensitively analyze multiple subcellular fractions and identify novel translocations without a priori knowledge. For example, Horner et al. investigated proteome changes in fractions of the ER, cytosol, and ER–mitochondrion-associated membranes (MAM) during HCV and Sendai virus infections. These subcellular compartments were isolated by density gradient ultracentrifugation, and their proteins were quantified by MS by using spectral counting to determine both changes in abundance and candidate translocations upon infection. In this case, translocations were detected by decreases in the abundance of a protein in one fraction with a simultaneous increase in abundance in another fraction (Fig. 2D). Their study revealed that MAVS protein-mediated antiviral signaling restructures the MAM by altering its protein composition, which includes protein translocations (28). Proteomic data from whole cells and subcellular fractions can also be integrated to identify translocations. Weekes et al., for example, quantified temporal HCMV-induced changes both in whole-cell lysates and at the cell surface by using isobaric tags and quantitative MS (29). The decrease in cell surface protein levels, in conjunction with an increase in whole-cell protein levels, was used to predict intracellular protein sequestration, as shown for CD155, a host factor known to be translocated by HCMV to evade natural killer cells.
As it becomes increasingly evident that viral infections alter multiple organelles concomitantly, a global view of proteome organization is necessary to better understand the virus-host interplay. Spatial proteomic methods, including protein correlation profiling, localization of organelle proteins by isotope tagging (LOPIT), and dynamic organellar maps, can generate protein localization profiles for multiple subcellular compartments (16, 17). These methods use the signature distributions of organelles upon density gradient centrifugation and analysis of the density fractions by MS. A protein's subcellular localization is determined by matching its distribution profile to that of a set of organelle marker proteins. Although some organelles may not be fully resolved because of similarities in their densities (e.g., ER and Golgi apparatus vesicles), these methods attempt to model all major subcellular compartments. Thus, in contrast to methods producing discrete fractions, they minimize false assignments from contaminant fractions and provide a full picture of cellular organization. Recently, an extension of these methods (Fig. 2E) was used to determine the localization and abundance of host and viral proteins across the HCMV replication cycle (7). A challenge that has to be taken into account in such studies is that infection may trigger alterations of organelle marker distribution, a problem that impacts most approaches that rely on organelle markers for subcellular assignment (e.g., colocalization microscopy). This can complicate the assignment of proteins to discrete organelles during infection. Therefore, the careful selection and validation of adequate markers in the context of infection is a first step that is important for the success of this approach. Spatial proteomic analysis determined a global spatial reorganization of the cell proteome, illustrated by temporally regulated changes in abundance and translocation of host proteins, such as the expected MICA translocation (27) and the previously unrecognized MYO18A translocation (7). Additionally, numerous HCMV proteins (nearly 100 proteins out of the ∼200 estimated HCMV protein-coding genes) are targeted to organelles, including the mitochondria, Golgi apparatus, ER, and lysosomes. Among these were previously uncharacterized viral proteins (e.g., pUL13), which will provide valuable insight into mechanisms of virus-induced organelle remodeling.
To fully understand how the viral life cycle modulates the host's spatial organization, several protein properties need to be considered in parallel with subcellular localization. These include protein interactions, posttranslational modifications, abundance, and turnover rate, to name a few. Some of the upcoming challenges and research opportunities that we foresee are in the integration of different methods to monitor these protein properties in both subcellular space and time of infection and in the analysis of increasingly large and complex multidimensional protein data sets.
PERSPECTIVE AND FUTURE DIRECTIONS
Spatial regulation of the host and viral proteome is an important aspect of virus-host interactions. Evolution in a compartmentalized host has required viruses to localize viral proteins to appropriate compartments and develop strategies to, first, access the host machinery and building blocks necessary for viral particle assembly and, second, regulate host pathways directed by organelles. As tools are developed to assess protein localization with greater detail and scope, the number of host proteins found to be spatially regulated by viruses continues to increase, as well as the array of viruses for which this type of regulation is observed. Proteomic analyses of multiple organelles have begun to describe extensive spatial reorganization of host proteins during infection (7, 25, 26, 28). Importantly, the viral life cycle is a dynamic process and characterizing cellular reorganization in both time and space is a key step in painting the full picture of viral infection. Investigations of virus-induced spatial cell remodeling, facilitated by further developments in microscopy and spatial proteomics, promise to provide an exciting new perspective on the biology and pathogenicity of viral infection.
ACKNOWLEDGMENTS
Many scientists have contributed to organelle studies, and we apologize for not being able to credit all of these important studies.
We are grateful for funding from the NIH (R01 GM114141 and R01 HL127640 to I.M.C.), a Mallinckrodt Scholar Award, and a Dodds Fellowship to P.M.J.B.
There is no conflict of interest regarding the publication of this article.
REFERENCES
- 1.Alwine JC. 2012. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog 8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatel-Chaix L, Bartenschlager R. 2014. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside—caught in the web. J Virol 88:5907–5911. doi: 10.1128/JVI.03404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid M, Speiseder T, Dobner T, Gonzalez RA. 2014. DNA virus replication compartments. J Virol 88:1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. 2009. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5:318–328. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bäuerlein FJB, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grünewald K. 2015. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163:1692–1701. doi: 10.1016/j.cell.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 7.Jean Beltran PM, Mathias RA, Cristea IM. 2016. A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst 3:361–373. doi: 10.1016/j.cels.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJM, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogue IB, Scherer J, Enquist LW. 2016. Exocytosis of alphaherpesvirus virions, light particles, and glycoproteins uses constitutive secretory mechanisms. mBio 7:e00820-16. doi: 10.1128/mBio.00820-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colberg-Poley AM, Patterson GH, Salka K, Bhuvanendran S, Yang D, Jaiswal JK. 2015. Superresolution imaging of viral protein trafficking. Med Microbiol Immunol 204:449–460. doi: 10.1007/s00430-015-0395-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, Koshiba T. 2014. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun 5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- 12.Seo JY, Yaneva R, Hinson ER, Cresswell P. 2011. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E, Seiradake E, Jones EY, Davis I, Grünewald K, Kaufmann R. 2015. Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins. Sci Rep 5:9583. doi: 10.1038/srep09583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J. 2017. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greco TM, Diner BA, Cristea IM. 2014. The impact of mass spectrometry-based proteomics on fundamental discoveries in virology. Annu Rev Virol 1:581–604. doi: 10.1146/annurev-virology-031413-085527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christoforou A, Mulvey CM, Breckels LM, Geladaki A, Hurrell T, Hayward PC, Naake T, Gatto L, Viner R, Arias AM, Lilley KS. 2016. A draft map of the mouse pluripotent stem cell spatial proteome. Nat Commun 7:8992. doi: 10.1038/ncomms9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itzhak DN, Tyanova S, Cox J, Borner GH. 2016. Global, quantitative and dynamic mapping of protein subcellular localization. eLife 5:e16950. doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. 2016. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc 11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kammouni W, Wood H, Saleh A, Appolinario CM, Fernyhough P, Jackson AC. 2015. Rabies virus phosphoprotein interacts with mitochondrial complex I and induces mitochondrial dysfunction and oxidative stress. J Neurovirol 21:370–382. doi: 10.1007/s13365-015-0320-8. [DOI] [PubMed] [Google Scholar]
- 20.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. 2016. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiz DA, Glassbrook JE, Pellett PE. 2016. Protein-protein interactions suggest novel activities of human cytomegalovirus tegument protein pUL103. J Virol 90:7798–7810. doi: 10.1128/JVI.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeBlasio SL, Chavez JD, Alexander MM, Ramsey J, Eng JK, Mahoney J, Gray SM, Bruce JE, Cilia M. 2015. Visualization of host-polerovirus interaction topologies using protein interaction reporter technology. J Virol 90:1973–1987. doi: 10.1128/JVI.01706-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweppe DK, Chavez JD, Lee CF, Caudal A, Kruse SE, Stuppard R, Marcinek DJ, Shadel GS, Tian R, Bruce JE. 2017. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A 114:1732–1737. doi: 10.1073/pnas.1617220114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward AM, Calvert ME, Read LR, Kang S, Levitt BE, Dimopoulos G, Bradrick SS, Gunaratne J, Garcia-Blanco MA. 2016. The Golgi associated ERI3 is a flavivirus host factor. Sci Rep 6:34379. doi: 10.1038/srep34379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudleski-O'Regan N, Greco TM, Cristea IM, Shenk T. 2012. Increased expression of LDL receptor-related protein 1 during human cytomegalovirus infection reduces virion cholesterol and infectivity. Cell Host Microbe 12:86–96. doi: 10.1016/j.chom.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varjak M, Saul S, Arike L, Lulla A, Peil L, Merits A. 2013. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. J Virol 87:10295–10312. doi: 10.1128/JVI.01105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fielding CA, Aicheler R, Stanton RJ, Wang ECY, Han S, Seirafian S, Davies J, McSharry BP, Weekes MP, Antrobus PR, Prod'homme V, Blanchet FP, Sugrue D, Cuff S, Roberts D, Davison AJ, Lehner PJ, Wilkinson GWG, Tomasec P. 2014. Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog 10:e1004058. doi: 10.1371/journal.ppat.1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horner SM, Wilkins C, Badil S, Iskarpatyoti J, Gale M Jr. 2015. Proteomic analysis of mitochondrion-associated ER membranes (MAM) during RNA virus infection reveals dynamic changes in protein and organelle trafficking. PLoS One 10:e0117963. doi: 10.1371/journal.pone.0117963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang ECY, Aicheler R, Murrell I, Wilkinson GWG, Lehner PJ, Gygi SP. 2014. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]