ABSTRACT
Coxsackievirus A16 (CV-A16; Picornaviridae) is an enterovirus (EV) type associated with hand, foot, and mouth disease (HFMD) in children. To investigate the spatial spread of CV-A16, we used viral sequence data sampled during a prospective sentinel surveillance of HFMD in France (2010 to 2014) and phylogenetic reconstruction. A data set of 168 VP1 sequences was assembled with 416 publicly available sequences of various geographic origins. The CV-A16 sequences reported were assigned to two clades, genogroup B and a previously uncharacterized clade D. The time origins of clades B and D were assessed in 1978 (1973 to 1981) and 2004 (2001 to 2007), respectively. The shape of the global CV-A16 phylogeny indicated worldwide cocirculation of genetically distinct virus lineages over time and across geographic regions. Phylogenetic tree topologies and Bayes factor analysis indicated virus migration. Virus transportation events in clade B within Europe and Asia and between countries of the two geographic regions were assessed. The sustained transmission of clade D viruses over 4 years was analyzed at the township level in France and traced back to Peru in South America. Comparative genomics provided evidence of recombination between CV-A16 clades B and D and suggested an intertype recombinant origin for clade D. Time-resolved phylogenies and HFMD surveillance data indicated that CV-A16 persistence is sustained by continuing virus migration at different geographic scales, from community transmission to virus transportation between distant countries. The results showed a significant impact of virus movements on the epidemiological dynamics of HFMD that could have implications for disease prevention.
IMPORTANCE Coxsackievirus A16 is one of the most prevalent enterovirus types in hand, foot, and mouth disease outbreaks reported in Southeast Asia. This study is based on epidemiological and viral data on HFMD caused by CV-A16 in a European country. The phylogeographic data complemented the syndromic surveillance with virus migration patterns between geographic regions in France. The results show how viral evolutionary dynamics and global virus spread interact to shape the worldwide pattern of an EV disease. CV-A16 transmission is driven by movements of infected individuals at different geographic levels: within a country (local dynamics), between neighboring countries (regional dynamics), and between distant countries (transcontinental dynamics). The results are consistent with our earlier data on EV-A71 and confirm the epidemiological interconnection of Asia and Europe with regard to EV infections.
KEYWORDS: enterovirus, molecular epidemiology, pediatric infectious disease
INTRODUCTION
Hand, foot, and mouth disease (HFMD), a common childhood illness, is caused by a large array of enteroviruses (EVs) found worldwide, notably those derived from a taxonomic species designated EV-A (1). It is a mucocutaneous disease characterized by a self-limiting course in most children under 5 years of age (2). HFMD outbreaks have been reported frequently throughout East and Southeast Asia since 1997, the year of the first large outbreak during which fatal infections caused by EV-A71 were reported in Malaysia (3). Several issues arising from the transmission of EV-A71 have been explored in both clinical and observational studies (4) and in epidemiological investigations (5). The virus is neurotropic and responsible for severe neurological and cardiopulmonary outcomes, which develop as complications of HFMD (2).
Coxsackievirus A16 (CV-A16) is another type within the EV-A species and is characterized by a high prevalence in HFMD outbreaks in Asia (6). Unlike EV-A71, CV-A16 has occasionally been associated with severe health issues (7–10). It was first isolated in South Africa in 1951, and the first outbreak of CV-A16 infections was reported in Canada in 1957 (11). It was jointly reported with EV-A71 during the large-scale outbreaks documented throughout the Asia-Pacific region (12). Sporadic CV-A16 cases have been documented in Spain (2010 to 2012), and two CV-A16 outbreaks have been reported elsewhere in Europe over the last 20 years (13, 14).
Current hypotheses to explain the burden of HFMD in East and Southeast Asia include the involvement of a large diversity of antigenic EV types and genogroups within EV types (15, 16), differing immunological patterns of different age groups within the human population (including waning of maternal immunity [17]), and crowding of susceptible children, which promotes interpersonal transmission (http://www.wpro.who.int/emerging_diseases/documents/HFMDGuidance/en/). Other factors are involved in the dynamics of HFMD and EV spread, either environmental, such as mean temperature and relative humidity (18), or epidemiological, such as the incubation period (19), the rate of symptomatic infections (19), and the interindividual transmission rate (20). Despite attempts to marshal evidence that would explain the epidemiological features of HFMD in East and Southeast Asia, our understanding of the mechanisms involved remains fairly rudimentary. The occurrence of HFMD and persistence of the EV lineages involved in this disease have not generally been considered a result of spatial processes. In earlier studies, we suggested that virus migration had a major impact on the occurrence of acute meningitis caused by EV-A71 and other EV types (21, 22). In the present study, we addressed this question to investigate specifically the role of CV-A16 infections in the occurrence of HFMD.
HFMD is not under active epidemiological surveillance in Europe, and therefore, data are lacking. The yearly circulation patterns of EV types involved in HFMD are unknown, because surveillance is based on passive case reporting or the review of severe cases associated with neurologic complications (23, 24). Since 2010, we have taken steps to help improve HFMD surveillance in France via prospective investigation of clinical and virological data (25, 26). Local surveillance was performed until 2013 and extended to the whole country in 2014, an initiative that has provided new epidemiological data on infections caused notably by species A EV types, including CV-A16. We used viral sequences from this 5-year period (2010 to 2014) of surveillance and assembled molecular sequence data sets of CV-A16 isolates sampled worldwide since the 1980s. The molecular analyses provided insights into the evolutionary features and circulation of this virus at local and global geographic scales.
RESULTS
Epidemiological surveillance of HFMD in France.
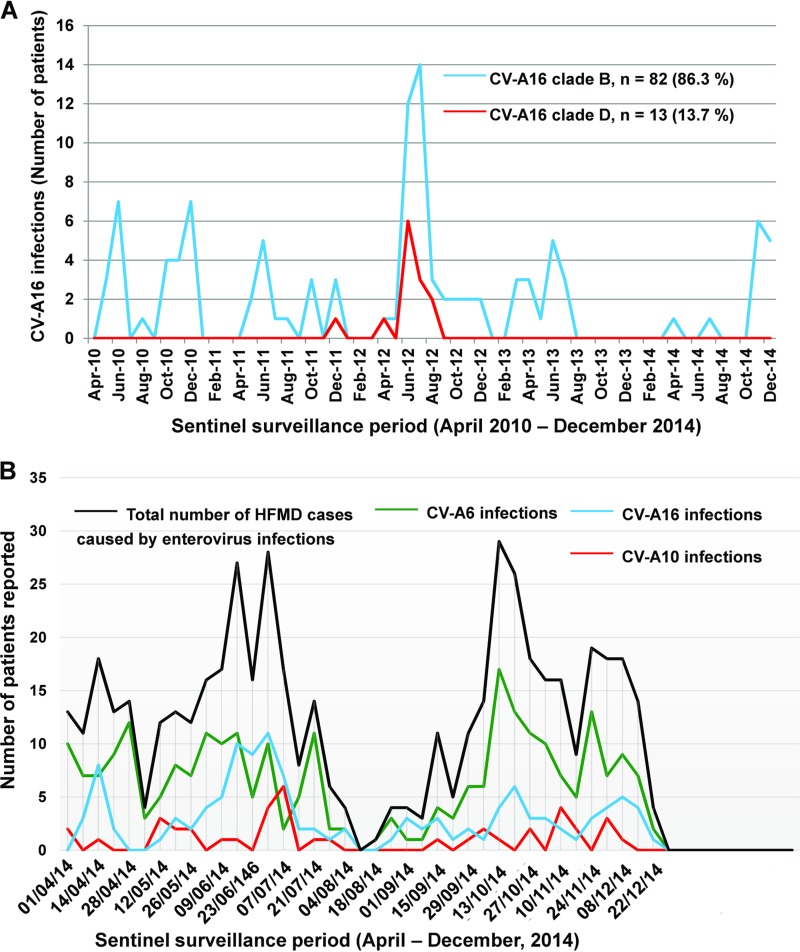
A total of 218 children (median age, 2.4 years; range, 2 months to 10 years) were recorded with HFMD manifestations associated with CV-A16 infections during two longitudinal, prospective, community-based surveillance studies in France, including 96 children (43.6%) living in the Clermont-Ferrand urban area, central France, between 1 April 2010 and 31 December 2014 and 122 children (56.4%) recruited in other French cities between 1 April and 31 December 2014. The monthly distribution of infections showed an increase in June and July 2012, a period during which CV-A16 represented 38.5% of all laboratory-confirmed EV infections reported in association with HFMD manifestations (Fig. 1A). CV-A16 infections accounted for 24.4% of all HFMD cases caused by EVs during the national investigation in 2014. The HFMD cases caused by CV-A16 were documented throughout the period, with an increase in CV-A16 infections observed during weeks 25 to 27 in 2014 (Fig. 1B).
FIG 1.
Coxsackievirus A16 infections reported among HFMD cases in France with two sentinel networks. (A) Data (monthly distribution of CV-A16 infections) from a sentinel surveillance reporting network of pediatricians implemented in the city of Clermont-Ferrand (France). Coxsackievirus A16 infections were reported prospectively in the pediatric community between 1 April 2010 and 31 December 2014 (31). A total of 218 EV infections were associated with a clinical diagnosis of HFMD in children, and 95 cases were caused by CV-A16 over the study period. (B) Data (weekly distribution of enterovirus infections) from a sentinel surveillance reporting network of pediatricians implemented throughout France, including Clermont-Ferrand. The clinical cases of HFMD were reported prospectively between 1 April and 31 December 2014. The CV-A16 infections reported over the same period and shown in panel A are included in the national data in panel B. A total of 529 children reported with a clinical diagnosis of HFMD tested positive for an EV infection. Of these children, 129 (24.4%) had a CV-A16 infection. The three main EV types identified in children reported with HFMD were, in decreasing order, CV-A6, CV-A16, and CV-A10.
Genetic diversity of CV-A16 estimated with the VP1 and 3CD sequence data.
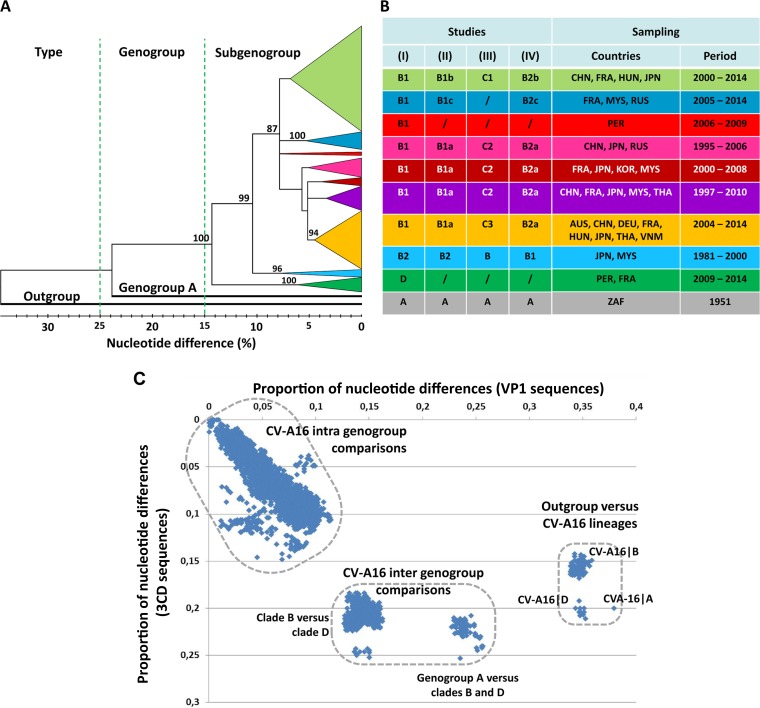
The diversity of the CV-A16 population was investigated by analysis of complete gene sequences (891 nucleotides [nt]) of the VP1 capsid protein, which harbors the main antigenic sites in EVs. The nucleotide sequences were determined in isolates from 128/218 (59%) patients reported during the two epidemiological studies. Only one sequence was analyzed from duplicate data. Sequences determined in isolates from 40 patients during routine clinical workup of EV infections between 2003 and 2014 in Austria (n = 3), France (n = 23), Germany (n = 8), and Hungary (n = 6) were also included. The overall 168 sequences were compared with 416 publicly available sequences reported in 20 countries (see below). The phylogenetic relationships determined with the VP1 sequences (Fig. 2A) showed that the two CV-A16 clades (large phylogenetic groups) previously reported as genogroups A and B (27, 28) had more than 23% nucleotide differences. Genogroup A was represented by a single sequence (G-10∣South Africa∣1951), while genogroup B included multiple phylogenetic subgroups below the 10% p-distance threshold. A number of CV-A16 VP1 sequences were assigned to a genogroup C in another study (29) but in fact clustered within distinct subgroups in the actual genogroup B (Fig. 2B) (30). A CV-A16 genogroup C was also reported on the basis of analyses of VP4 gene sequences but was not confirmed by investigation of the VP1 sequences (31, 32). A number of discordances in the assignments of CV-A16 phylogenetic groups/subgroups were described earlier by Mizuta et al. (33). In the present study, the tree provided phylogenetic evidence of a distinct clade at the 14.4% p-distance threshold. This clade comprised 36 sequences sampled in France between 2011 and 2014 and 1 in Peru in 2009 (accession number KF956720) (34). An exhaustive analysis of the CV-A16 VP1 sequences publicly available between 2015 and January 2017 revealed no report of additional sequence data that could be assigned to this previously unreported clade, which was designated by the letter D.
FIG 2.
CV-A16 genetic diversity assessed with the VP1 gene sequences. (A) Phylogenetic tree based on complete VP1 coding sequences of 573 CV-A16 sequences. The tree was inferred with the neighbor-joining method from genetic distances calculated with the p-distance algorithm. The tree topology was assessed with 1,000 bootstrap pseudoreplicates. The bootstrap values are shown only for the main phylogenetic clusters. The branches of consistent phylogenetic clusters were compressed for more clarity. The clade(s) (genogroup[s]), geographic origin(s), and isolation date(s) of virus strains are indicated for each cluster. The CV-A16 strain G10 (South Africa, 1951) is the only sample assigned to clade A (CV-A16∣A). The EV-A71 BrCr sequence was used as an outgroup. Distance thresholds used for demarcation of phylogenetic clusters are indicated with dashed lines. (B) Variations in the designations of CV-A16 phylogenetic groups (genogroups and subgenogroups) in the literature. (Studies I to IV) The table shows the correspondence between phylogenetic data reported in the present study (I) and the CV-A16 groups described in the following other studies: Zhang et al. (27) and Sun et al. (28) (II), Li et al. (31) and Iwai et al. (29) (III), and Zong et al. (30) (IV). A number of discrepancies in the designations of the main CV-A16 phylogenetic groups are indicated./,not applicable; AUS, Australia; CHN, China; DEU, Germany; FRA, France; HUN, Hungary; JPN, Japan; KOR, South Korea; MYS, Malaysia; PER, Peru; ZAF, South Africa; RUS, Russia; THA, Thailand; VNM, Vietnam. (C) Scatterplot comparing the pairwise nucleotide distances calculated with the VP1 and 3CD gene sequences of a sample of 181 CV-A16. The sample comprised 111 strains of the present study and 70 strains for which sequence data are publicly available. The main genetic clusters are delimited by dashed lines.
A scatterplot of pairwise nucleotide p-distances was drawn for a subset of 181 virus strains (this study, n = 111; GenBank, n = 70) to compare the VP1 structural gene (P1 genomic region) and a nonstructural locus (3CD, in the P3 region). Genetic distances among molecular sequences were distributed within three clusters, indicating extended genetic diversity in CV-A16 sequence populations (Fig. 2C). The pattern showed that the previously unreported clade was genetically divergent in the genomic region P3 and equally distant from genogroups A and B.
Evolutionary history of CV-A16 estimated with the VP1 sequence data set.
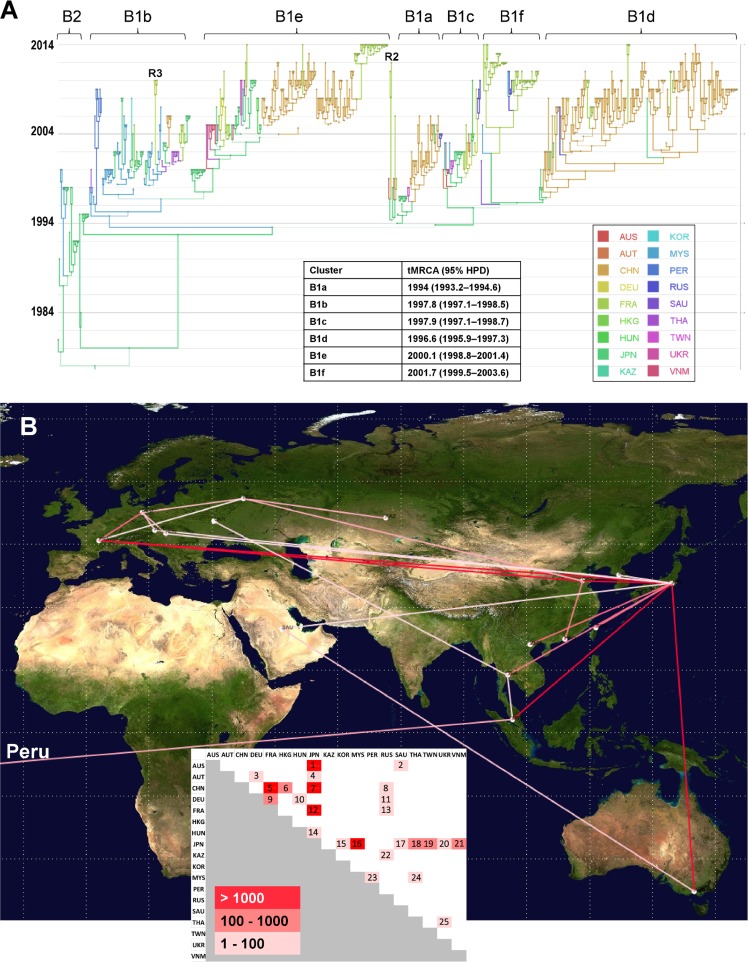
The global history of CV-A16 was reconstructed through a phylogenetic analysis of VP1 sequences sampled worldwide to investigate the temporal transmission of virus strains and the evolutionary origin of clade D. The CV-A16 genealogy depicted eight main clusters supported by posterior probability (PP) values of >0.9, of which seven belonged to genogroup B (Fig. 3A). The remaining one was clade D (Fig. 4A). The time span of sequence sampling covered the three most recent decades, and the time of origin of CV-A16 was dated 1907 (95% highest posterior probability density [HPD] interval, 1860 to 1945). The predominant B1 cluster arose in 1992 (95% HPD, 1990 to 1994), and the minor B2 cluster in 1978 (95% HPD, 1973 to 1981). The shape of the whole tree was typical of cocirculation of multiple virus lineages over time and across geographic regions. This pattern was also seen for individual lineages (notably, B1b and B1d to B1f). The phylogenetic structure of lineages B1a, B1d, and B1e also indicated replacement of virus lineages over time. The overall topology suggested different transmission histories for the individual B1 lineages at a worldwide scale.
FIG 3.
Phylogeography of coxsackievirus A16 genogroup B. (A) Temporal distribution of lineages. Time is indicated on the left as calendar years in the MCC tree reconstructed with a Bayesian MCMC analysis (discrete phylogeographic model). The branches and tree nodes are colored according to the geographic location that had the highest probability. The size of each circle represents the location's probability. Line width indicates the posterior probability of the corresponding lineage (thick lines indicate high posterior probability values). The time origins (TMRCA) of the main clusters are indicated in the inset table. The tree topology was used to assess the most probable virus migration events between countries shown in panel B. R2, probable intertypic recombinant strain (see Fig. 5A); R3, probable intratypic recombinant strain (see Fig. 5C); AUS, Australia; AUT, Austria; CHN, China; DEU, Germany; FRA, France; HKG, Hong Kong; HUN, Hungary; JPN, Japan; KAZ, Kazakhstan; KOR, South Korea; MYS, Malaysia; PER, Peru; RUS, Russia; SAU, Saudi Arabia; THA, Thailand; TWN, Taiwan; UKR, Ukraine; VNM, Vietnam. (B) Patterns of spatial spread assessed among sampling countries. The sampling countries are indicated with filled white circles. The lines connecting countries are colored according to the heat map (inset) showing Bayes factor (BF) values estimated between two geographic locations. Map source: Google, 2016.
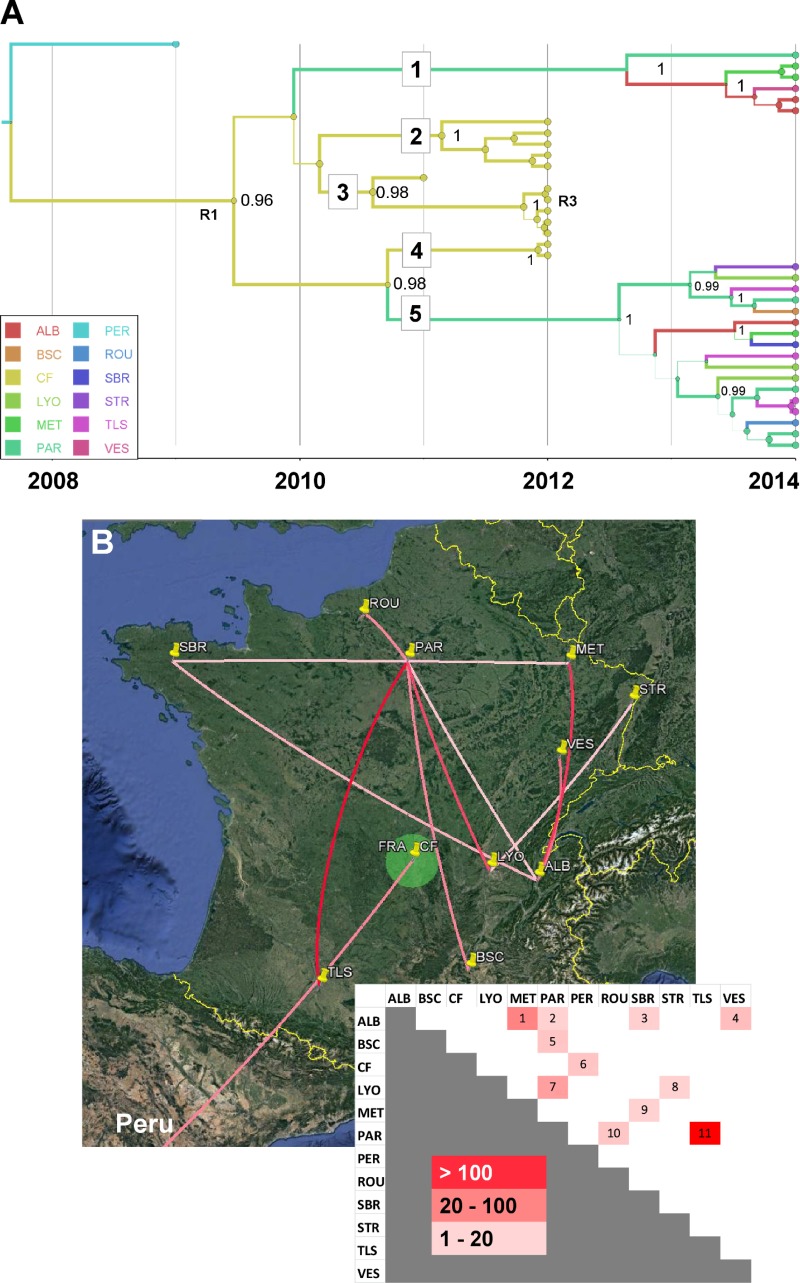
FIG 4.
Phylogeography of coxsackievirus A16 clade D. Temporal distribution of lineages (A) and pattern of spatial spread assessed among French cities (B). Map source: Google, 2016. See the legend to Fig. 3 for details. R1 indicates a probable intertypic recombination event associated with the emergence of genogroup D and R3 an intratypic recombinant (see Fig. 5C). ALB, Aix les Bains; BSC, Bagnols sur Ceze; CF, Clermont-Ferrand; LYO, Lyon; MET, Metz; PAR, Paris; PER, Peru; ROU, Rouen; SBR, Saint-Brieuc; STR, Strasbourg; TLS, Toulouse; VES, Vesoul.
The genealogy of the previously unreported clade D was distinct from that of genogroup B, but its precise evolutionary origin remained elusive because of insufficiently dense virus sampling in the 1990s and early 2000s. The time to the most recent common ancestor (TMRCA) of clade D was 2006 (95% HPD, 2004 to 2008), and the phylogeny of this clade showed five virus lineages, which depicted virus transmission between French cities after 2009 (see below).
Spatial processes of CV-A16 transportation at different geographic scales.
A model that explicitly incorporated the countries where the CV-A16 sequences were sampled was used to investigate virus spread between 18 different geographic locations for clade B (Fig. 3A). The analysis provided phylogenetic evidence of 25 virus transportation events with statistical significance assessed by Bayes factor values of >3 (Fig. 3B). The geographic patterns of virus spread assessed do not imply direct transmission between sampled countries. Japan was the country most frequently involved in virus transportation, with 12 distinct geographic locations (Australia, Austria, China, France, Hungary, South Korea, Malaysia, Saudi Arabia, Thailand, Taiwan, Ukraine, and Vietnam). France was assessed in three long-distance virus transportation events (with China, Russia, and Japan) and one with Germany, which was also involved in short-distance virus transportation events (with Austria and Hungary) and a long-distance virus transportation event, with Russia. In the analysis, Peru was assessed as being connected with Malaysia.
The transportation of CV-A16 clade D viruses between 11 cities sampled in France was analyzed. The 23 viral sequences of lineages 1 and 5 had 10 different sampling origins (Fig. 4A). The diversity of lineages indicated active virus spread in 2013 and 2014 all over the country and provided evidence of 11 virus transportation events (Fig. 4B). The p-distances within lineages ranged between 1% nucleotide differences (lineage 1) and 1.6% nucleotide differences (lineage 5), and the sequences within each of the two lineages had a recent common ancestor (TMRCA of <1.5 year before 2014), which suggested distinct transmission chains. In contrast, the viral sequences of lineages 2, 3, and 4 were sampled in a single geographic location (Clermont-Ferrand) in 2011 and 2012. The phylogenetic pattern was consistent with the hypothesis of long-term virus circulation in the general population living in this area between 2009 and 2012.
Genetic origins of different CV-A16 clades and lineages.
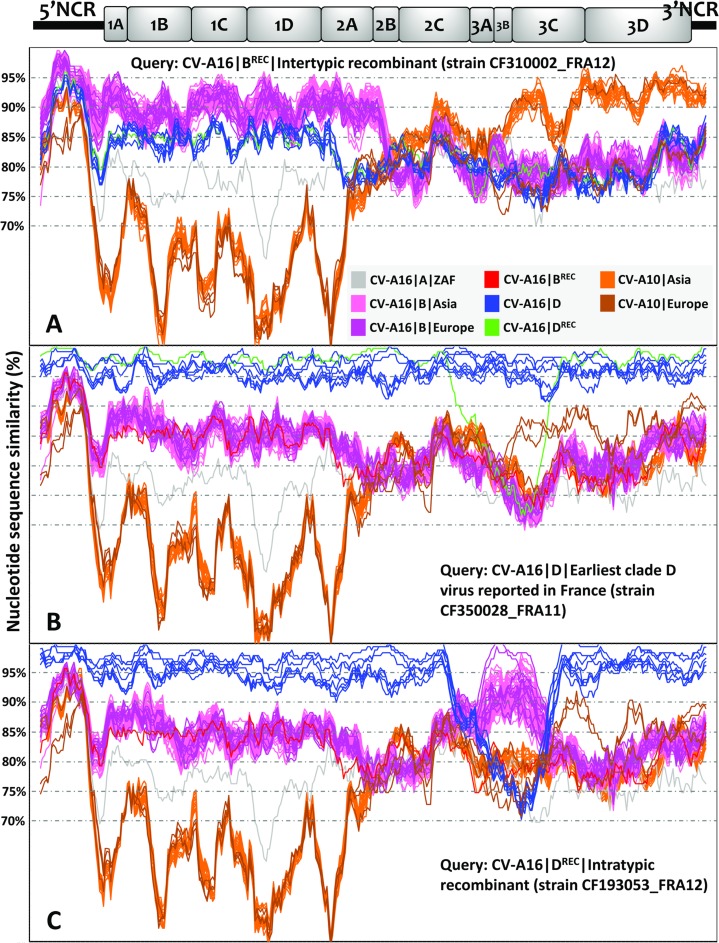
The genetic origin of CV-A16 clade D was investigated by analysis of the complete genomes of 23 CV-A16 (genogroup B, n = 13; clade D, n = 10) and 3 coxsackievirus A10 (CV-A10) isolates recovered in patients with HFMD (France, 2010). CV-A10 isolates were identified in an exploratory analysis as close genetic relatives of CV-A16 among a sample of EV-A genomes (data not shown). The 28 genomes were compared with publicly available genomes of 101 CV-A16 and 28 CV-A10 isolates (see Table S1 in the supplemental material). The plots shown in Fig. 5 indicated substantial variations in nucleotide similarity along the genomes between clades A, B, and D. The CV-A16 clade B genomes sampled in Asia between 1997 and 2014 and in Europe between 2003 and 2012 had substantial similarities. An exception was a recombinant genome (CV-A16∣BREC∣CF310002_FRA12) whose 3′ half was more similar to CV-A10 genomes sampled in Asia than to CV-A16 clade B genomes sampled in France (Fig. 5A). The recombination analysis of the CV-A16∣BREC genome with the RDP program (35) provided statistical evidence of a recombination breakpoint at position 3550 (2A gene) with six out of seven methods (P value range, 7.7 × 10−16 to 2.1 × 10−67). The limited nucleotide similarity over genes 2B to 3C indicated that the relatedness with the CV-A10 lineages was ancient or was inherited via a complex pathway of recombination events.
FIG 5.
Nucleotide similarities between CV-A16 and CV-A10 full-length genomes. The similarity plots were obtained with a data set of 118 CV-A16 genomes (including 24 determined in this study) and 31 CV-A10 genomes (including 3 determined in this study). A sliding window of 200 nucleotides moving in steps of 20 nucleotides was used in the analyses. The sequences chosen as queries are as follows: CV-A16∣B∣CF310002∣FRA12 (accession number LT617110, example of intertypic recombination) (A); CV-A16∣D∣CF350028∣FRA11 (accession number LT617105, earliest representative of clade D) (B); and CV-A16∣D∣CF193053∣FRA12 (accession number LT617109, example of intratypic recombination) (C). A schematic diagram of the CV-A16 genome (x axis) indicates the approximate boundaries of untranslated regions and gene sequences. The color coding for CV-A16 genogroups is gray for genogroup A, pink for genogroup B, and blue for genogroup D, except that the CV-A16∣CF193053∣FRA12 sequence is in green; CV-A10 sequences are shown in orange and brown. NCR, noncoding region.
All the CV-A16 clade D genomes but one were strictly colinear with one another from the 5′ noncoding region to the 3′ end (Fig. 5B). The region located between the 3B gene and the 3′ end of these genomes displayed high nucleotide similarity with a set of 2 CV-A10 genomes collected in France in 2010, but the relatedness was only distant. The mean nucleotide similarity between CV-A16 clade B and D genomes was 83%. An incongruent clade D genome (CV-A16∣DREC∣CF193053_FRA12) had >95% nucleotide similarity with clade B genomes sampled in France and Germany (Fig. 5C). The precise boundaries of the recombinant segment (genes 2C, 3A, 3B, and 3C) were assessed with six out of seven RDP methods as being at nucleotide positions 4890 and 5593 (P value range, 3.0 × 10−27 to 1.2 × 10−92), which confirmed the occurrence of intratype recombination.
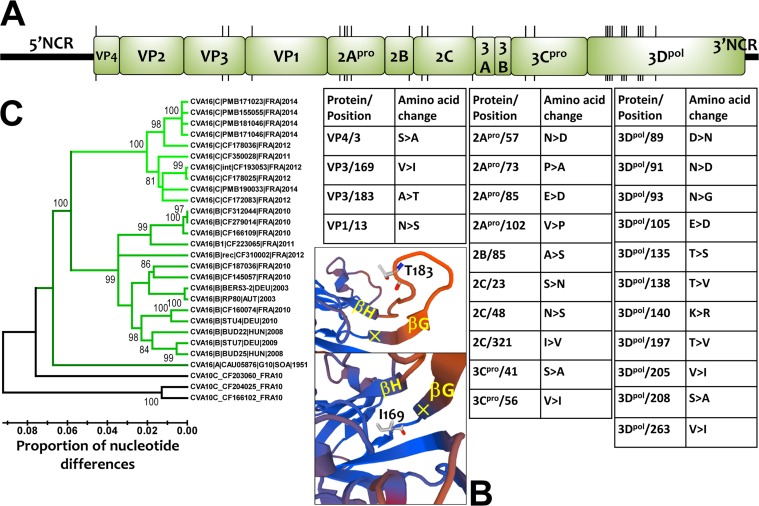
The comparative analysis of the open reading frame (ORF) sequences showed 25 amino acid changes between CV-A16 clades B and D, most of which (n = 21) were scattered in the nonstructural proteins (Fig. 6A). Notably, the 2Apro and 3Dpol viral proteins had changes at 4 and 11 amino acid positions, respectively. Molecular modeling with the coordinates of a crystal structure of CV-A16 (PDB identifier [ID] 5C9A) (36) showed that two amino acid changes were located within the VP3 capsid protein. One was exposed at the virion's top surface between β-sheets G and H, and the other embedded within the capsid at the base of β-sheet G (Fig. 6B). The two amino acid differences between clades B and D in the VP4 and VP1 proteins were located inside the virus particle and were not involved in the antigenic features of the virus. The nucleotide sequences of the 5′ noncoding region were heterogeneous within clades B and D (Fig. 6C). Clade A was equally distant from clades B (13.6% nucleotide differences) and D (13.3%), and clades B and D differed by 11.6% nucleotide variations.
FIG 6.
Genomic and amino acid variations among CV-A16 clade D viruses. (A) Locations of codons bearing amino acid changes between CV-A16 clade B and D viruses. The precise locations and the amino acid changes (genogroup B→clade D) are indicated in the tables. (B) Three-dimensional model of the VP3 protein reconstructed for a clade D virus (CF350028∣FRA11; accession number LT617105) showing two amino acid changes in the GH loop (top) and the base of the GH loop (bottom). (C) Phylogenetic tree based on the complete sequences of the 5′ untranslated region of CV-A16, showing the monophyletic topologies of CV-A16 clades B and D and the intraclade diversity. The sequences of CV-A10 were used as an outgroup; the tree was inferred with the neighbor-joining method.
DISCUSSION
Combining data from longitudinal HFMD surveillance and global phylogenetic analyses of viral sequences, we show that the spatial migration dynamics of CV-A16 was a major determinant in the epidemiological pattern seen in France. European countries were epidemiologically connected with distant geographic locations in Asia and South America.
The study expands the diversity of CV-A16 to a clade whose epidemiological origin was traced back to Peru. Earlier comparative analyses of the VP4 and VP1 protein genes suggested that CV-A16 strains circulating worldwide belonged to three genogroups, designated A, B, and C (27–33). Our study revealed the recent emergence and effective spread of a previously undescribed genogroup that we suggest be designated by the letter D. The strains of genogroup D cocirculated with those of genogroup B in France over the period studied. Both genogroups had a mean evolutionary rate of 4.5 × 10−3 substitutions/site/year, close to the rates determined for other EV types (37). Molecular comparisons of nearly complete genomes showed that amino acid residues in the antigenic sites reported for genogroup B (36) were similar to those in genogroup D. The amino acid variation at position 183 in the VP3 capsid protein (Ala in genogroup B versus Thr in genogroup D) was the only exception. This position is located within the GH loop, a structure reported to form an antigenic site with the VP1 GH loop in an adjacent protomer of the viral capsid (38). The other two amino acid changes found in the VP4 and VP1 proteins were unlikely to be involved in antigenic features because of their internal location within the capsid. In addition, the genomic comparisons showed the recombinant origin of genogroup D. The emergence of genogroup D may have resulted from genetic features acquired through recombination, as suggested for EV-A71 (39, 40). Further studies are needed to determine whether the concurrent circulation dynamics of CV-A16 genogroups B and D in the general population are determined by antigenic properties and/or factors in the 5′ untranslated region (UTR) and nonstructural proteins.
The earliest strain of CV-A16 genogroup D was reported in 2009 in Iquitos, Loreto region, northeastern Peru, a city located in the Amazon rainforest (34). A closely related strain was reported in France 2 years later, which suggests long-range virus transportation. The intercity virus transmission events inferred from the phylogeny of this genogroup and the detection of an intratype recombinant strain provided evidence of an active spread of the virus throughout France between 2009 and 2014. The five lineages of genogroup D had more than 3% nucleotide variations. This could indicate that different transmission chains persisted over the years if we assume a maximum threshold of 1 to 2% nucleotide differences to establish an epidemiological link among EV strains (41). Consistent with this hypothesis, the continuing HFMD surveillance provided epidemiological evidence that a number of cases reported in France in 2016 were caused by genogroup D viruses (data not shown).
Although different CV-A16 lineages were identified in France, they were not associated with HFMD outbreaks and the HFMD manifestations caused by viruses of genogroups B and D were clinically indistinguishable. Accounting for less than 25% of all HFMD cases over the surveillance period, CV-A16 was not a predominant cause of the disease among children, other than a temporary increase in the infection frequency during the summer of 2012. The CV-A16 transmission was in sharp contrast with the epidemic pattern of CV-A6 over the same period (25, 26). In countries of East and Southeast Asia, CV-A16 has been associated with large HFMD outbreaks in the pediatric population (6, 42). Children documented with HFMD in our study had a median age of 2.4 years. This epidemiological pattern was consistent with the data of a seroepidemiology study performed in Germany in 2006, in which a rate of neutralizing antibodies against CV-A16 of only 27% was reported in children aged 1 to 4 years (43). The CV-A16 strains sampled over the sole year 2014 in France were unlikely to have been epidemiologically related because they were assigned to five phylogenetically distinct lineages, three in genogroup B and two in genogroup D. This finding is consistent with the distinct evolutionary pathways of genogroup B lineages as assessed by their time origins (large TMRCA range, 1995 to 2002) and the diversity of geographic sampling locations. Overall, the data indicating that HFMD cases could arise through multiple introductions of distinct genetic lineages during a same year may be relevant for disease outbreak modeling from surveillance data (20). Modeling HFMD outbreaks on the assumption of only one index case may not reflect epidemiological reality.
The occurrence of HFMD cases in France was associated with extensive CV-A16 migration at three geographic scales: (i) within the country, (ii) between France and neighboring countries, and (iii) between France and distant countries. The first pattern was exemplified by the intercity spread of CV-A16 genogroup D over 3 years after the initial report in Peru (34). The second pattern was assessed by cross-border virus exchanges between Germany and France but also within Europe between Germany and Austria and Hungary. The spread of CV-A16 was also accelerated by migration to distant countries between Europe and Asia or South America, as shown by transcontinental transportation events inferred between France and China, Japan, Russia, and Peru. CV-A16 migration was also assessed between Japan and continental China and Taiwan, Kazakhstan, and Russia, as well as Australia and Japan and the Kingdom of Saudi Arabia. Although the CV-A16 sequences sampled in Japan represented only 13% of the VP1 data set analyzed in the present study, this country was involved in 48% of the transportation events assessed for genogroup B viruses. This is consistent with epidemiological data reporting a high seasonal incidence of CV-A16 infections in Japan in 2007, 2008, and 2011 (32, 33; http://www.nih.go.jp/niid/en/iasr-e.html). The spread of virus strains between distant geographic areas deserves special attention as a cause of outbreaks, as reported for poliomyelitis upsurges in population communities with low levels of vaccine-induced immunity (44).
Challenges with data collection impose certain limitations on the conclusions that can be drawn from our study. Notably, the global virus transportation events are underestimated because there are scarce CV-A16 sequence data available for Africa, the Americas, and Europe. Despite this inherent bias, the phylogenetic patterns provided key underlying determinants for HFMD dynamics at a worldwide scale and for how EVs can persist during seasonal troughs. The role of recombination in the evolutionary dynamics of CV-A16 might have been underestimated because complete genomes were investigated in limited numbers.
CV-A16 transmission is driven by movements of infected individuals at different geographic levels, and the epidemiological patterns of HFMD are shaped by viral evolutionary dynamics and global virus spread. This finding and the evidence of widespread circulation of a recombinant CV-A16 variant could have implications for disease prevention, because Asia is epidemiologically interconnected with distant geographic areas. Accordingly, HFMD and other EV-associated diseases need global surveillance. Developing effective control strategies against HFMD will require a comprehensive understanding of transmission dynamics within the susceptible host population in countries where the disease burden is widespread but also in those where it is not recognized as a significant public health concern.
MATERIALS AND METHODS
Longitudinal community-based studies in France.
The CV-A16 strains were sampled during a prospective HFMD surveillance in French children whose attending pediatricians practiced in the urban area of Clermont-Ferrand (April 2010 to December 2014) and in other French cities (April to December 2014). The criteria for including children in the studies encompassed acute fever (body temperature of >37.5°C) with mucocutaneous symptoms such as oral ulcers on the tongue and buccal mucosa with vesicular rash on the hands, feet, knees, or buttocks. Demographic and clinical information for each patient enrolled, which included the children's age, sex, and medical status relating to mouth ulcers, vesicles on hands and feet, and fever, were recorded in an anonymized standard form and reviewed retrospectively. Throat swabs were obtained by the examining pediatricians, batched, and sent to our laboratory in viral transport medium. The EV strains were identified with molecular typing methods described earlier (25). Briefly, partial sequences of the 1D gene encoding the VP1 capsid protein were amplified from throat swabs and compared with reference sequences by BLAST analysis.
Written informed consent was obtained from all participants or guardians on behalf of the child participants.
Gene amplification and nucleotide sequencing of PCR products.
The complete 1D/VP1 gene sequence was amplified with either a single round (amplicon length of 1,200 bp) or seminested (2,240 bp) PCR assay. The 3CD locus sequence was amplified with a single round (amplicon length of 1,176 bp) using two primers, HEVAS3CD and HEVAR3CD, as previously described (45). The complete viral genomes were determined through two gene amplification stages performed with the Phusion Flash polymerase (Thermo Scientific) and oligonucleotide primers (Table 1). The 5′ part of the viral genomes (from the 5′ noncoding region to the 2C gene, 4,450 nt) was amplified with the IFT7R5NC and HEVAR2C primers and the 3′ part (2C gene to 3′ noncoding region, 3,000 nt) with the HEVAR2C and HEVA3NC primers. The following amplification conditions were used: one cycle of 10 s at 98°C, 41 cycles consisting of 5 s at 98°C and 70 s at 72°C, and a final cycle of 2 min at 72°C. The sequencing reactions were performed by primer walking with the ABI Prism BigDye terminator version 1.1 cycle sequencing kit (Applied Biosystems). After a purification step with the NucleoSEQ kit (Macherey-Nagel), the sequencing products were analyzed with an ABI 3500 genetic analyzer (Applied Biosystems).
TABLE 1.
Oligonucleotide primers used in gene amplification reactions
| Primer | Genomic region | Nucleotide sequence (5′→3′)a |
|---|---|---|
| IFT7R5NC | 5′ noncoding region | TTAAAACAGCCTGTGGGTTG |
| 5NCS663 | 5′ noncoding region | GCGGAACCGACTACTTTGGGTGTCCGTGTTTC |
| HEVAS1495 | VP3 protein coding sequence | GGTNATNCCNTGGATAAGYAACAC |
| CA16-1D-S1 | VP1 protein coding sequence | TGYCCGAAYAAYATGATGGGCAC |
| 1D-6-S1 | VP1 protein coding sequence | RCACGTCAGGGCGTGGRTAC |
| HEVAR2807 | 2A protease coding sequence | CYACNGCNCARGGNTGYGACACGA |
| HEVAR2C | 2C protein coding sequence | CATGCAGTTCAAGAGCAARCACCG |
| HEVAS2C | 2C protein coding sequence | CATGCAGTTCAAGAGCAARCACCG |
| 3CD-9-R1 | 3C protease coding sequence | GAGCACATGCCMTCAATG |
| HEVAS3CD | 3C protease coding sequence | GACCARGGNCAYTTYACNATGYTRGG |
| HEVAR3CD | 3D polymerase coding sequence | GGAGYAARYTACCRATYCTA |
| HEVA3NC | 3′ noncoding region | GTGGGGGTAAATTTGTTATAACCAGAATAGC |
Nucleotides are designated according to the standard IUPAC code.
Data collection and compilation of nucleotide sequence data sets.
The CV-A16 nucleotide sequences determined were compared with publicly available sequence data (as of July 2016). For the VP1 data set, sequences <891 nt long were discarded and only those with fully specified dates (year) and countries of origin were used. The complete genome data set was constructed by collating all CV-A16 genomes available. The 3CD sequence data set was derived from the complete genome data set. The sequence data sets were constructed with the BioEdit version 7.2.5 software (http://www.mbio.ncsu.edu/bioedit/bioedit.html). All the sequence data sets are available upon request.
Genetic diversity analysis.
The diversity of CV-A16 lineages was analyzed with the neighbor-joining phylogenetic method available in MEGA5.01 (46). The proportions of nucleotide differences were estimated in pairwise comparisons (p-distance model), and the phylogenetic relatedness was represented with a tree. Statistical consistency of tree nodes was estimated by the bootstrap method (1,000 random samplings).
Phylogenetic reconstruction of CV-A16 phylogeny and geographic spread of virus strains.
The CV-A16 genealogical tree was reconstructed with the BEAST version 1.8.4 program (47). An uncorrelated lognormal prior distribution of nucleotide substitution rates among lineages was used for the molecular clock model. The evolutionary history was modeled with the general time reversible (GTR) model as the nucleotide substitution model. The phylogenetic parameters were coestimated in different runs of a Markov chain Monte Carlo (MCMC) process involving 200 × 106 generations. The Tracer version 1.5 program (http://tree.bio.ed.ac.uk/software/tracer/) was used to check for convergence of parameter estimates (operator effective sample size of >200). The trees estimated with the MCMC procedure were sampled to obtain a final 20,000 trees. The mean estimates and 95% highest posterior probability density (HPD) intervals calculated for each operator were compiled by analyzing the BEAST output files with the Tracer version 1.5 program. Maximum clade credibility (MCC) trees were calculated with the TreeAnnotator version 1.8.4 program (http://beast.bio.ed.ac.uk/treeannotator), and topological support was assessed by estimating the values of the posterior probability (PP) density of each node.
The countries where the virus strains were collected were used as discrete character states to estimate changes of geographic locations in the phylogenetic trees (48). The geographic locations of the ancestral nodes were coestimated with the phylogeny by a discrete phylogeographic diffusion model (symmetrical substitution model). Bayesian stochastic search variable selection was also used for identifying the statistically significant transition rates between locations. Information was summarized in MCC trees, with the branches colored according to geographic locations. Significant virus transportation events (migration) were identified with the Bayes factor test implemented in the Bayesian stochastic search variable selection (BSSVS) procedure (48).
Recombination analysis.
Patterns of nucleotide similarity between a large sample of complete genomes, including different EV-A types, were explored with the SimPlot version 3.5.1 program (49), using a sliding window of 200 nt moving in steps of 20 nt. The molecular and phylogenetic signals indicative of possible recombination events within and between CV-A16 and CV-A10 genomes were further analyzed with the RDP version 4.56 computer program (35). The nucleotide sequences were analyzed by default methods (RDP, Geneconv, Bootscan, MaxChi, Chimera, SiScan, and Phylpro), and to exclude the possibility of false-positive detection, only putative recombinant signals assessed with five methods were considered.
Molecular modeling of the CV-A16 VP3 protein.
Structure-based in silico prediction of the VP3 capsid protein was carried out with the SWISS-MODEL, an automated protein structure homology-modeling server (50). An automatic search for a template was performed with the VP3 sequences of CV-A16 strains RP80∣AUT∣2003 (clade B) and CF350028∣FRA∣2011 (clade D). The PDB entry with ID 5C9A (36) was used in subsequent modeling.
Accession number(s).
The nucleotide sequences of CV-A16 were deposited in GenBank under accession numbers LT577699 to LT577844 (VP1 gene) and LT577845 to LT577930 (partial 3CD gene). The complete viral genomes were deposited under accession numbers LT617091 to LT617115 (CV-A16) and LT617116 to LT617118 (CV-A10).
Supplementary Material
ACKNOWLEDGMENTS
HFMD French Study Network. This study was made possible by the close involvement of all ambulatory pediatricians who took part in the sentinel surveillance system on behalf of the Association Française de Pédiatrie Ambulatoire (AFPA, http://www.afpa.org), including Anne-Sophie Belloin-Perraud, Pierre Blanc, Patrice Bouissou, Marie-Annick Burgess, Marie-Edith Burthey, Michelle Carsenti, Marie-Christine Chauvel, Robert Cohen, Caroline Colombet, François Corrard, Liliane Cret, Veronique Dagrenat-Taleb, Simon Dib, Philippe Franchineau, Cécile Guiheneuf, Jean-Louis Guillon, Sylvie Hubinois, Pierre Jarry, Flaviane Kampf-Maupu, Fabienne Kochert, Caroline Laffort, Anne-Louise Lambert, Francine Lecailler, Hélène Le Scornet, Murielle Louvel, Nicolas Mathieu, Jean-François Maurer, Bruno Mazauric, Agnès Mercier, Frédéric Mestre, Solange Moore-Wipf, Sylvie Pauliat-Desbordes, Florence Pichot-Jallas, Saholy Razafinarivo Schoreisz, Céline Rollet-Lautraite, Sylvie Sandid, Isabelle Sartelet, Simone Saumureau, Philippe Simon, Abderrahim Terrache, Georges Thiebault, Jean-Pierre Tiberghien, Eric Van Melkebeke, Sandrine Vernoux, François Vié le Sage, Martine Wagner-Vaucard, and Andreas Werner, and the Association des Pédiatres de la Région Auvergne—Formation Médicale Continue, including Frédérique Mestre, Véronique Desvignes, Michel Gannat, Béatrice Delcros, Juliette Binaud-Hadj, Michel Magnin, Alma Morin, Brigitte Bourbonnais, Philippe Franchineau, Geneviève Binet-Denier, Marie-Laure Herbelin-Wagner, Anne Piollet, Marie Annick Brotte-Miossec, Michelle Debost, Béatrice Nemesin, Philippe Dieterlen, and Sophie Jeancenelle.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
We are indebted to Gisela Enders, director of Laboratory Professor Gisela Enders & Kollegen MVZ (Stuttgart, Germany) for help and constant involvement in our European molecular epidemiology studies. We acknowledge the technical contributions of Gwendoline Jugie, Nathalie Rodde, and Isabelle Simon for helpful assistance with virus genotyping. We thank Jeffrey Watts for help in preparing the English manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00630-17.
REFERENCES
- 1.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 2.Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. 2010. Clinical features, diagnosis and management of human enterovirus 71 infection. Lancet Neurol 9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 3.Lum LC, Wong KT, Lam SK, Chua KB, Goh AY. 1998. Neurogenic pulmonary oedema and enterovirus 71 encephalomyelitis. Lancet 352:1391. doi: 10.1016/S0140-6736(05)60789-1. [DOI] [PubMed] [Google Scholar]
- 4.Chang LY, King CC, Hsu KH, Ning HC, Tsao KC, Li CC, Huang YC, Shih SR, Chiou ST, Chen PY, Chang HJ, Lin TY. 2002. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics 109:e88. doi: 10.1542/peds.109.6.e88. [DOI] [PubMed] [Google Scholar]
- 5.Yang B, Wu P, Wu JT, Lau EH, Leung GM, Yu H, Cowling BJ. 2015. Seroprevalence of enterovirus 71 antibody among children in China: a systematic review and meta-analysis. Pediatr Infect Dis J 34:1399–1406. doi: 10.1097/INF.0000000000000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao Q, Wang Y, Yao X, Bian L, Wu X, Xu M, Liang Z. 2014. Coxsackievirus A16: epidemiology, diagnosis, and vaccine. Hum Vaccin Immunother 10:360–367. doi: 10.4161/hv.27087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright HT Jr, Landing BH, Lennette EH, McAllister RM. 1963. Fatal infection in an infant associated with Coxsackie virus group A, type 16. N Engl J Med 268:1041–1044. doi: 10.1056/NEJM196305092681904. [DOI] [PubMed] [Google Scholar]
- 8.Wang CY, Li Lu F, Wu MH, Lee CY, Huang LM. 2004. Fatal coxsackievirus A16 infection. Pediatr Infect Dis J 23:275–276. doi: 10.1097/01.inf.0000115950.63906.78. [DOI] [PubMed] [Google Scholar]
- 9.Legay F, Lévêque N, Gacouin A, Tattevin P, Bouet J, Thomas R, Chomelt JJ. 2007. Fatal coxsackievirus A-16 pneumonitis in adult. Emerg Infect Dis 13:1084–1086. doi: 10.3201/eid1307.070295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astrup BS, Johnsen IB, Engsbro AL. 2016. The role of coxsackievirus A16 in a case of sudden unexplained death in an infant—a SUDI case. Forensic Sci Int 259:e9–e13. doi: 10.1016/j.forsciint.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Robinson CR, Doane FW, Rhodes AJ. 1958. Report of an outbreak of febrile illness with pharyngeal lesions and exanthem: Toronto, summer 1957; isolation of group A Coxsackie virus. Can Med Assoc J 79:615–621. [PMC free article] [PubMed] [Google Scholar]
- 12.Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, Chang Z, Liu F, Fang VJ, Zheng Y, Cowling BJ, Varma JK, Farrar JJ, Leung GM, Yu H. 2014. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis 14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabrerizo M, Tarragó D, Muñoz-Almagro C, Del Amo E, Domínguez-Gil M, Eiros JM, López-Miragaya I, Pérez C, Reina J, Otero A, González I, Echevarría JE, Trallero G. 2014. Molecular epidemiology of enterovirus 71, coxsackievirus A16 and A6 associated with hand, foot and mouth disease in Spain. Clin Microbiol Infect 20:O150–O156. doi: 10.1111/1469-0691.12361. [DOI] [PubMed] [Google Scholar]
- 14.Ljubin-Sternak S, Slavic-Vrzic V, Vilibić-Čavlek T, Aleraj B, Gjenero-Margan I. 2011. Outbreak of hand, foot and mouth disease caused by Coxsackie A16 virus in a childcare centre in Croatia, February to March 2011. Euro Surveill 16(21):pii=19875 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19875. [PubMed] [Google Scholar]
- 15.Klein M, Chong P. 2015. Is a multivalent hand, foot, and mouth disease vaccine feasible? Hum Vaccin Immunother 11:2688–2704. doi: 10.1080/21645515.2015.1049780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bessaud M, Razafindratsimandresy R, Nougairède A, Joffret ML, Deshpande JM, Dubot-Pérès A, Héraud JM, de Lamballerie X, Delpeyroux F, Bailly JL. 2014. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS One 9:e90624. doi: 10.1371/journal.pone.0090624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu FC, Liang ZL, Meng FY, Zeng Y, Mao QY, Chu K, Song XF, Yao X, Li JX, Ji H, Zhang YJ, Li L, Pan HX, Xu K, Dai WM, Zhang WW, Deng F, Wang H, Wang JZ. 2012. Retrospective study of the incidence of HFMD and seroepidemiology of antibodies against EV71 and CoxA16 in prenatal women and their infants. PLoS One 7:e37206. doi: 10.1371/journal.pone.0037206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Cao K, Zhang Y, Fang L, Li X, Xu Q, Huang F, Tao L, Guo J, Gao Q, Guo X. 2016. Different effects of meteorological factors on hand, foot and mouth disease in various climates: a spatial panel data model analysis. BMC Infect Dis 16:233. doi: 10.1186/s12879-016-1560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh WM, Bogich T, Siegel K, Jin J, Chong EY, Tan CY, Chen MI, Horby P, Cook AR. 2016. The epidemiology of hand, foot and mouth disease in Asia: a systematic review and analysis. Pediatr Infect Dis J 35:e285–e300. doi: 10.1097/INF.0000000000001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi S, Liao Q, Van Boeckel TP, Xing W, Sun J, Hsiao VY, Metcalf CJ, Chang Z, Liu F, Zhang J, Wu JT, Cowling BJ, Leung GM, Farrar JJ, van Doorn HR, Grenfell BT, Yu H. 2016. Hand, foot, and mouth disease in China: modeling epidemic dynamics of enterovirus serotypes and implications for vaccination. PLoS Med 13:e1001958. doi: 10.1371/journal.pmed.1001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassel C, Mirand A, Lukashev A, TerletskaiaLadwig E, Farkas A, Schuffenecker I, Diedrich S, Huemer HP, Archimbaud C, Peigue-Lafeuille H, Henquell C, Bailly JL. 2015. Transmission patterns of human enterovirus 71 to, from and among European countries, 2003 to 2013. Euro Surveill 20(34):pii=30005. doi: 10.2807/1560-7917.ES.2015.20.34.30005. [DOI] [PubMed] [Google Scholar]
- 22.Othman I, Mirand A, Slama I, Mastouri M, Peigue-Lafeuille H, Aouni M, Bailly JL. 2015. Enterovirus migration patterns between France and Tunisia. PLoS One 10:e0145674. doi: 10.1371/journal.pone.0145674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuffenecker I, Mirand A, Antona D, Henquell C, Chomel JJ, Archimbaud C, Billaud G, Peigue-Lafeuille H, Lina B, Bailly JL. 2011. Epidemiology of human enterovirus 71 infections in France, 2000-2009. J Clin Virol 50:50–56. doi: 10.1016/j.jcv.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Böttcher S, Obermeier PE, Neubauer K, Diedrich S, Laboratory Network for Enterovirus Diagnostics. 2016. Recombinant enterovirus A71 subgenogroup C1 strains, Germany, 2015. Emerg Infect Dis 22:1843–1846. doi: 10.3201/eid2210.160357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirand A, Henquell C, Archimbaud C, Ughetto S, Antona D, Bailly JL, Peigue-Lafeuille H. 2012. Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect 18:E110–E118. doi: 10.1111/j.1469-0691.2012.03789.x. [DOI] [PubMed] [Google Scholar]
- 26.Mirand A, le Sage FV, Pereira B, Cohen R, Levy C, Archimbaud C, Peigue-Lafeuille H, Bailly JL, Henquell C. 2016. Ambulatory pediatric surveillance of hand, foot and mouth disease as signal of an outbreak of coxsackievirus A6 infections, France, 2014-2015. Emerg Infect Dis 22:1884–1893. doi: 10.3201/eid2211.160590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang D, Yan D, Zhu S, Liu J, Wang H, Zhao S, Yu D, Nan L, An J, Chen L, An H, Xu A, Xu W. 2010. Molecular evidence of persistent epidemic and evolution of subgenotype B1 coxsackievirus A16-associated hand, foot, and mouth disease in China. J Clin Microbiol 48:619–622. doi: 10.1128/JCM.02338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun T, Liu Y, Zhang Y, Zhou L. 2014. Molecular phylogeny of coxsackievirus A16. J Clin Microbiol 52:3829–3830. doi: 10.1128/JCM.01330-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwai M, Masaki A, Hasegawa S, Obara M, Horimoto E, Nakamura K, Tanaka Y, Endo K, Tanaka K, Ueda J, Shiraki K, Kurata T, Takizawa T. 2009. Genetic changes of coxsackievirus A16 and enterovirus 71 isolated from hand, foot, and mouth disease patients in Toyama, Japan between 1981 and 2007. Jpn J Infect Dis 62:254–259. [PubMed] [Google Scholar]
- 30.Zong W, He Y, Yu S, Yang H, Xian H, Liao Y, Hu G. 2011. Molecular phylogeny of Coxsackievirus A16 in Shenzhen, China, from 2005 to 2009. J Clin Microbiol 49:1659–1661. doi: 10.1128/JCM.00010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, He Y, Yang H, Zhu J, Xu X, Dong J, Zhu Y, Jin Q. 2005. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J Clin Microbiol 43:3835–3839. doi: 10.1128/JCM.43.8.3835-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosoya M, Kawasaki Y, Sato M, Honzumi K, Hayashi A, Hiroshima T, Ishiko H, Kato K, Suzuki H. 2007. Genetic diversity of coxsackievirus A16 associated with hand, foot, and mouth disease epidemics in Japan from 1983 to 2003. J Clin Microbiol 45:112–120. doi: 10.1128/JCM.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuta K, Abiko C, Aoki Y, Ikeda T, Matsuzaki Y, Hongo S, Itagaki T, Katsushima N, Ohmi A, Nishimura H, Ahiko T. 2013. Molecular epidemiology of Coxsackievirus A16 strains isolated from children in Yamagata, Japan between 1988 and 2011. Microbiol Immunol 57:400–405. doi: 10.1111/1348-0421.12041. [DOI] [PubMed] [Google Scholar]
- 34.Carrion G, Huaman JL, Silva M, Ampuero JS, Paz I, Ocaña VR, Laguna-Torres VA, Hontz RD. 2016. Molecular epidemiology of coxsackievirus A16 strains from four sentinel surveillance sites in Peru. Int J Infect Dis 52:83–85. doi: 10.1016/j.ijid.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He YQ, Chen L, Xu WB, Yang H, Wang HZ, Zong WP, Xian HX, Chen HL, Yao XJ, Hu ZL, Luo M, Zhang HL, Ma HW, Cheng JQ, Feng QJ, Zhao DJ. 2013. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus A6- and coxsackievirus A10-associated hand, foot, and mouth disease infections from 2008 to 2012 in Shenzhen, China. J Clin Microbiol 51:3560–3566. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren J, Wang X, Zhu L, Hu Z, Gao Q, Yang P, Li X, Wang J, Shen X, Fry EE, Rao Z, Stuart DI. 2015. Structures of coxsackievirus A16 capsids with native antigenicity: implications for particle expansion, receptor binding, and immunogenicity. J Virol 89:10500–10511. doi: 10.1128/JVI.01102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang SW, Hsu YW, Smith DJ, Kiang D, Tsai HP, Lin KH, Wang SM, Liu CC, Su IJ, Wang JR. 2009. Reemergence of enterovirus 71 in 2008 in Taiwan: dynamics of genetic and antigenic evolution from 1998 to 2008. J Clin Microbiol 47:3653–3662. doi: 10.1128/JCM.00630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McWilliam Leitch EC, Cabrerizo M, Cardosa J, Harvala H, Ivanova OE, Koike S, Kroes AC, Lukashev A, Perera D, Roivainen M, Susi P, Trallero G, Evans DJ, Simmonds P. 2012. The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol 86:2676–2685. doi: 10.1128/JVI.06065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorba J, Campagnoli R, De L, Kew O. 2008. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J Virol 82:4429–4440. doi: 10.1128/JVI.02354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Wu S, Xiong Y, Li T, Wen Z, Yan M, Qin K, Liu Y, Wu J. 2014. Co-circulation and genomic recombination of coxsackievirus A16 and enterovirus 71 during a large outbreak of hand, foot, and mouth disease in Central China. PLoS One 9:e96051. doi: 10.1371/journal.pone.0096051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabenau HF, Richter M, Doerr HW. 2010. Hand, foot and mouth disease: seroprevalence of coxsackie A16 and enterovirus 71 in Germany. Med Microbiol Immunol 199:45–51. doi: 10.1007/s00430-009-0133-6. [DOI] [PubMed] [Google Scholar]
- 44.Gumede N, Jorba J, Deshpande J, Pallansch M, Yogolelo R, Muyembe-Tamfum JJ, Kew O, Venter M, Burns CC. 2014. Phylogeny of imported and reestablished wild polioviruses in the Democratic Republic of the Congo from 2006 to 2011. J Infect Dis 210:S361–S367. doi: 10.1093/infdis/jiu375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirand A, Schuffenecker I, Henquell C, Billaud G, Jugie G, Falcon D, Mahul A, Archimbaud C, Terletskaia-Ladwig E, Diedrich S, Huemer HP, Enders M, Lina B, Peigue-Lafeuille H, Bailly JL. 2010. Phylogenetic evidence for a recent spread of two populations of human enterovirus 71 in European countries. J Gen Virol 91:2263–2277. doi: 10.1099/vir.0.021741-0. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.