ABSTRACT
Mitochondria are crucial to proper neuronal function and overall brain health. Mitochondrial dysfunction within the brain has been observed in many neurodegenerative diseases, including prion disease. Several markers of decreased mitochondrial activity during prion infection have been reported, yet the bioenergetic respiratory status of mitochondria from prion-infected animals is unknown. Here we show that clinically ill transgenic mice overexpressing hamster prion protein (Tg7) infected with the hamster prion strain 263K suffer from a severe deficit in mitochondrial oxygen consumption in response to the respiratory complex II substrate succinate. Characterization of the mitochondrial proteome of purified brain mitochondria from infected and uninfected Tg7 mice showed significant differences in the relative abundance of key mitochondrial electron transport proteins in 263K-infected animals relative to that in controls. Our results suggest that at clinical stages of prion infection, dysregulation of respiratory chain proteins may lead to impairment of mitochondrial respiration in the brain.
IMPORTANCE Mitochondrial dysfunction is present in most major neurodegenerative diseases, and some studies have suggested that mitochondrial processes may be altered during prion disease. Here we show that hamster prion-infected transgenic mice overexpressing the hamster prion protein (Tg7 mice) suffer from mitochondrial respiratory deficits. Tg7 mice infected with the 263K hamster prion strain have little or no signs of mitochondrial dysfunction at the disease midpoint but suffer from a severe deficit in mitochondrial respiration at the clinical phase of disease. A proteomic analysis of the isolated brain mitochondria from clinically affected animals showed that several proteins involved in electron transport, mitochondrial dynamics, and mitochondrial protein synthesis were dysregulated. These results suggest that mitochondrial dysfunction, possibly exacerbated by prion protein overexpression, occurs at late stages during 263K prion disease and that this dysfunction may be the result of dysregulation of mitochondrial proteins.
KEYWORDS: mitochondrial metabolism, neurodegeneration, prions, scrapie, transmissible spongiform encephalopathy
INTRODUCTION
Mammalian prion diseases are fatal, transmissible neurodegenerative diseases in which the infectious agent is a misfolded, insoluble, protease-resistant form (PrPSc) of the normally soluble and protease-sensitive cellular prion protein (PrPC) (1). Pathogenesis in prion diseases is associated with accumulation of PrPSc as either amyloid or nonamyloid aggregates in the brain that can correlate with large-scale cell loss and gliosis. The precise mechanisms underlying cell death and ultimately organismal death in prion diseases are not yet understood. Some studies have shown that PrPSc can be toxic to neuronal cells and trigger endoplasmic reticulum (ER) stress ultimately followed by apoptosis (2). Arguing against the direct toxicity of protease-resistant PrPSc, clinical disease and death can occur in its absence, perhaps via small soluble PrPSc oligomers (3–7). It is also possible that loss of function of PrPC may be a factor in the spongiform changes seen in the brain following prion infection (8, 9), although conditionally ablating or suppressing PrPC expression after early clinical signs can reverse, instead of exacerbate, the disease process (10).
Synaptic degeneration associated with changes in PrPC may also be an important aspect of the neuropathological changes associated with clinical prion infection. At the initial stages of disease, infected animals display changes in behavior that correlate with the synaptic degeneration preceding neuronal death (11–13). PrPC can be enriched in axonal processes, neuronal synapses, and neuromuscular junctions, where its localization correlates well with synaptic proteins such as synaptophysin (14, 15). The conversion of synaptic PrPC to PrPSc as the disease progresses has been proposed as the cause of axonal degeneration and downstream neuronal cell death (14, 16, 17). Mechanistically, it has been suggested that mitochondrial dysfunction may actually precede synaptic degeneration, potentially making it a very early event in prion disease (18).
Mitochondrial dysfunction has been observed in many neurodegenerative diseases, including Parkinson's disease (PD), Alzheimer's disease (AD), Huntington's disease, and amyotrophic lateral sclerosis (19, 20). Bioenergetics deficits have even been suggested to precede neurodegeneration in mouse models of AD, and amyloid beta (Aβ) protein is actively imported to mitochondria where it localizes with mitochondrial cristae and likely induces oxidative stress and synaptic degeneration (21–23). Mutations within PARKIN and PINK1, two proteins that regulate mitochondrial quality control, have been implicated in PD (24), and energy deficits in animal models lacking these proteins have been reported, supporting a role for them in PD pathology (25). There is also some evidence for mitochondrial dysfunction in hamster (26) and mouse (18, 27) prion disease models. A study by Choi and colleagues reported decreased enzymatic activity of electron transport chain (ETC) proteins and increased oxidative stress in the brain mitochondria of 263K-infected hamsters (26). A more recent study by Ansoleaga and colleagues reported downregulation of mRNA and protein levels of several crucial mitochondrial metabolic proteins in postmortem tissues of Creutzfeldt-Jakob disease (CJD) patients (28). To the best of our knowledge, however, dysfunction of mitochondrial respiration in prion disease models has not yet been characterized.
Mitochondrial respiration involves the ETC, which is composed of several multiprotein complexes (I to V) embedded within the inner mitochondrial membrane that participate in oxidative phosphorylation. During oxidative phosphorylation, a series of redox reactions drive a proton gradient across the inner mitochondrial membrane, leading to a mitochondrial membrane potential that is critical to maintaining mitochondrial function and viability. Complexes I, III, and IV are primarily responsible for pumping protons across the inner membrane to maintain this proton gradient. Complex V (ATP synthase) is a highly complex molecular turbine motor that converts the potential energy of the proton gradient across the inner mitochondrial membrane to the chemical energy needed for the conjugation of inorganic phosphate to ADP, generating much of the energy for the cells in the form of ATP. Finally, complex II (succinate dehydrogenase) is unique due to its coupling to the citric acid cycle. Complex II can contribute to the NADH pool through the oxidation of succinate to fumarate as well as contribute electrons directly to the ETC, although no protons are translocated during this process. Mitochondrial bioenergetics can be measured in both live cells and purified mitochondria and can be used to determine the energy deficits or level of oxidative stress that may ultimately lead to cell death.
In the current study, we have analyzed mitochondrial bioenergetics in a transgenic mouse model (Tg7) of hamster prion infection. Ex vivo analysis of ADP- and succinate-induced oxygen uptake in mitochondria purified from the brains of hamster prion-infected and mock-infected animals revealed severe bioenergetics deficits at the clinical stage of prion disease. Interestingly, we also identified significant differences in the relative abundances of several mitochondrial proteins in infected versus mock-infected animals, including proteins belonging to complex I and complex V of the ETC. Further analysis of posttranslational modifications (PTMs) of mitochondrial proteins revealed modest but significant differences in the deamidation state of several ETC proteins in infected versus uninfected animals. Taken together, our results demonstrate that prion infection can lead to mitochondrial deficits which may, in turn, contribute to the neurodegeneration observed in these diseases.
RESULTS
Prion disease in hamster 263K-infected Tg7 mice.
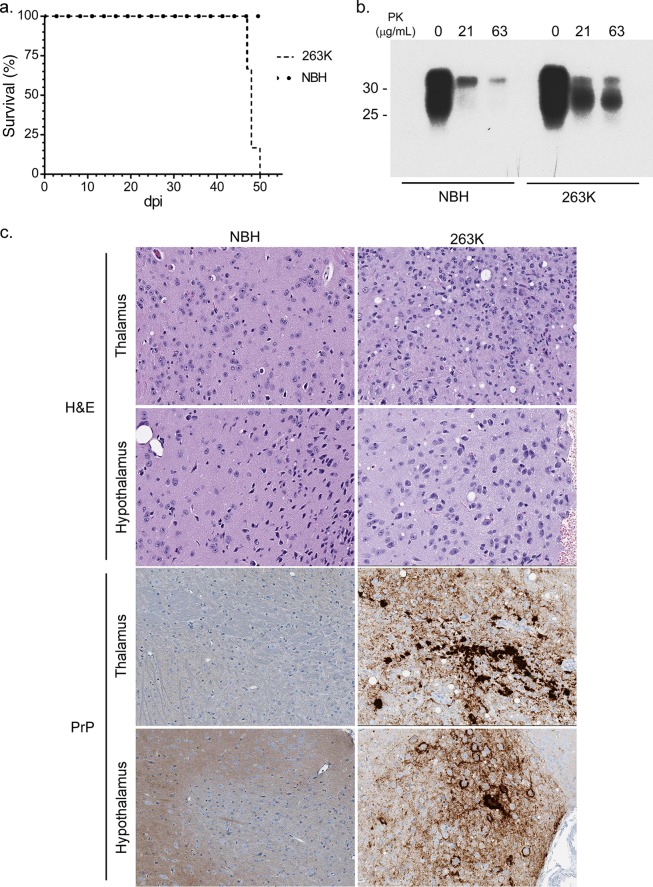
Tg7 mice inoculated with prion strain 263K (Tg7Sc mice) developed clinical illness and were euthanized at 46 to 49 days, while age-matched control Tg7 mice inoculated with normal brain homogenate (Tg7NBH) showed no signs of disease (Fig. 1a). Tg7Sc mice were positive for PrPSc as assessed by proteinase K (PK) treatment of brain homogenates and subsequent immunoblotting with the anti-PrP mouse monoclonal 3F4 antibody. Representative immunoblots are shown for Tg7Sc and Tg7NBH samples at 48 days postinoculation (dpi), when Tg7Sc mice were at the clinical stage of disease (Fig. 1b). Histological analysis confirmed spongiform change and PrPSc deposition in the brains of clinically positive Tg7Sc mice, while Tg7NBH mice showed no pathology (Fig. 1c).
FIG 1.
Biochemical and histological examination of 263K hamster prion-infected Tg7 mice. (a) Survival curves for 263K hamster prion-infected (Tg7Sc) and normal brain homogenate-inoculated age-matched (Tg7NBH) Tg7 mice. The time to clinical disease was 48 ± 1.2 dpi for Tg7Sc mice (n = 6). Tg7NBH mice (n = 6) remained healthy for the duration of the experiment. (b) Brain tissue lysate from clinically ill (48 dpi) Tg7Sc mice and Tg7NBH was PK treated and probed with the anti-PrP mouse monoclonal 3F4 antibody. Tg7Sc mice were positive for protease-resistant PrPSc. Tg7NBH mouse samples were negative for PrPSc, with a nonspecific band at approximately 35 kDa. Molecular mass markers are shown on the left. (c) Sagittal brain sections from Tg7NBH and Tg7Sc mice were immunostained with the anti-PrP rabbit polyclonal antibody EP1802Y. Representative PrPSc staining and neuropathology in Tg7NBH (48 dpi) and Tg7Sc (48 dpi) mice are shown.
Tg7 mice infected with hamster 263K prions suffer from severe deficits in oxygen uptake during clinical stages of disease.
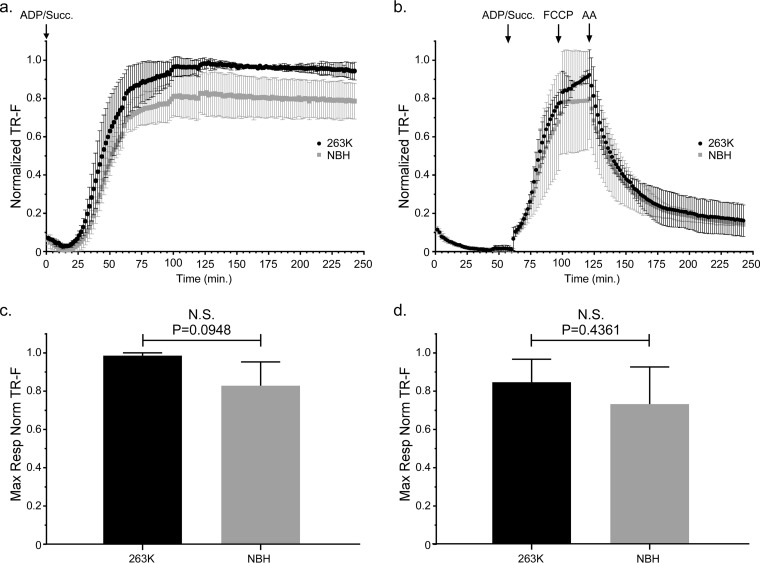
In order to assess mitochondrial function in 263K-infected Tg7 mice, we isolated mitochondria from the brains of Tg7Sc and Tg7NBH mice for ex vivo functional analysis. We first analyzed mitochondria from brains taken at approximately 20 dpi, which represents the approximate midpoint of the disease incubation period. When oxygen consumption was measured with succinate and saturating ADP provided from the beginning of the assay, mitochondria from Tg7Sc mice appeared to have a higher respiratory rate than those from Tg7NBH mice (Fig. 2a). However, statistical analysis revealed that the difference in the maximal respiratory responses of Tg7Sc and Tg7NBH mice under these conditions was not significantly different (P = 0.0948 [Fig. 2c]).
FIG 2.
Mitochondrial oxygen consumption is unaltered in 263K prion-infected Tg7 mice at the midpoint of prion disease incubation. Isolated brain mitochondria from Tg7NBH and Tg7Sc mice euthanized at 20 dpi were assayed for their ability to take up oxygen in response to the complex II substrate succinate. (a) Saturating concentrations of ADP and succinate were added to each well at the beginning of the assay (arrow at 0 min, ADP/Succ.). A single Tg7Sc and Tg7NBH brain was assayed in each experiment. For each individual experiment (n = 3), fluorescence intensity was normalized to the maximum fluorescence value of the oxygen reactive probe MitoXpress within that experiment. Error bars represent the normalized standard deviations for each time point. (b) Isolated brain mitochondria from Tg7NBH and Tg7Sc mice were allowed to equilibrate in the absence of substrate in order to obtain a basal level of oxygen uptake. Saturating concentrations of ADP and succinate (arrow at 60 min. ADP/Succ.) were added to wells containing mitochondria. The proton ionophore and oxidative phosphorylation uncoupler FCCP was then added to each well containing isolated mitochondria (arrow at 115 min, FCCP) to obtain maximal uncoupled respiratory rates. Lastly, the complex III inhibitor antimycin A was added (arrow at 135 min) to shut down electron transport. Data were normalized as for panel a. (c) Maximal respiration for panel a was determined by comparing the mean of the maximal values for each normalized value. (d) Maximal respiration for panel b was determined by comparing the mean of the maximal values for each normalized value. For panels c and d, an unpaired two-tailed Student t test was performed to determine significance. The normalized mean is derived from the average of 3 technical replicates for 3 experimental animals. TR-F, time-resolved fluorescence; N.S., not significant.
A second assay was done in which, in order to establish a respiratory response curve for both baseline mitochondrial oxygen consumption and the rate of oxygen consumption (i.e., mitochondrial respiration), the addition of succinate and ADP was delayed for approximately 1 h to allow the mitochondria to exhaust any endogenous substrate. Baseline rates of oxygen consumption as well as oxygen uptake were similar for mitochondria from Tg7NBH and Tg7Sc mouse brain 20 dpi (Fig. 2b). Treatment of mitochondria isolated from Tg7NBH and Tg7Sc mice with the oxidative phosphorylation uncoupler trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP) did not appear to stimulate increased oxygen uptake in Tg7NBH mouse mitochondria, while Tg7Sc mouse mitochondria did respond by increasing their oxygen consumption rate. Addition of the complex III inhibitor antimycin A (AA) led to the expected drop in oxygen consumption (Fig. 2b). Although Tg7Sc mouse mitochondria appeared to have a slightly higher maximal respiratory potential than Tg7NBH mouse mitochondria, statistical analysis of the respiratory rates of mitochondria from Tg7Sc and Tg7NBH mice at 20 dpi again revealed no significant difference (P = 0.43 [Fig. 2d]). Thus, there appeared to be no significant respiratory impairment of brain mitochondria at the midpoint of the 263K incubation period in Tg7 mice.
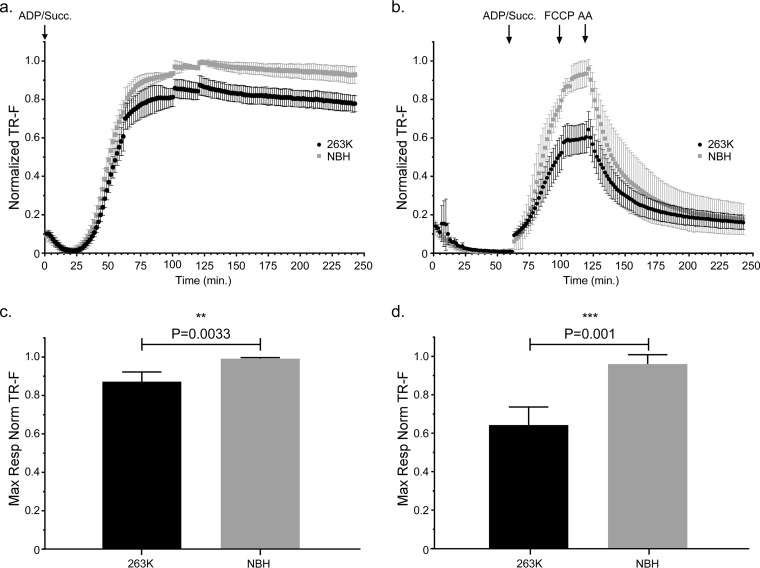
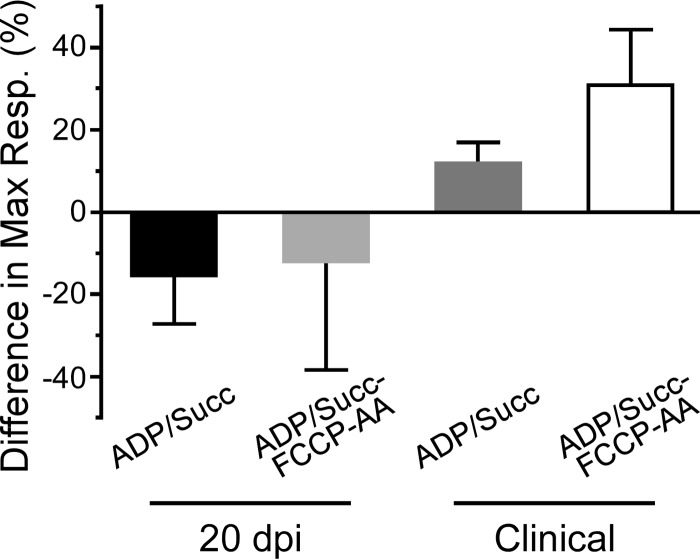
In contrast to the mitochondrial bioenergetics data at 20 dpi, we noted that mitochondria from Tg7Sc mice at the clinical stage of disease appeared to have decreased maximal respiration compared to that of mitochondria from Tg7NBH age-matched controls when succinate and saturating ADP were provided at the beginning of the assay (Fig. 3a). When succinate, ADP, FCCP, and AA were added during the respiratory response assay, mitochondria from clinically ill Tg7Sc mice clearly showed a severe defect in their ability to take up oxygen in response to ADP and succinate (Fig. 3b). Interestingly, Tg7Sc mitochondria appeared to have a muted response to the uncoupling agent FCCP, similar to Tg7NBH mice at 20 dpi. Differences in the maximal respiration for both assays (Fig. 3a and b) was confirmed by a statistical analysis of the maximal respiratory rate which revealed a highly significant decrease in the respiratory potential of mitochondria from Tg7Sc mice (Fig. 3c, P = 0.003, and 3d, P = 0.001). Analysis of the differential maximum respiratory rates of mitochondria from Tg7Sc mice and age-matched Tg7NBH mice also revealed that depending on assay conditions, clinically ill mice suffered from 12% and 35% decreases in their maximum oxygen consumption compared to that of NBH-inoculated control animals (Fig. 4). Consistent with the data in Fig. 2, mitochondria from Tg7Sc mice appeared to have a very modest but not significant increase in their respiratory potential at 20 days postinfection compared to Tg7NBH mice (Fig. 4). Cumulatively, these results demonstrate that mitochondrial bioenergetics deficits correlate with clinical prion disease in 263K prion-infected Tg7 mice.
FIG 3.
Mitochondrial oxygen consumption is significantly decreased in clinically positive 263K prion-infected Tg7 mice. Isolated brain mitochondria from Tg7Sc mice euthanized at clinical stages of prion disease (48 dpi ± 1.2 dpi) or Tg7NBH-inoculated, age-matched control mice were assayed for their ability to take up oxygen in response to the complex II substrate succinate. (a) Saturating concentrations of ADP and succinate were added to each well at the beginning of the assay (arrow at 0 min, ADP/Succ.). Normalized fluorescence intensity of the oxygen reactive probe MitoXpress is shown. Data were normalized as described in the legend to Fig. 2. Error bars represent the normalized standard deviations for each time point. (b) Isolated brain mitochondria from Tg7NBH and Tg7Sc mice were allowed to equilibrate in the absence of substrate in order to obtain a basal level of oxygen uptake. Saturating concentrations of ADP and succinate (arrow at 60 min, ADP/Succ.) were added to wells containing mitochondria. The proton ionophore and oxidative phosphorylation uncoupler FCCP was then added to each well containing isolated mitochondria (arrow at 115 min, FCCP) to obtain maximal uncoupled respiratory rates. Lastly, the complex III inhibitor antimycin A was added (arrow at 135 min) to shut down electron transport. (c) Maximal respiration for panel a was determined by comparing the mean of the maximal values for each normalized value. (d) Maximal respiration for panel B was determined by comparing the mean of the maximal values for each normalized value. For panels c and d, an unpaired two-tailed Student t test was performed to determine significance. The normalized mean is derived from the average of 3 technical replicates for 4 experimental animals. Significance is denoted by the P value given above the bars.
FIG 4.
Altered maximal mitochondrial respiration rates in clinically positive 263K prion-infected Tg7 mice. Differences in the maximal respiration rate were calculated by subtracting the difference in maximal respiration between Tg7NBH and Tg7Sc animals for each time point and experimental condition. The difference in the normalized mean of the maximal respiratory values for Tg7NBH and Tg7Sc (maximal respiratory value for Tg7NBH − maximal respiratory value for Tg7Sc × 100) is represented by each bar (n = the average of 3 technical replicates of either 3 (20 dpi) or 4 (clinical) mice. Error bars represent the standard deviations.
Variable polarization of brain-derived mitochondria from Tg7 mice.
The ETC drives a proton gradient across the inner mitochondrial membrane by pumping protons into the intermembrane space, resulting in a transmembrane potential. Thus, any disruption in ETC-driven proton pumping could result in mitochondrial membrane depolarization and a loss of the mitochondrial membrane potential. This, in turn, could affect mitochondrial function. To assess mitochondrial membrane potential, mitochondria isolated from clinically ill Tg7Sc mice and age-matched Tg7NBH mice were stained with the membrane potential-sensitive dye JC-1 and analyzed by flow cytometry. When green fluorescent JC-1 crosses a polarized membrane, it can aggregate and its fluorescence spectrum shifts to bright red. Accumulation of red fluorescent JC-1 is thus indicative of mitochondrial polarization and a healthy membrane potential maintained by the pumping of protons across the inner membrane via the proteins of the ETC.
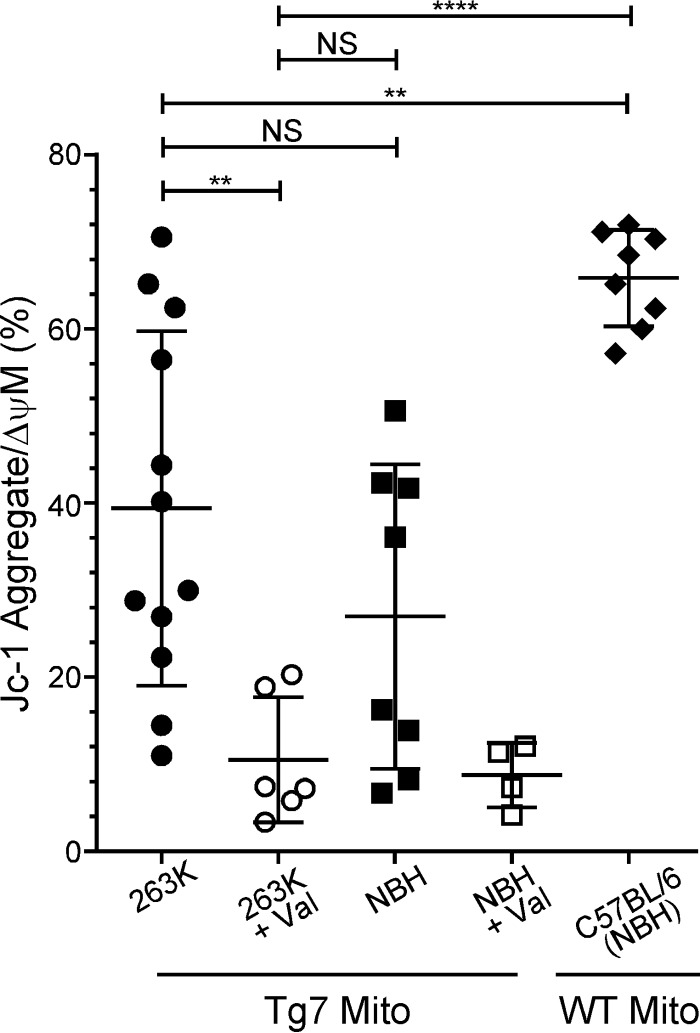
Surprisingly, mitochondria isolated from both clinically ill Tg7Sc mice and age-matched Tg7NBH mice showed a widely varying range of membrane polarization, with mitochondria from some animals appearing to be polarized with relatively high proportions of red JC-1 aggregate (>60%) and other animals appearing to have completely depolarized mitochondria, similar to the valinomycin-treated controls (≤20% JC-1 bright red) (Fig. 5). To determine whether these large differences between animals were due to the Tg7 mouse or significant variability within our assay, we isolated mitochondria from wild-type (WT) C57BL/6 mice 150 days after inoculation with normal mouse brain homogenate. Mitochondria isolated from wild-type C57BL/6 mice were much more uniform in their polarization state, suggesting that the wide variability in mitochondrial polarization seen in Tg7 mice was not an artifact of the assay itself.
FIG 5.
Tg7 mice have various degrees of membrane polarization regardless of prion infection. Isolated brain mitochondria from Tg7Sc mice euthanized at clinical stages of prion disease (n = 6 mice in duplicate), Tg7NBH-inoculated, age-matched control mice (n = 4 mice in duplicate), or NBH-inoculated WT C57BL/6 mice (n = 4 mice in duplicate) were assayed for their mitochondrial membrane potential using the membrane potential-sensitive dye JC-1. For valinomycin-treated samples, each symbol represents an individual mouse. Mito, mitochondria; Val, valinomycin. Significance was determined using a one-way analysis of variance (ANOVA) with Bonferroni's multiple-comparison test and a 95% confidence threshold. Error bars represent standard deviations of the mean. NS, not significant. **, P ≤ 0.005; ****, P ≤ 0.0001.
Mitochondria from C57BL/6 mice clustered toward the high end of the red JC-1 gate, with ≥60% of the mitochondrial population fluorescing bright red (Fig. 5). We therefore defined a population of polarized mitochondria as one in which greater than 60% of the population fluoresced with bright red JC-1 aggregate. Of note, no Tg7NBH samples reached this threshold, while multiple Tg7Sc samples did. Overall, the percentage of polarized mitochondria in both Tg7Sc and Tg7NBH mice was significantly different from that of wild-type C57BL/6, mice with mitochondria from some Tg7 mice appearing to be completely depolarized (Fig. 5). These results suggest that even in the absence of prion infection, the Tg7 mouse may be predisposed to mitochondrial dysfunction.
MS analysis reveals dysregulation of the mitochondrial proteome in 263K-infected Tg7 mice.
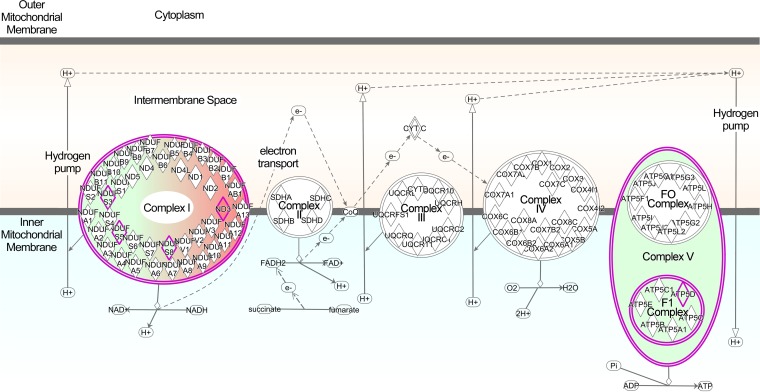
In order to determine whether mitochondrial deficits in oxygen uptake were associated with changes to mitochondrial proteins, we characterized the mitochondrial proteome by mass spectrometry (MS). A semiquantitative analysis based upon spectral counts (i.e., the total number of peptides identified for a given protein) was used to compare the relative apparent abundances of mitochondrial proteins in 263K-infected and NBH-infected Tg7 mice. Approximately 411 mitochondrial proteins were strongly identified (i.e., more than 5 spectral counts for Tg7NBH or Tg7Sc), and 22 were shown to be differentially expressed in Tg7Sc and Tg7NBH mice (Table 1). Several complex I proteins were significantly downregulated in Tg7Sc (Fig. 6 and Table 1), and there was a nearly 3-fold decrease in the delta subunit of the complex V ATP synthase. In contrast, NADH-ubiquinone oxidoreductase chain 3 in complex I was upregulated in Tg7Sc mice.
TABLE 1.
Changes in relative abundance of mitochondrial proteins in hamster prion-infected Tg7 mice
| Protein name | Protein identifier | Tg7NBH SpCa | Tg7Sc SpCa | Fold Δb | P valuec |
|---|---|---|---|---|---|
| NADH-ubiquinone oxidoreductase chain 3 | P03899 | 0 ± 0 | 6.2 ± 1 | 18.5 | <0.001 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 1 | Q9CQA6 | 1.0 ± 1.7 | 6.4 ± 1.6 | 6.7 | 0.01 |
| Mitofusin 1 | Q811U4 | 2.2 ± 2.0 | 7.2 ± 1.4 | 3.3 | 0.02 |
| Glycine amidinotransferase | Q9D964 | 28.3 ± 5.7 | 91.2 ± 28.8 | 3.2 | 0.02 |
| Translocase subunit Tim8 A | Q9WVA2 | 2.1 ± 1.9 | 6.1 ± 0.3 | 2.9 | 0.02 |
| Monoacylglycerol lipase ABHD12 | Q99LR1 | 14.2 ± 5.1 | 27.2 ± 4.3 | 1.9 | 0.03 |
| Sarcosine dehydrogenase | Q99LB7 | 7.5 ± 1.2 | 11.9 ± 1.4 | 1.6 | 0.01 |
| Valyl-tRNA synthetase | Q3U2A8 | 10 ± 0.4 | 15 ± 2.7 | 1.5 | 0.03 |
| NLR family member X1 | Q3TL44 | 18.2 ± 2.1 | 25.4 ± 1.2 | 1.4 | 0.007 |
| Mitoguardin 2 | Q8BK03 | 11.7 ± 2.2 | 15.9 ± 0.8 | 1.4 | 0.04 |
| 28S ribosomal protein S9 | Q9D7N3 | 20.1 ± 2.1 | 25.2 ± 1.1 | 1.3 | 0.02 |
| NADH dehydrogenase iron-sulfur protein 3 | Q9DCT2 | 285.2 ± 15.9 | 246.4 ± 14.8 | −1.2 | 0.04 |
| [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1 | Q3UV70 | 36.3 ± 1.7 | 30.1 ± 1.6 | −1.2 | 0.009 |
| Kynurenine-oxoglutarate transaminase 3 | Q71RI9 | 22.4 ± 2.2 | 17.6 ± 0.9 | −1.3 | 0.02 |
| NADH dehydrogenase iron-sulfur protein 5 | Q99LY9 | 73.9 ± 4.5 | 51.6 ± 8.4 | −1.4 | 0.02 |
| Prohibitin | P67778 | 256.6 ± 26.1 | 176.8 ± 3.7 | −1.5 | 0.006 |
| Citrate lyase subunit beta-like protein | Q8R4N0 | 36.3 ± 4.6 | 24.3 ± 2.7 | −1.5 | 0.02 |
| Probable prolyl-tRNA synthetase | Q8CFI5 | 16.1 ± 1.7 | 10.4 ± 1.5 | −1.5 | 0.01 |
| NADH dehydrogenase iron-sulfur protein 8 | Q8K3J1 | 66.2 ± 14.9 | 40.7 ± 5.1 | −1.6 | 0.05 |
| Tryptophanyl-tRNA synthetase | Q9CYK1 | 12.0 ± 0.7 | 5.8 ± 2.4 | −2.1 | 0.01 |
| ATP synthase subunit delta | Q9D3D9 | 18.7 ± 6.5 | 6.5 ± 3.7 | −2.9 | 0.05 |
| Choline dehydrogenase | Q8BJ64 | 6.3 ± 1.3 | 1.2 ± 2.1 | −5.2 | 0.02 |
Average number of peptides (spectral counts or SpC) ± standard deviation. The average is derived from LC-MS/MS analysis of mitochondria extracted from 3 individual mouse brains. SpCs were derived using the Byonic software package.
Fold change was calculated from the average normalized SpC values from each group and indicates the change in relative abundance in Tg7Sc mice compared to Tg7NBH mice. If either group had an average SpC of 0, the value of 1 was assigned to obtain an accurate fold change.
P values were calculated using the Student t test to compare SpC in Tg7NBH mice to SpC in Tg7Sc mice.
FIG 6.
Ingenuity Pathway Analysis diagram of differentially regulated ETC proteins. Proteins in Tg7Sc mice that were altered in abundance are indicated by diamonds. Green indicates a decrease in the relative abundance of the protein compared to that in Tg7NBH mice, while red indicates an increase in relative abundance. Highly significant differences were observed in components of complex I and ATP synthase (complex V). Abbreviations: NDUS3, NADH dehydrogenase iron-sulfur protein 3; NDUS5, NADH dehydrogenase iron-sulfur protein 5; NDUS8, NADH dehydrogenase iron-sulfur protein 8; ND3, NADH-ubiquinone oxidoreductase chain 3; ATP5D, ATP synthase subunit delta.
Several proteins that regulate mitochondrial dynamics were significantly altered in abundance in Tg7Sc mitochondria (mitoguardin 2, mitofusin, and prohibitin), suggesting that mitochondrial fission and fusion may also be adversely impacted at clinical stages of 263K prion disease. In Tg7Sc mitochondria, there were also differences in the relative abundances of two tRNA synthases compared to those in TgNBH mice (Table 1), a possible indication of changes in the translation of mitochondrial mRNA. Finally, we also characterized differential posttranslational modifications by mass spectrometry and detected some changes to mitochondrial proteins in Tg7Sc mouse mitochondria compared to Tg7NBH mouse mitochondria (Table 2). In particular, there was a 30-fold increase in the deamidation of the 28S ribosomal protein S34, again suggesting alterations in the translation of mitochondrial mRNA.
TABLE 2.
Relative abundance of posttranslational modifications to mitochondrial proteins in hamster prion-infected Tg7 mice
| Protein name | Protein identifier | PTMa | Fold Δb | P valuec |
|---|---|---|---|---|
| 28S ribosomal protein S34 | Q9JIK9 | Deamidation (NQ) | 24.9 | 0.002 |
| NADH dehydrogenase 1 α subcomplex subunit 9 | Q9DC69 | Pyro-glu from Q | 5.0 | 0.02 |
| Isoleucyl-tRNA synthetase | Q8BIJ6 | Pyro-glu from Q | 4.1 | 0.02 |
| Sideroflexin-1 | Q99JR1 | Acetylation (N-term) | 2.1 | 0.01 |
| Voltage-dependent anion-selective channel protein 2 | Q60930 | Acetylation (N-term) | 1.7 | 0.003 |
| Mitochondrial inner membrane protein | Q8CAQ8 | Pyro-glu from Q | 1.7 | 0.01 |
| Cytochrome b-c1 complex subunit 2 | Q9DB77 | Deamidation (NQ) | −1.3 | 0.003 |
| Sideroflexin 1 | Q99JR1 | Oxidation (M) | −1.3 | 0.03 |
| NADH dehydrogenase 1 α subcomplex subunit 12 | Q7TMF3 | Acetylation (N-term) | −1.4 | 0.003 |
| Glutamate dehydrogenase 1 | P26443 | Deamidation (NQ) | −1.6 | 0.005 |
| Cytochrome c1 heme protein | Q9D0M3 | Deamidation (NQ) | −1.6 | 0.02 |
| ATP synthase subunit α | Q03265 | Deamidation (NQ) | −1.8 | 0.03 |
| NADH dehydrogenase iron-sulfur protein 3 | Q9DCT2 | Pyro-glu from Q | −1.9 | 0.005 |
| Ca2+-binding mitochondrial carrier protein Aralar1 | Q8BH59 | Deamidation (NQ) | −1.9 | 0.04 |
| ATP synthase subunit α | Q03265 | Sodium adduct | −2.2 | 0.03 |
| Ca2+-binding mitochondrial carrier protein Aralar1 | Q8BH59 | Deamidation (NQ) | −2.4 | 0.03 |
| ATP synthase subunit α | Q03265 | Formylation | −2.4 | 0.003 |
| NADH dehydrogenase iron-sulfur protein 2 | Q91WD5 | Oxidation (M) | −2.5 | 0.01 |
| Mitochondrial inner membrane protein | Q8CAQ8 | Pyro-glu from Q | −2.7 | 0.03 |
| Cytochrome c oxidase subunit 4 isoform 1 | P19783 | Deamidation (NQ) | −3.3 | 0.04 |
Posttranslational modification (PTM) identified on a single peptide from the protein identified. The amino acids altered are indicated. A protein occurring multiple times in the list indicates that more than one peptide with a PTM was identified. SpCs were derived using the PEAKS Studio version 8 software package. M, methionine; Q, glutamine; NQ, asparagine and glutamine; N-term, N-terminal amino group of peptide.
Fold change was calculated from the average normalized SpC values from each group and indicates the change in relative abundance in Tg7Sc mice from that in Tg7NBH mice. If either group had an average SpC of 0, the value of 1 was assigned to obtain an accurate fold change.
P values were calculated using the Student t test to compare SpC in Tg7NBH mice to SpC in Tg7Sc mice.
To confirm the mass spectrometry results, the relative expression level of two mitochondrial proteins that were changed in abundance in 263K-infected Tg7 mice was assessed by Western blotting. Since there was not enough material remaining of the purified mitochondrial samples used for mass spectrometry, whole-brain homogenates from three 263K- and three NBH-inoculated Tg7 mice was used for Western blot analysis. In agreement with the mass spectrometry data, expression levels of choline dehydrogenase were reduced and NADH-ubiquinone oxidoreductase chain 3 increased in brain homogenates of 263K-infected Tg7 mice compared to NBH-inoculated mice (Fig. 7). The relative difference in abundance did not appear to be as high as that observed by mass spectrometry (Table 1), but this was likely due to the use of brain homogenate for the Western blot analysis versus purified mitochondria for the mass spectrometry analysis. Overall, the mass spectrometry and immunoblot data are consistent with the experimental data showing that clinically ill Tg7Sc mice suffer from mitochondrial bioenergetics deficits as well as the hypothesis that dysregulation of key ETC proteins can lead to mitochondrial dysfunction in prion-infected mice.
FIG 7.

Altered expression of mitochondrial proteins choline dehydrogenase and NADH-ubiquinone oxidoreductase chain 3 in the brains of 263K-inoculated Tg7 mice. Each lane represents one Tg7 mouse inoculated with normal brain homogenate or hamster 263K prions. For each lane, 500 μg of brain homogenate was loaded. A total of three mice were analyzed. (a) Choline dehydrogenase (CHDH). (b) NADH-ubiquinone oxidoreductase chain 3 (NDUFS3). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. The molecular mass of the protein is given on the right.
DISCUSSION
A previous study correlated deficits in mitochondrial enzymes with clinical illness in 263K prion-infected hamsters. However, whole mitochondria were not assessed ex vivo for functionality (26). Here we show that mitochondria isolated from clinically ill 263K-inoculated Tg7 mice suffer from a significant defect in their respiration ability in response to the complex II substrate succinate (Fig. 3). Using mass spectrometry, we noted significant downregulation of several complex I proteins and the delta subunit of ATP synthase in Tg7Sc mice compared to Tg7NBH mice. These data show that dysregulation of mitochondrial ETC proteins at clinical stages of prion disease accompany severe bioenergetics deficits in 263K-infected Tg7 mice which may contribute to organismal death. Our results are consistent with other studies that have reported altered mitochondrial protein and mRNA levels in both prion-infected mice and human CJD subjects (28, 29).
Tg7 mice do not express normal mouse PrPC but instead overexpress hamster PrPC at approximately 4 to 5 times the normal level (30, 31). While it is known that increased PrPC expression levels correlate with shorter incubation periods preceding clinical illness (32), it is also possible that other factors may make the Tg7 mouse inherently more susceptible to hamster prion infection. Our experiments examining mitochondrial membrane polarization surprisingly revealed that Tg7 mice vary greatly in their mitochondrial membrane potential compared to wild-type C57BL/6 mice. Furthermore, although mitochondria from some Tg7Sc mice did appear hyperpolarized compared to those from Tg7NBH control mice, 263K prion infection did not appear to significantly change the mitochondrial membrane potential of Tg7 mice compared to that of uninfected mice. The ETC maintains the differential membrane potential between the inner and outer mitochondrial membranes, and dysfunction or dysregulation of mitochondrial electron transport can result in loss of the mitochondrial membrane potential and mitochondrial function. Our data suggest that Tg7 mice suffer from an inherent defect in mitochondrial membrane potential which may make them more prone to mitochondrial dysfunction during 263K prion infection. We recently reported that PrPC can be found under homeostatic conditions in mitochondria of uninfected wild-type mice and transgenic mice overexpressing PrPC and that the relative amounts of PrPC in the mitochondria correlated with the PrPC expression level (33). This leads to the intriguing, but speculative, possibility that association of PrPC with mitochondria could affect mitochondrial function and that overexpression of PrPC could exacerbate any dysfunction. If this is the case, it is possible that the more rapid disease course in 263K-infected Tg7 mice is a consequence of PrPC overexpression leading not only to more rapid deposition of pathogenic levels of PrPSc but also to increased levels of mitochondrial dysfunction.
The net flow of electrons through the mitochondrial ETC culminates in the consumption of molecular oxygen as it is reduced to H2O and complex I, complex III, and complex IV pump protons, generating a chemiosmotic gradient between the inner membrane and matrix that drives ATP synthesis from ADP and inorganic phosphate. Analysis of the oxygen consumption rate of mitochondria can thus provide crucial information regarding both the physiological status and energy-producing capabilities of the mitochondrion. We analyzed the oxygen consumption capabilities of isolated mitochondria from Tg7Sc and Tg7NBH mice at 20 days and the clinical endpoint to determine whether 263K prion infection induces changes in mitochondrial respiration (Fig. 2 and 3). We chose the more restricted respiratory substrate succinate due to its unique ability to directly provide electrons to coenzyme Q10 following oxidation by complex II while, through its oxidation to fumarate, still contributing to generation of NADH utilized by complex I. At 20 days postinfection, Tg7Sc and Tg7NBH mice appeared to have virtually identical respiratory profiles both in terms of maximal oxygen consumption in the presence of saturating ADP plus succinate and in respiratory response curves where ADP and succinate addition was delayed. We did note moderate increases in oxygen consumption by Tg7Sc mice at 20 dpi in both assays, but this did not reach significance. It is possible that at 20 dpi mitochondrial damage is just beginning to accumulate and that Tg7Sc mitochondria compensate for this damage by increasing oxygen consumption. However, at clinical time points Tg7Sc mice were significantly impaired in their ability to take up oxygen (Fig. 3 and 4). Deficiencies of this magnitude suggested to us that alterations in the chemical properties or absolute abundance of ETC proteins may be a major contributing factor to mitochondrial dysfunction. Indeed, we found highly significant downregulation of several crucial ETC proteins. Particularly telling were several dysregulated proteins of complex I and downregulation of the delta subunit of ATP synthase (Fig. 6). These observations in of themselves substantiate our observations that mitochondria from Tg7Sc mice are significantly impaired in their respiratory ability.
We also observed a 34% decrease in the relative abundance of prohibitin in Tg7Sc mice. Prohibitins can form multiprotein ring-like structures in the inner mitochondrial membrane, where they may act as chaperones for ETC proteins or regulate aspects of mitochondrial morphology (34). A decrease in prohibitin protein in the mitochondrion during prion disease might therefore lead to destabilization of the ETC. Also of interest, we noted dysregulation of two key proteins involved in mitochondrial fusion and fission, a process often referred to as mitochondrial dynamics. Mitochondrial dynamics is a carefully controlled process which ensures that mitochondrial DNA is properly segregated, membranes are partitioned between mitochondria appropriately, and cristae are properly restructured. We noted a 30% increase in the relative abundance of mitoguardin 2 in Tg7Sc animals (Table 1). Mitoguardin 2 is localized on the outer mitochondrial membrane and is thought to regulate mitochondrial fusion via modulation of phospholipid metabolism through facilitating the generation of PLD6/MitoPLD dimers. It was recently shown that mitoguardin 2 is essential for neuronal homeostasis and regulating mitochondrial fusion (35). Interestingly, we also noted a significant upregulation of mitofusin in Tg7Sc mice (Table 1). Mitofusins are GTPases localized to the outer mitochondrial membrane, where they are essential for mitochondrial fusion in mammals (36). Thus, our proteomic analysis suggests dysfunction of mitochondrial fusion at clinical stages of 263K hamster prion disease which could also significantly impair mitochondrial function.
Other studies have reported dysregulation of superoxide dismutase (SOD) enzymes or oxidative stress during prion disease (26, 29, 37), and we hypothesized that changes in the oxidation status of ETC proteins would be observed in Tg7Sc mice, leading us to perform a PTM analysis of Tg7Sc and Tg7NBH mice. However, we did not detect any significant differences in oxidized methionines or other PTMs indicative of oxidation damage to ETC proteins. Nor did we see any evidence of dysregulation of antioxidant enzymes in the current study. It is possible that the short incubation period of the 263K hamster prion strain in the Tg7 mouse (∼48 days) may not provide sufficient time for oxidative damage to accumulate to a detectable level on ETC protein moieties. We did, however, note significant decreases in the NQ deamidation profile of several complex I, III, and V proteins (Table 2). It is possible that this PTM negatively affects electron transport. However, more work is needed to interpret these observations.
MATERIALS AND METHODS
Ethics statement.
The animal experimental protocol was reviewed and approved by the Rocky Mountain Laboratories Animal Care and Use Committee (animal study protocol 2014-006). The Rocky Mountain Laboratories are fully accredited by the American Association for Laboratory Animal Care and this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (38).
Prion infection of transgenic mice.
Mice overexpressing hamster PrPC on a PrP-null background (Tg7) were originally described by Race et al. (31). Tg7 mice express 4- to 5-fold the normal level of PrPC (30) and are susceptible to the 263K hamster scrapie agent, developing a clinical spongiform encephalopathy within 45 to 50 days following intracranial (i.c.) inoculation. Tg7 mice were inoculated i.c. either with 50 μl of 263K scrapie brain homogenate from hamsters containing 2 × 109.2 infectious doses/g of brain tissue or with an equivalent volume/mass of hamster normal brain homogenate (NBH). Tg7 mice inoculated with 263K are referred to here as Tg7Sc, while Tg7 mice inoculated with normal brain homogenate are referred to as Tg7NBH.
Immunoblotting.
Brains were harvested and frozen in liquid nitrogen until processed. To make a 20% brain homogenate, 1 ml of phosphate-buffered saline (PBS; pH 7.4) was added to 200 mg of brain tissue and processed in a Mini-beadbeater (Biospec Products) with two 30-s runs. The sample was then vortexed for 1 min, followed by sonication for 2 min. For PrP Western blots, 10 μl of 20% brain homogenate was transferred to a clean tube containing 4.8 μl of lysis buffer (10% Triton X-100, 10% deoxycholate, 1 M Tris-HCl [pH 8.3], and 0, 21, or 63 mg/ml of proteinase K [PK]). Samples were incubated for 30 min in a circulating water bath at 37°C. PK activity was quenched by the addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM. The samples were then resuspended in 50 μl of 4× lithium dodecyl sulfate sample buffer (LDS; Life Technologies) containing 2.5 μl of 14.3 M β-mercaptoethanol. Approximately 10 μl of the sample was loaded onto 4 to 12% NuPAGE gels (Life Technologies) and then run at 160 V for 55 min in 2-(N-morpholino)ethanesulfonic acid (MES) running buffer (Life Technologies). For assessment of choline dehydrogenase and NADH-ubiquinone oxidoreductase chain 3, 20% brain homogenates were directly combined with LDS sample buffer as described above and approximately 10 μl, corresponding to 500 μg of protein, was loaded onto a 4 to 12% NuPAGE gel.
Gels were blotted onto polyvinylidene difluoride (PVDF) membranes by wet transfer in transfer buffer (Towbin's buffer: 25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol, 0.025 to 0.1% SDS) at 90 V for 75 min, and the membranes were blocked by incubation with 5% powdered milk in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20) for 1 h. For PrP blots, the mouse monoclonal anti-PrP antibody 3F4 biotin conjugate (Biolegend) was diluted 1:25,000 in TBST and then incubated with the membranes overnight with rocking at 4°C. Choline dehydrogenase was probed for using a rabbit polyclonal antibody (Novus Bio) diluted at 1:2,000 in TBST, and NADH-ubiquinone oxidoreductase chain 3 was probed for using a rabbit polyclonal antibody (Novus Bio) diluted at 1,000 in TBST. Membranes were washed 3 times for 5 min each with gentle agitation following primary antibody incubation. For PrP blots, biotin horseradish peroxidase (HRP) conjugate was diluted 1:25,000 and incubated with the membranes for 2 h at room temperature with gentle agitation. For choline dehydrogenase and NADH-ubiquinone oxidoreductase chain 3 blots, a sheep anti-rabbit HRP-conjugated secondary antibody was diluted 1:6,500 in TBST and incubated with the membrane for 2 h at room temperature with gentle agitation. Following 6 5-min washes, all blots were developed and protein was detected with ECL Prime (GE Healthcare) per the manufacturer's instructions.
Isolation of mitochondria.
The Miltenyi Mitochondria Extraction Kit for mouse tissue (Miltenyi Biotech) and the Miltenyi Mitochondria Isolation Kit for mouse tissue (Miltenyi Biotech) were used for brain tissue preparation and subsequent mitochondrial isolation as described previously (33). Each mitochondrial isolation was derived from 200 mg of starting brain tissue.
Oxygen uptake and bioenergetics.
Oxygen consumption was measured ex vivo in isolated mitochondria using the oxygen-sensitive phosphorescent probe MitoXpress (Cayman Chemical Company). The MitoXpress probe has a peak excitation wavelength of 380 nm and peak emission at 650 nm, and filters corresponding to these wavelengths were used for the assay. Time-resolved fluorescence (TR-F) measurements were conducted using a delay of 30 μs and gate integration time of 100 μs. Mitochondria were isolated as described above and resuspended to a final concentration of 20 mg/ml of protein in mitochondrial measurement buffer (MMB; 250 mM sucrose, 15 mM KCl−, 1 mM EGTA, 5 mM MgCl2, 30 mM K2HPO4 [pH 7.4]). To control for assay variability, respiration experiments were always conducted with a paired sample from one Tg7Sc mouse for every Tg7NBH mouse. Respiration assays with paired Tg7Sc and Tg7NBH mice were separated by days to weeks in time by staggering animal inoculation times.
For each sample, 0.2 mg (10 μl) of mitochondrial suspension was added to each of six wells of an optical-bottom microplate containing 100 μl of MMB. The complex II substrate succinate [(CH2)2(CO2H)2] (25 mM) and 1.65 mM ADP were added to three of six wells. All wells containing sample were overlaid with 100 μl of mineral oil and placed in a FLUOstar OMEGA plate reader (BMG Labtech), and the assay was performed at 37°C, with readings taken every 75 s. To determine the mitochondrial response to the substrate and ADP, the proton ionophore trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP) uncoupling agent, and the antimycin A (AA) electron transport inhibitor, the remaining three wells for each sample had reagents added to them as the assay progressed as follows. At approximately 60 min, succinate and ADP were added to final concentrations of 25 mM and 1.65 mM, respectively, to initiate mitochondrial respiration and oxygen uptake. To assess the maximal respiratory potential of mitochondria, at 100 min the uncoupling agent FCCP was added to each well at a final concentration of 50 μM in order to disrupt ATP synthesis and increase oxygen consumption. Finally, at 122 min, the complex III-specific electron transport inhibitor AA was added at a final concentration of 2.5 μM to shut down mitochondrial respiration and oxygen uptake. Readings continued for a total assay time of 242 min. Each biological sample and condition was run in triplicate. Each experiment included a Tg7Sc and Tg7NBH mouse which were assayed at the same time. Experiments were normalized relative to each other using Tg7Sc and Tg7NBH mouse pairs and the following formula: (Tr-F value/maximum Tr-F value) × 100.
Mitochondrial membrane potential.
To assess membrane potential in isolated mitochondria, we utilized the membrane potential sensitive dye JC-1 and the Isolated Mitochondria staining kit (Sigma-Aldrich). JC-1 staining solution was prepared according to the manufacturer's instructions. Mitochondria were prepared as described above (2 mg/ml) and transferred to 5-ml polystyrene tubes. Approximately 2 mg of mitochondria (as determined by protein content) was added to 320 μl of either buffer alone (unstained), JC-1 solution (stained), or JC-1 solution containing 0.5 μg/ml of valinomycin to depolarize mitochondria. Samples were run in duplicate on a BD Accuri flow cytometer, and 10,000 events were recorded for each sample. Gating was empirically determined using mitochondria from wild-type C57BL/6 NBH-infected mice as a positive control for polarized mitochondria and valinomycin treatment of all Tg7Sc and Tg7NBH mouse samples as a control for depolarization. Data were analyzed using FlowJo v10. The average (n = 2) proportion of the mitochondrial population fluorescing bright red corresponds to high membrane potential indicative of ETC functional homeostasis.
Histology.
Mice were euthanized and the brains removed and placed in 10% neutral buffered formalin. Brains were embedded in paraffin blocks and sectioned sagitally. Hematoxylin and eosin (H&E) staining was carried out on a Leica Autostainer XL according to the manufacturer's instructions. For detection of PrP using the anti-PrP mouse monoclonal antibody 3F4, fresh sections were cut from paraffin blocks and deparaffinized. Antigen retrieval was performed using a Biocare decloaking chamber in citrate buffer (pH 6.0) at 120°C for 20 min. Staining was completed on a Ventana Discovery XT using the 3F4 biotinylated monoclonal antibody at a dilution of 1:50 for 60 min. For staining using the anti-PrP rabbit monoclonal antibody EP1802Y (GeneTex), antigen retrieval was carried out using the extended CC1 protocol (cell conditioning buffer containing Tris-Borate-EDTA [pH 8.0], ∼100 min at 100°C). Antigen retrieval and staining were performed entirely online with the automated Discovery XT staining system (Ventana Medical Systems) using a DABMap detection kit and hematoxylin counterstain. The primary antibody EP1802Y was used at a dilution of 1:6,000 in antibody dilution buffer (Ventana ADB250) and applied for 60 min at 37°C. The secondary antibody was a biotinylated goat anti-rabbit Ig applied for 32 min at 37°C (Biogenex ready-to-use Super Sensitive Rabbit Link).
Trypsin–endoproteinase Lys-C digestion of highly enriched mitochondrial samples.
Pelleted mitochondrial proteins were processed using methodology based upon the filter-aided sample preparation (FASP) procedure (39). Each mitochondrial pellet, normalized to 200 μg of total protein as determined by the bicinchoninic acid (BCA) assay, was solubilized in 50 μl of denaturing buffer (40 mM Tris [pH 10.4], 7 M urea, 2 M thiourea, 1% C7BzO detergent). Dithiothreitol (DTT) was then added from a 1 M stock solution to a final concentration of 15 mM, and the sample was rotated at 37°C for 30 min on an Eppendorf thermomixer. Iodoacetamide (IAA; Pierce) from a freshly made aqueous stock solution (375 mM) was added to the mitochondrial sample to a final concentration of 75 mM. Alkylation proceeded at room temperature in the dark for 20 min, and the reaction was quenched by the addition of DTT to a final concentration of 150 mM. This mixture was diluted with 250 μl of freshly prepared Tris-urea (TU) buffer and then added to an Amicon 30,000-molecular-weight-cutoff (MWCO) spin filter (Fisher) which was centrifuged at 14,000 × g for 15 min with a rotor temperature of 20°C. The filtrate was set aside and the centrifugation procedure was repeated twice more with 250 μl of TU buffer and then twice more using freshly made 50 mM ammonium bicarbonate (ABC) buffer (pH 8). Finally, the samples were diluted in 50 mM ABC buffer containing 5 μg of a trypsin–endoproteinase Lys-C mixture (Promega). Each spin filter was equipped with a fresh receiving vial, sealed with Parafilm, and then incubated for approximately 16 h in a water bath set to 37°C. The enzymatic digest was subjected to centrifugation at 14,000 × g for 5 min, and trypsin-digested peptides were collected as the filtrate. An additional 50 μl of 500 mM NaCl solution was added to the spin filter, and filtrate was again collected by centrifugation. This solution was dried by speed vacuum and then further fractionated into 4 separate fractions using the high-pH reversed-phase peptide fractionation kit (Thermo Fisher) per the directions of the manufacturer. The final samples were again dried by speed vacuum and stored at −20°C until needed.
Mass spectrometry data acquisition.
Mitochondrial samples were digested with a trypsin–endoproteinase Lys-C mixture as described above, resulting in 2 groups (normal brain homogenate inoculated and 263K scrapie agent inoculated) with 3 biological replicates representing each group. Individual replicates were further fractionated into 4 fractions as described above, resulting in a total of 24 individual samples which were then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an Agilent 1200 series nanoflow high-performance liquid chromatograph (HPLC) coupled to a chip-cube nanospray source connected to an Agilent 6550 iFunnel quadrupole time of flight (Q-TOF) system (Agilent Technologies). Samples were loaded onto a 40-nl trapping column using 3% acetonitrile (ACN)–0.1% formic acid (FA) and then separated on a 75-μm by 150-mm analytical column (Agilent G4240-62006-ZORBAX 300SB-C18). Separation was achieved with a reversed-phase step gradient at 0.35 μl/min starting with 97% buffer A (0.1% FA in water) to 10% buffer B (90% ACN–0.1% FA in water) in 2 min, then 30% buffer B in 105 min, 40% buffer B in 120 min, and then holding at 90% buffer B from 125 to 130 min and back to 97% buffer A by 135 min. The survey scan was done with positive ion polarity with an m/z range from 300 to 1,700 at a scan rate of 8 spectra/second and an MS/MS scan rate of 3 spectra/second with a maximum of 20 precursor ions selected per cycle.
Protein identification by tandem mass spectrometry.
Raw LC-MS/MS data were searched using PEAKS Studio version 8. Data were processed using a parent mass error tolerance of 20 ppm and a fragment mass error of 0.4 Da. Cysteine was searched with a fixed modification of 57.02, and variable modifications were set for methionine oxidation (+15.99), N-terminal acetylation (+42.01), and deamidation (NQ, +0.98). The database was Swiss-Prot (http://www.uniprot.org) filtered for mouse taxonomy with the exception of PrP, for which the hamster sequence (Swiss-Prot number P04273) was inserted in order to reflect the hamster PrP sequence of the Tg7 mouse. Initial search results gave a total of 609,522 MS scans and 710,832 MS/MS scans, resulting in 291,698 peptide-spectrum matches with peptide −10 log P values of ≥20.7 and protein −10 log P values of ≥20, corresponding to a false-discovery rate (FDR) of 0.1%. The remaining MS/MS spectra were then searched against this initial protein list for known PTMs using the PTM search module within PEAKS, yielding a total of 1,466 proteins with 13,814 unique peptide sequences.
To validate the PEAKS search engine results, the same raw data were searched using Byonic software (Protein Metrics) using the Preview module to optimize search parameters. A precursor mass tolerance of 30 ppm and a fragment mass tolerance of 40 ppm were used with trypsin specificity and a maximum of 2 missed cleavages allowed. These search results yielded a total of 1,907 individual protein identifications with a predominance of mitochondrial proteins, most of which overlapped with the preceding search results.
Mass spectrometry quantitation and statistics.
Six spectral count (SpC) samples consisting of two groups with three biological replicates in each group were normalized using the scaling method separately for peptide and protein counts. Fold change was calculated from the average normalized values from each group, where if either group had an average of zero, the value of one was assigned to obtain an accurate fold change. P values were calculated using the Student t test between the two groups. Posttranslational modifications (PTMs) were summarized from the replicates with any PTM from one, two, or all three replicates being reported. Resulting data were analyzed through the use of Qiagen's Ingenuity Pathway Analysis (IPA).
ACKNOWLEDGMENTS
We thank Karin Peterson, Byron Caughey, and Allison Kraus for critically evaluating the manuscript and Anita Mora, Austin Athman, and Ryan Kissinger for technical assistance in preparation of the figures. We also gratefully acknowledge Nancy Kurtz and Lori Lubke for assisting with histological preparation of tissue.
This work was supported by the National Institutes of Health intramural research program (AI000752-20).
REFERENCES
- 1.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. 1998. Prion protein biology. Cell 93:337–348. [DOI] [PubMed] [Google Scholar]
- 2.Soto C. 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci 4:49–60. [DOI] [PubMed] [Google Scholar]
- 3.Lasmézas CI, Deslys JP, Robain O, Jaegly A, Beringue V, Peyrin JM, Fournier JG, Hauw JJ, Rossier J, Dormont D. 1997. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 4.Piccardo P, Manson JC, King D, Ghetti B, Barron RM. 2007. Accumulation of prion protein in the brain that is not associated with transmissible disease. Proc Natl Acad Sci U S A 104:4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J, Wollmann R, Lindquist S. 2002. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 6.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. 1998. A transmembrane form of the prion protein in neurodegnerative disease. Science 279:827–834. [DOI] [PubMed] [Google Scholar]
- 7.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. 2005. The most infectious prion protein particles. Nature 437:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hetz C, Maundrell K, Soto C. 2003. Is loss of function of the prion protein the cause of prion disorders? Trends Mol Med 9:237–243. [DOI] [PubMed] [Google Scholar]
- 9.Westergard L, Christensen HM, Harris DA. 2007. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta 1772:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302:871–874. [DOI] [PubMed] [Google Scholar]
- 11.Sisková Z, Page A, O'Connor V, Perry VH. 2009. Degenerating synaptic boutons in prion disease: microglia activation without synaptic stripping. Am J Pathol 175:1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffrey M, Halliday WG, Bell J, Johnston AR, MacLeod NK, Ingham C, Sayers AR, Brown DA, Fraser JR. 2000. Synapse loss associated with abnormal PrP precedes neuronal degeneration in the scrapie-infected murine hippocampus. Neuropathol Appl Neurobiol 26:41–54. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham C, Deacon R, Wells H, Boche D, Waters S, Diniz CP, Scott H, Rawlins JN, Perry VH. 2003. Synaptic changes characterize early behavioural signs in the ME7 model of murine prion disease. Eur J Neurosci 17:2147–2155. [DOI] [PubMed] [Google Scholar]
- 14.Russelakis-Carneiro M, Hetz C, Maundrell K, Soto C. 2004. Prion replication alters the distribution of synaptophysin and caveolin 1 in neuronal lipid rafts. Am J Pathol 165:1839–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Askanas V, Bilak M, Engel WK, Leclerc A, Tome F. 1993. Prion protein is strongly immunolocalized at the postsynaptic domain of human normal neuromuscular junctions. Neurosci Lett 159:111–114. [DOI] [PubMed] [Google Scholar]
- 16.Kitamoto T, Shin RW, Doh-ura K, Tomokane N, Miyazono M, Muramoto T, Tateishi J. 1992. Abnormal isoform of prion proteins accumulates in the synaptic structures of the central nervous system in patients with Creutzfeldt-Jakob disease. Am J Pathol 140:1285–1294. [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier JG, Escaig-Haye F, Billette de Villemeur T, Robain O. 1995. Ultrastructural localization of cellular prion protein (PrPc) in synaptic boutons of normal hamster hippocampus. C R Acad Sci III 318:339–344. [PubMed] [Google Scholar]
- 18.Sisková Z, Mahad DJ, Pudney C, Campbell G, Cadogan M, Asuni A, O'Connor V, Perry VH. 2010. Morphological and functional abnormalities in mitochondria associated with synaptic degeneration in prion disease. Am J Pathol 177:1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin MT, Beal MF. 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795. [DOI] [PubMed] [Google Scholar]
- 20.Chiti F, Dobson CM. 2006. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366. [DOI] [PubMed] [Google Scholar]
- 21.Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. 2009. Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A 106:14670–14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E, Ankarcrona M. 2008. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci U S A 105:13145–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy PH, Beal MF. 2008. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer's disease. Trends Mol Med 14:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durcan TM, Fon EA. 2015. The three ‘P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev 29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Requejo-Aguilar R, Bolanos JP. 2016. Mitochondrial control of cell bioenergetics in Parkinson's disease. Free Radic Biol Med 100:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SI, Ju WK, Choi EK, Kim J, Lea HZ, Carp RI, Wisniewski HM, Kim YS. 1998. Mitochondrial dysfunction induced by oxidative stress in the brains of hamsters infected with the 263 K scrapie agent. Acta Neuropathol 96:279–286. doi: 10.1007/s004010050895. [DOI] [PubMed] [Google Scholar]
- 27.Choi HS, Choi YG, Shin HY, Oh JM, Park JH, Kim JI, Carp RI, Choi EK, Kim YS. 2014. Dysfunction of mitochondrial dynamics in the brains of scrapie-infected mice. Biochem Biophys Res Commun 448:157–162. doi: 10.1016/j.bbrc.2014.04.069. [DOI] [PubMed] [Google Scholar]
- 28.Ansoleaga B, Garcia-Esparcia P, Llorens F, Hernandez-Ortega K, Carmona Tech M, Antonio Del Rio J, Zerr I, Ferrer I. 12 June 2016. Altered mitochondria, protein synthesis machinery, and purine metabolism are molecular contributors to the pathogenesis of Creutzfeldt-Jakob disease. J Neuropathol Exp Neurol doi: 10.1093/jnen/nlw048. [DOI] [PubMed] [Google Scholar]
- 29.Moore RA, Sturdevant DE, Chesebro B, Priola SA. 2014. Proteomics analysis of amyloid and nonamyloid prion disease phenotypes reveals both common and divergent mechanisms of neuropathogenesis. J Proteome Res 13:4620–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kercher L, Favara C, Chan C-C, Race R, Chesebro B. 2004. Differences in scrapie-induced pathology of the retina and brain in transgenic mice that express hamster prion protein in neurons, astrocytes, or multiple cell types. Am J Pathol 165:2055–2067. doi: 10.1016/S0002-9440(10)63256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Race R, Oldstone M, Chesebro B. 2000. Entry versus blockade of brain infection following oral or intraperitoneal scrapie administration: role of prion protein expression in peripheral nerves and spleen. J Virol 74:828–833. doi: 10.1128/JVI.74.2.828-833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prusiner S, Scott M, Foster D, Pan K, Groth D, Mirenda C, Torchia M, Yang S, Serban D, Carlson GA, Hoppe PC, Westaway D, DeArmond SJ. 1990. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63:673–686. doi: 10.1016/0092-8674(90)90134-Z. [DOI] [PubMed] [Google Scholar]
- 33.Faris R, Moore RA, Ward A, Race B, Dorward DW, Hollister JR, Fischer ER, Priola SA. 2017. Cellular prion protein is present in mitochondria of healthy mice. Sci Rep 7:41556. doi: 10.1038/srep41556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatsuta T, Model K, Langer T. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol Biol Cell 16:248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Liu X, Bai J, Tian X, Zhao X, Liu W, Duan X, Shang W, Fan HY, Tong C. 2016. Mitoguardin regulates mitochondrial fusion through MitoPLD and is required for neuronal homeostasis. Mol Cell 61:111–124. [DOI] [PubMed] [Google Scholar]
- 36.Santel A, Fuller MT. 2001. Control of mitochondrial morphology by a human mitofusin. J Cell Sci 114:867–874. [DOI] [PubMed] [Google Scholar]
- 37.Milhavet O, McMahon HE, Rachidi W, Nishida N, Katamine S, Mange A, Arlotto M, Casanova D, Riondel J, Favier A, Lehmann S. 2000. Prion infection impairs the cellular response to oxidative stress. Proc Natl Acad Sci U S A 97:13937–13942. doi: 10.1073/pnas.250289197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 39.Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]