ABSTRACT
Enterovirus D68 (EV-D68) is one of the many nonpolio enteroviruses that cause mild to severe respiratory illness. The nonstructural protein 3Dpol is an RNA-dependent RNA polymerase (RdRP) of EV-D68 which plays a critical role in the replication of the viral genome and represents a promising drug target. Here, we report the first three-dimensional crystal structure of the RdRP from EV-D68 in complex with the substrate GTP to 2.3-Å resolution. The RdRP structure is similar to structures of other viral RdRPs, where the three domains, termed the palm, fingers, and thumb, form a structure resembling a cupped right hand. Particularly, an N-terminal fragment (Gly1 to Phe30) bridges the fingers and the thumb domains, which accounts for the enhanced stability of the full-length enzyme over the truncation mutant, as assessed by our thermal shift assays and the dynamic light scattering studies. Additionally, the GTP molecule bound proximal to the active site interacts with both the palm and fingers domains to stabilize the core structure of 3Dpol. Interestingly, using limited proteolysis assays, we found that different nucleoside triphosphates (NTPs) stabilize the polymerase structure by various degrees, with GTP and CTP being the most and least stabilizing nucleosides, respectively. Lastly, we derived a model of the core structure of 3Dpol stabilized by GTP, according to our proteolytic studies. The biochemical and biophysical characterizations conducted in this study help us to understand the stability of EV-D68-3Dpol, which may extend to other RdRPs as well.
IMPORTANCE Enterovirus D68 (EV-D68) is an emerging viral pathogen, which caused sporadic infections around the world. In recent years, epidemiology studies have reported an increasing number of patients with respiratory diseases globally due to the EV-D68 infection. Moreover, the infection has been associated with acute flaccid paralysis and cranial nerve dysfunction in children. However, there are no vaccines and antiviral treatments specifically targeting the virus to date. In this study, we solved the crystal structure of the RNA-dependent RNA polymerase of EV-D68 and carried out systematic biophysical and biochemical characterizations on the overall and local structural stability of the wild-type (WT) enzyme and several variants, which yields a clear view on the structure-activity relationship of the EV-D68 RNA polymerase.
KEYWORDS: crystal structure, thermostability, RNA-dependent RNA polymerase, 3Dpol
INTRODUCTION
Enterovirus D68 (EV-D68) belongs to the Picornaviridae family of small nonenveloped, cytoplasmic, plus-strand RNA viruses, which includes poliovirus, foot-and-mouth disease virus, hepatitis A virus, coxsackievirus, and rhinovirus. EV-D68 typically causes mild to severe respiratory illness, but the symptoms range from minor to flu-like symptoms to debilitating illness and polio-like symptoms. Some people with severe respiratory illness may require hospitalization and intensive care. The EV-D68 virus can often be found in an infected person's respiratory secretions, such as saliva, sputum, or nasal mucus. As a result, it likely spreads between people through coughs, sneezes, or touches. EV-D68 was first isolated in California in 1962, where it gave rise to various degrees of respiratory illness (1, 2). In 2014, a national EV-D68 outbreak occurred, with 1,153 cases reported from the United States and 2,287 cases confirmed worldwide. There were possibly additional mild infections for which people did not seek medical treatment. Aside from causing severe respiratory illness, this outbreak led to 14 deaths in the United States and 3 deaths in Canada (3, 4). In addition, EV-D68 has been implicated in cases of a polio-like disorder called acute flaccid myelitis. EV-D68 is particularly disabling in young children, as well as in the very weak, as serious complications have frequently been reported for infants and for those with weakened immune systems. For example, EV-D68 was found in two of five children during a 2012-2013 cluster of polio-like disease cases in California (5). Currently, therapeutic treatments of EV-D68 are limited because there are no vaccines or antiviral drugs against this virus (6).
RNA-dependent RNA polymerases (RdRPs) play a key role in viral genome replication. In positive-sense RNA viruses like EV-D68, RdRPs replicate the infecting positive-sense genomes into the minus-sense genomes. These minus-sense replicates serve as the templates for positive-sense genomes, which are in turn used for translation or are packed into new virions.
To date, the three-dimensional structures of RdRPs from five families of RNA viruses have been reported. These viruses are the following: (i) the Picornaviridae members poliovirus (Protein Data Bank [PDB] accession numbers 1RDR and 1RA6) (7–12) and EV71 (PDB code 3N6M) (13), (ii) the Caliciviridae family member rabbit hemorrhagic disease virus (PDB codes 1KHV and 1KHW) (14, 15), (iii) Flaviviridae family member hepatitis C virus (PDB codes 1CSJ and 1C2P) (16–19), (iv) Cystoviridae family member phage ϕ6 (PDB codes 1HHS and 1UVI) (20, 21), and (v) Reoviridae family member reovirus (PDB code 1MUK) (22). These RdRPs share similar overall structures that resemble a cupped right hand, and the structural domains were named accordingly: fingers, palm, and thumb. The fingers domain can be further divided into the index, ring, middle, and pinky domains. In addition to the overall structural similarity, all RdRPs display six conserved sequence motifs, termed A to F. Motifs A to D are shared between the single-subunit RNA and DNA-directed polymerases, while motifs E and F are unique to RdRPs and reverse transcriptases (RTs). Additionally, all but one of these sequence motifs (motif F) are found in the palm domain, which houses the active site in single-subunit polymerases and represents the most conserved region. Motif F is found in the fingers domain, and in conjunction with motif D from the palm domain, it contains several basic residues that help to form the nucleotide entry tunnel. In contrast, the thumb domain lacks any of the conserved sequence motifs and interacts with the fingers domain to “close” the hand. It is considered to form the path that guides the template nucleic acid through the polymerases (23).
Thompson et al. studied the enhanced stability of the poliovirus polymerase, brought about by the binding of nucleoside triphosphates (NTPs) as well as by fingers-thumb interactions. They showed that the interactions between the fingers and thumb domains are important for the structural integrity and function of poliovirus 3Dpol (polio-3Dpol). In addition, they demonstrated that binding of the NTPs stabilizes the polymerase structure by providing bridging interactions between the palm and fingers domain (11).
Since the EV-D68 outbreak in 2014, there has been an urgent need for EV-D68 research. Nonstructural viral proteins are often conserved and essential to viral functions, making themselves ideal targets for antiviral therapy. For example, zidovudine and nevirapine, which are widely used as anti-HIV drugs, are based on reverse transcriptase while antiviral inhibitors like valopicitabine directly target the RNA polymerase of the hepatitis C virus. We here report the first crystal structure of the 3Dpol in complex with GTP. In addition, we employed the dynamic light scattering (DLS) and differential scanning fluorimetry (DSF) methods to measure the overall thermostability of the full-length enzyme as well as several variants, including its N-terminally truncated mutant, and confirmed the importance of the internal interactions between the domains to protein stability. We also characterized polymerase stability in the presence of various NTPs, whose binding leads to differentially increased thermostability and local compactness. Taken together, these data indicated that the enclosed active site is important for the structural integrity of the enzyme. Due to the structural conservation among other picornaviral polymerases, it is very likely that our results and characterization methods on EV-D68-3Dpol can also be applied to other viral RdRPs.
RESULTS
Overall structure and active site of EV-D68-3Dpol.
The target protein was purified by a three-step purification protocol, yielding ∼20 mg of 3Dpol per liter of LB culture. The first encoded amino acid is glycine, which has been reported to be important for the activity of Picornaviridae RdRPs (10). In order to express our target protein heterologously in Escherichia coli, a methionine was added before the starting glycine. The methionine residue can be removed by the combined actions of deformylase and methionine aminopeptidase (24–26), as suggested by the clear electron density of a homogeneous glycine presence at the N terminus in the crystal structure. The identity of the purified protein was confirmed by Western blot analysis (data not shown).
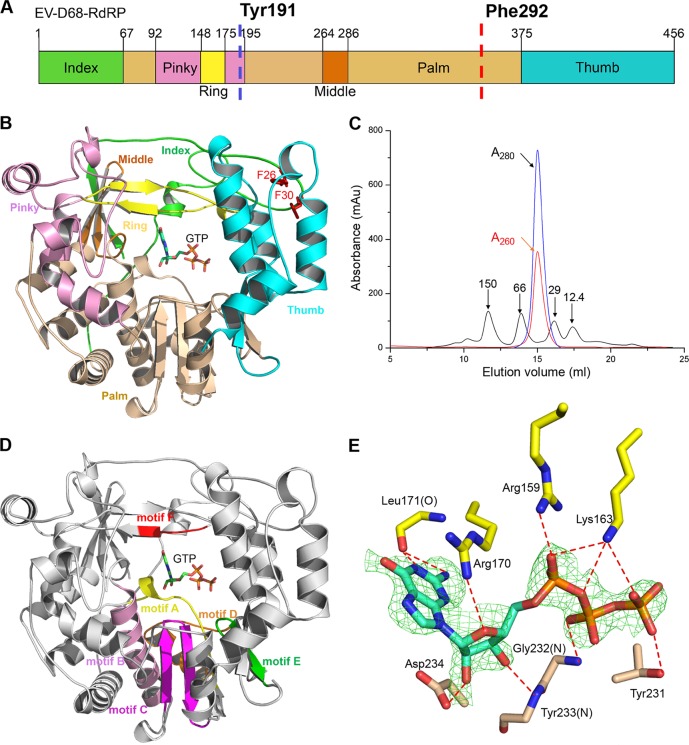
The crystal structure of 3Dpol in complex with GTP was subsequently determined to 2.3 Å (Table 1), and it displays a conserved right-handed structure similar to structures of other RdRPs in the Picornaviridae family. There is only a monomer in the asymmetric unit, comprising the three domains, fingers, palm, and thumb (Fig. 1A and B). Size exclusion chromatography suggested that EV-D68-3Dpol forms a monomer in solution as well (Fig. 1C). In addition, the UV absorbance at 260 nm indicates that there are no copurified nucleic acids or nucleosides present in the protein sample.
TABLE 1.
Diffraction data collection and refinement statistics
| Parameter | Value for the parametera |
|---|---|
| Diffraction source | Oxford Diffraction Xcalibur Nova diffractometer |
| Wavelength (Å) | 1.54 |
| Temp (K) | 100 |
| Detector | Oxford Onyx CCD |
| Crystal detector distance (mm) | 65 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 60 |
| Space group | P21 |
| Cell parameters | |
| a, b, c (Å) | 55.9, 81.44, 58.45 |
| α, β, γ (°) | 90, 120, 90 |
| Resolution range (Å) | 26.21–2.30 (2.42–2.30) |
| Total no. of reflections | 69,524 (9,918) |
| No. of unique reflections | 21,405 (3,156) |
| Completeness (%) | 98.1 (100) |
| Redundancy | 3.2 (3.1) |
| I/σ(I) | 6.0 (1.1) |
| Rmergeb | 0.201 (0.939) |
| Rrimc | 0.132 (0.631) |
| Resolution range (Å) | 26.21–2.30 (2.40–2.30) |
| No. of reflections | 21,378 |
| Rwork/Rfreed | 0.224/0.267 |
| No. of non-H atoms | |
| Protein | 3,613 |
| Ligand | 32e |
| Water | 243 |
| RMS deviation | |
| Bond length (Å) | 0.003 |
| Bond angle (°) | 0.564 |
| Avg B factor (Å2) | |
| Protein | 35.41 |
| Ligand | 51.49e |
| Water | 32.96 |
| Ramachandran plot (%) | |
| Favored regions | 98.03 |
| Allowed regions | 1.97 |
Values in parentheses are for the highest-resolution shell.
Rmerge = Σ |(I − < I >)|/σ(I), where I is the observed intensity.
rim, redundancy-independent merging.
Rwork = Σhkl ‖Fo| − |Fc‖/Σhkl |Fo|, calculated from working data set. Rfree is calculated from 5.0% of data.
GTP.
FIG 1.
The overall structure of the EV-D68-3Dpol. (A) The domain organization of EV-D68-3Dpol. The division of domains is based on other RdRP structures in the Picornaviridae family, and each domain is color coded as shown. Two predicated proteolysis sites are indicated by dashed vertical lines, and the corresponding residues are indicated. (B) The right-hand arrangement of the overall structure of 3Dpol. The color scheme is the same as shown in panel A. The GTP molecule and the two phenylalanines are shown as sticks. (C) The gel filtration running trace of EV-D68-3Dpol with a Superdex 200 10/300 Increase column, superimposed on the trace of four standard protein markers. The blue and red curves represent the UV absorbances, as indicated. (D) The six conserved motifs (motifs A to F) in the structure: motif A (residues 225 to 237), motif B (residues 289 to 306), motif C (residues 317 to 332), motif D (residues 336 to 353), motif E (residues 366 to 376), and motif F (residues 168 to 172). (E) The simulated Fo − Fc omit electron density map of bound GTP (contoured at 2.5σ) and its interactions with surrounding residues at the active site. The hydrogen bonds are shown by the red dashed lines (distance of ≤3.35 Å). The surrounding residues are labeled. Au, arbitrary units.
The crystal structure of EV-D68-3Dpol shows features that are typical of single-subunit RdRPs. The first four of the six conserved motifs form the core structure of the palm domain, containing two α-helices (from motifs B and D) and a three-stranded antiparallel β-sheet (from motifs A and C). The fifth motif, E, harbors a β-sheet that interacts with both the N-terminal thumb domain and the core of the palm domain (Fig. 1D).
The fingers domain interacts with the palm domain through the bound GTP to stabilize the core structure. Residues Cys12 to Phe30 of the index finger domain interact with residues Arg398 to Thr400 from the thumb domain to form the NTP entry tunnel. Trp399 and two buried aromatic residues, Phe26 and Phe30, are conserved in poliovirus RdRP and were shown to be critical for the stability of the polymerase (11).
The simulated Fo − Fc omit electron density map shows clear density for GTP. Although 3Dpol is Mg2+ dependent, in our structure, there are no Mg2+ or other divalent metal ions bound nearby (27, 28). Since an RNA primer and template were not added prior to crystallization, we speculate that the Mg2+ ions may appear after the enzyme achieves a productive state during catalysis. GTP is surrounded by a network of hydrogen bonds and electrostatic interactions, which are important for its association with the enzyme and subsequent positioning. Most of these interactions are observed in the structure of the EV71-3Dpol-GTP complex (13). Specifically, the hydrogen bond between the side chain of Asp234 and the 2′ hydroxy group of GTP is crucial for polymerase activity, with a distance of 2.42 Å. The base moiety of GTP interacts with the guanidinium group of Arg170 from the fingers domain through a stacking interaction. Leu171 forms two hydrogen bonds with the base moiety, whereas Tyr233 forms a hydrogen bond with the 3′ hydroxy group using their backbone atoms. In addition to Asp234, the equivalent residues of Arg170 and Leu171 in poliovirus RdRP have also been reported to play vital roles in the binding of GTP in the active site (7). Additionally, Arg159 and Lys163 form salt bridge interactions with the triphosphate moiety of GTP, while Gly232 and Thr231 form two hydrogen bonds with the β-phosphate oxygens (Fig. 1E).
Comparison to other RdRPs.
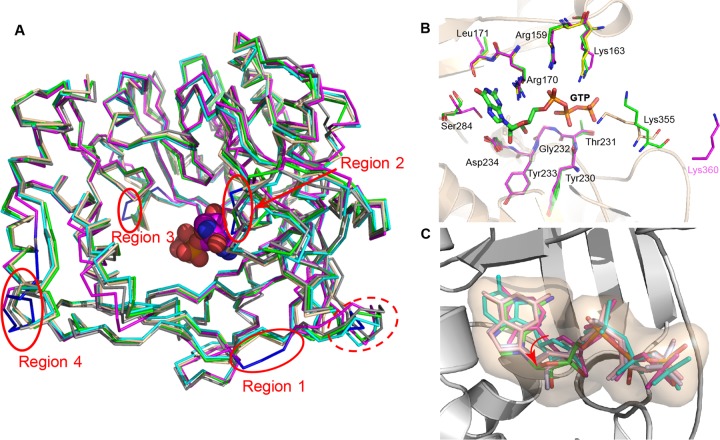
EV-D68-3Dpol shares high sequence identities with other Picornaviridae RdRPs: poliovirus (74%) (7, 10), coxsackievirus (74%) (12), human rhinovirus 16 (HRV-16) (57%) (9), and EV71 (63%) polymerases (13). To compare the overall and local differences between the EV-D68-3Dpol-GTP structure and homologous RdRP structures, we superimposed our structure (PDB 5XE0) (Fig. 2A, shown in wheat) with the polio-3Dpol-GTP complex (PDB 1RA7) (in green), HRV-16-3Dpol (PDB 1TP7) (in deep teal), coxsackievirus B3-3Dpol (PDB 3DDK) (in cyan), and the EV71-3Dpol-GTP complex (PDB 3N6M) (in magenta). All of these RdRPs closely resemble each other, with root mean square (RMS) deviations of ∼0.5 to 0.9 Å. One notable difference is that the fragment of Gly8 to Val11 in EV-D68-3Dpol forms a small loop, while this region is longer and forms a small helix in the other RdRPs (Fig. 2A). Among these structures, the EV71-3Dpol-GTP complex appears to be the most distant ortholog from the others in terms of structure. In particular, four regions in EV71-3Dpol-GTP with relatively large deviations stand out: region 1 (Asn18 to Pro20, in EV71-3Dpol numbering), region 2 (Gly290 to Gly293), region 3 (Asp359 to Ser361), and region 4 (Ser439 to Pro447). All of these local structural differences occur on loops. Of note, region 2 is close to the GTP-binding site. This fragment is rich in glycines and serines and represents a rather flexible region. These regions are quite different in their backbone positions in each polymerase and contain a central serine residue (Ser284 in EV-D68-3Dpol) (Fig. 2B, in wheat). In EV-D68-3Dpol-GTP, this serine is very close to the guanine ring of GTP, and its position is well defined by electron density. The large structural variations between each polymerase may not be caused by the binding of the GTP substrate as the serine in the EV71-3Dpol-GTP complex also adopts a distinct conformation (Ser289) (Fig. 2B, in magenta) while its guanine ring of GTP is very similar in position to that in our complex structure. Aside from Ser284, the other three residues contacting the phosphate groups, Arg170, Lys355, and Lys163, also have different orientations in our structure. Previously, Yang et al. demonstrated that the protonation state of the motif D lysine governs the open/closed states of the active site (29). In our complex structure, this particular lysine (Lys355) (Fig. 2B, in wheat) orientates differently from its counterpart in polio-3Dpol (Lys359) (in green). It points toward the phosphate of GTP although the electron density of the Lys355 side chain is weak. The flexibility of this residue may ensure the fast switch between the open and closed states of the active site. On the other hand, in the EV71-3Dpol-GTP complex structure, this lysine (Lys360) (Fig. 2B, in magenta) is far away from GTP (Fig. 2B).
FIG 2.
The structure comparison of EVD-68-3Dpol. (A) The superposition of EV-D68-3Dpol with polio-3Dpol (green), coxsackievirus-3Dpol (cyan), HRV-16-3Dpol (deep teal), and EV71-3Dpol (magenta). The N-terminal difference (residues Lys8 to Val11) of EV-D68-3Dpol is highlighted in the dashed red oval. The four local structural differences are shown in blue and indicated by red circles. GTP is shown as spheres. (B) The superposition of the active-site residues of EV-D68-3Dpol-GTP (PDB 5XE0; wheat), EV71-3Dpol-GTP (PDB 3N6M; magenta), and polio-3Dpol-GTP (PDB 1RA7; green). Only the residues of EV-D68-3Dpol and Lys360 from EV71-3Dpol are shown. GTP is shown in a stick model. (C) Superposition of GTP-bound EV-D68-3Dpol (PDB 5XE0; green) with polio-3Dpol bound by various NTPs (PDB 1RA7 in light blue; PDB 2IM2 in light magenta; PDB 2ILY in cyan) and EV71-3Dpol (PDB 3N6M in light pink). The rotation movement of the ribose ring of each NTP is indicated by the red arrow.
To check for possible differences in the NTP binding modes, we compared our EV-D68-3Dpol-GTP complex with the complexes of polio-3Dpol bound with various NTPs (PDB accession numbers 1RA7, 2IM2, 2ILY, and 2IM0) and the EV71-3Dpol-GTP complex (PDB 3N6M). Despite similar positions bound by each NTP, the GTP position moves closer to Asp234 (equivalent residue of Asp238 in EV71). Interestingly, the five-membered ribose ring of our GTP-bound structure (PDB 5EX0) rotates a small angle toward the active site, and its position is slightly “off” compared to the positions in other structures (Fig. 2C). This movement may be due to the strong interactions of the 2′ hydroxyl group with the terminal oxygen of the Asp234 carboxylate, which forms a short-distance hydrogen bond (∼2.4 Å). In addition, the bridging oxygens between the phosphates in GTP of EV-D68-3Dpol deviate from those in other structures, probably due to the differences in side chain orientation of Arg170, Lys355, and Lys163 (Fig. 2B).
Overall thermostability of EV-D68-3Dpol.
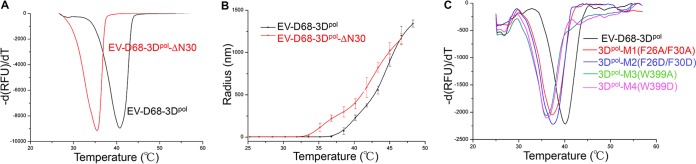
In our structure, the index finger domain protrudes into the thumb domain by insertion of Phe26 and Phe30 into the hydrophobic core at the top of the thumb domain. In addition to forming the first strand of the three-strand antiparallel β-sheet with the middle domain, the N-terminal 30 residues, which account for half of the index finger domain, also link the fingers and thumb domains. In particular, Trp399 inserts itself into the loop formed by Pro23 to Glu35 and stabilizes this region. Previous biochemical studies indicated that similar interactions play an important role in the in vitro activity of RdRP and in vivo viral infectivity (30). Using a thermal shift assay with SYPRO orange as the thermofluor, the overall thermostability of 3Dpol was investigated with DSF on a quantitative PCR (qPCR) machine. The results showed that the melting temperature (Tm) value of the full-length protein is ∼40.4°C. In contrast, an EV-D68-3Dpol mutant with a deletion of the first 30 residues (EV-D68-3Dpol-ΔN30) decreased the Tm value by ∼5.0 ± 0.2°C, underscoring the importance of the interactions between the thumb and ring finger domains for overall thermostability (Fig. 3A). DLS, a characterization technique of the aggregation state (or polydispersity) of protein in solution, also produced similar results. Apo-3Dpol has a radius of 3 nm as determined by DLS. As the temperature increases, the protein partially unfolds to form aggregates and eventually denatures, causing the measured particle size to increase dramatically (>100 nm). The temperatures that significantly changed the protein particle sizes of the full-length and truncated proteins were 38.5°C and 34.0°C, respectively (Fig. 3B). We further checked the destabilizing effects of the two double mutations F26A F30A and F26D F30D (the double mutations are on the full-length protein) (Table 2, mutants M1 and M2, respectively) by DSF and found that they reduced the Tm values by ∼3.0 ± 0.2°C. Similarly, the two single mutations W399A and W399D reduced the Tm values even further (to 36.0 and 36.3°C, respectively) (Fig. 3C; Table 2, M3 and M4). The points at which the protein particle sizes greatly change are an indication of the formation of aggregates in solution, suggesting partial denaturation. A further temperature rise leads to full unfolding of the protein. Taken together, these results indicated that the long loop of the fingers domain (Lys8 to Phe30) significantly contributes to the stability of 3Dpol by bringing the fingers and thumb domains together.
FIG 3.
The overall thermostability of EV-D68-3Dpol and its truncation mutant. (A) DSF measurements of the full-length EV-D68-3Dpol and the EV-D68-3Dpol-ΔN30 truncation mutant. (B) DLS measurements of the full-length EV-D68-3Dpol and the EV-D68-3Dpol-ΔN30 truncation mutant. (C) DSF measurements of four EV-D68-3Dpol mutants along with the WT. RFU, relative fluorescence units.
TABLE 2.
DSF summary of Tm values of EV-D68-3Dpol and its mutants
| Sample | Mutation(s) | Tm (°C) |
|---|---|---|
| EV-D68-3Dpol | 40.4 ± 0.2 | |
| EV-D68-3Dpol-ΔN30 | Deletion of Gly1 to Phe30 | 35.4 |
| EV-D68-3Dpol-M1 | F26A F30A | 37.4 ± 0.2 |
| EV-D68-3Dpol-M2 | F26D F30D | 37.5 ± 0.1 |
| EV-D68-3Dpol-M3 | W399A | 36.0 ± 0.2 |
| EV-D68-3Dpol-M4 | W399D | 36.3 ± 0.1 |
Thermostability enhancement by binding of NTPs.
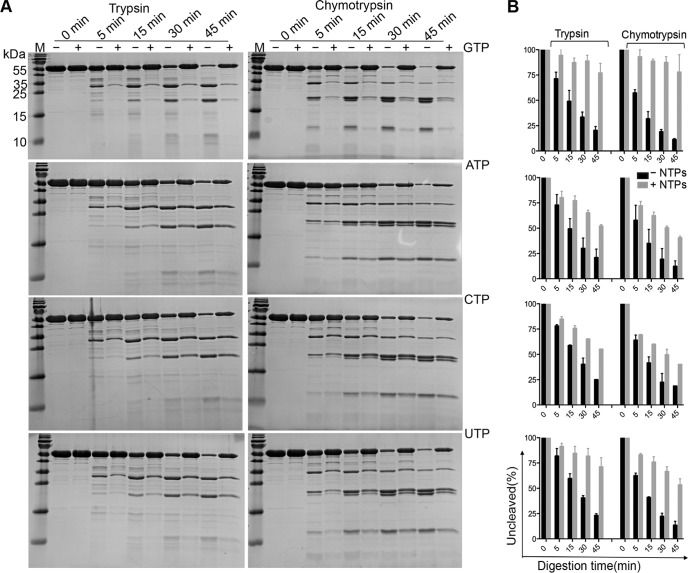
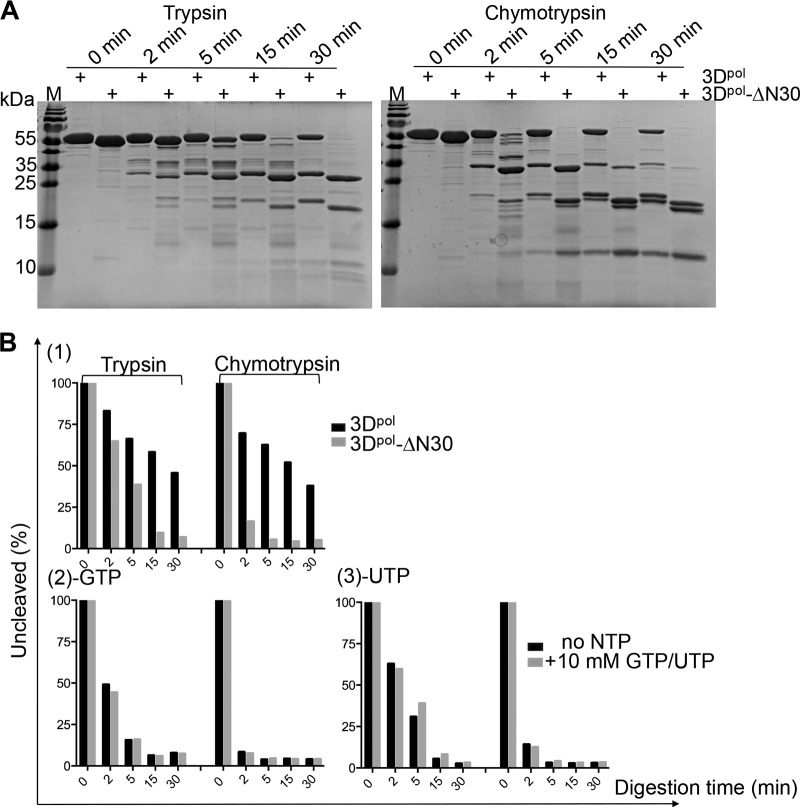
The bound GTP forms a total of 11 hydrogen bonds (≤3.35 Å) and two salt bridges with the protein, including two hydrogen bonds on the base ring and three on the 2′ hydroxy group of GTP. Previous structural analysis of polio-3Dpol by Thompson et al. indicated that the interactions with the NTPs stabilize the protein (11). To further investigate the importance of the interactions between the NTPs and EV-D68-3Dpol, we first measured the DLS profiles of 3Dpol before and after the binding of the NTPs. The results demonstrated that there was a 2.0 to 2.5°C (±0.5°C) boost in the unfolding temperature by each NTP compared to that of the apoprotein, consistent with the notion that the NTPs tend to stabilize the protein. However, GTP appears to have the largest enhancement effect, with a 2.5 ± 0.5°C increase (data not shown). The subtle but notable difference in performance by GTP motivated us to delve further into this subject, and we then performed limited proteolysis digestion on 3Dpol to probe the loose structural domains of the protein. Digestion by trypsin of the full-length protein generated three major fragments at ∼33, 22, and 12 kDa over a 45-min time course reaction, while chymotrypsin generated one extra band at ∼21 kDa (Fig. 4A). However, in the presence of 10 mM GTP, the protein was partially protected, which increased the percentages of the leftover protein at the end of the reaction from 21.9% (without GTP) to 76.9% (with GTP) for trypsin digestion and from 13.4% (without GTP) to 77.9% (with GTP) for chymotrypsin digestion (Fig. 4A and B). Similarly, digestion in the presence of 10 mM concentrations of other NTPs consistently showed protective effects but to different degrees. In summary, we observed the most and the least pronounced protection effects by GTP and CTP, respectively, and these digestion results were always consistent with the same proteases (Fig. 4). Additionally, the truncated protein EV-D68-3Dpol-ΔN30 showed a digestion pattern similar to that of the wild type (WT) but had a greater tendency to be degraded (Fig. 5A). As shown in Fig. 5B, the deletion mutant was barely protected by the binding of GTP or UTP, the two best stabilizers in the case of the full-length protein. Last, we tried to identify the degradation products from the proteolysis. Four polypeptide fragments were consistently generated by digestion with chymotrypsin. The digestion profiles suggested that the full-length protein (52.9 kDa) was first split into the 33- and 22-kDa polypeptides, and the former in turn gave rise to the 12- and 21-kDa fragments. Additionally, the truncation mutant also generated similar bands with slight size reductions, except for the 12-kDa band (Fig. 5A), and the size reduction roughly corresponded to the deletion of 30 residues. On the other hand, the 12-kDa fragment could be generated from both the full-length and truncated proteins. This result suggested that the major fragment at 33 kDa starts from the N terminus, while the 12-kDa fragment starts from the C terminus. Combined sequence and structural analyses suggested that the 33-kDa polypeptide roughly corresponds to the sum of the N-terminal palm domain and entire fingers domains (i.e., Gly1 to Phe292, with a calculated molecular mass of 32.5 kDa) (Fig. 1A), while the 12-kDa polypeptide corresponds to the sum of the N-terminal palm domain and the index, pinky, and ring domains (i.e., Tyr191 to Phe292, with a calculated molecular mass of 11.1 kDa) (Fig. 1A). Because 3Dpol is rich in aromatic residues (51 in total), it is difficult to pinpoint the exact digestion sites. It appears that the protease preferentially removes the C-terminal 175 residues (including the tag) and then cleaves after Tyr191 to further divide the N-terminal fragment into two polypeptides. The GTP-containing fragment is ∼22 kDa, which constitutes the core of the protein.
FIG 4.
The effects of the NTPs on the limited proteolysis profiles of EV-D68-3Dpol. (A) The digestion patterns of full-length EV-D68-3Dpol in the absence (−) or presence (+) of 10 mM NTPs (GTP, ATP, CTP, and UTP, in top-down order) by the two proteases in a 45-min time course digestion process. (B) The summary of the protective effects of GTP, ATP, CTP, and UTP, in parallel to the SDS-PAGE gel results shown on the left. The protein amount at time point 0 min was considered 100%.
FIG 5.
Comparison of the limited proteolysis patterns of EV-D68-3Dpol to those of the truncation mutant. (A) The digestion patterns of the truncation mutant and the full-length EV-D68-3Dpol. (B) Summary of the digestion of EV-D68-3Dpol-ΔN30 as well as of the full-length EV-D68-3Dpol without NTPs (graph 1), with GTP (graph 2), and with UTP (graph 3).
DISCUSSION
In this study, we report the first crystal structure of the EV-D68 RdRP in complex with GTP. RdRP is recognized as a potential target for antiviral therapy because it is a key enzyme for the replication of the viral RNA genome. The sequence and overall structure of EV-D68-3Dpol are similar to those of other RdRPs in the Picornaviridae family except for a few residues at the N terminus, which form a small loop instead of an α-helix. The conserved overall structure also preserves some important interactions between the domains. Residues Gly1 to Phe30 bring together the index, the middle finger, and the thumb domains by forming strong interactions within these domains. The DSF and DLS analyses indicated that the overall thermostability of the enzyme was reduced by 4.0 to 5.0°C (±0.2°C) due to the removal of this fragment. These results were consistent with the reports by Thompson et al. (11) and confirmed that the N terminus is critical to the overall stability and activity of the RdRPs.
GTP binds to the enzyme through a series of hydrogen bonds and electrostatic interactions. In turn, these interactions increase the thermostability of the enzyme, which has been demonstrated in previous work (31). Our DLS results showed that bound NTPs increase the initial unfolding temperatures by 2.0 to 2.5°C (±0.5°C). The structure-stabilizing function of the NTPs was confirmed by proteolysis analyses, and each NTP showed variable protection results against the proteases. This observation was also consistent with our cocrystallization trials of the protein with different NTPs. The GTP cocrystal gave the best diffraction result despite our attempts with all four NTPs, whose resolution extended to 2.3 Å. Here, we tried to offer an explanation by comparing our structure with known polio-3Dpol-NTP complex structures determined by Thompson et al. (11). Although it was difficult to quantitate the interactions in terms of energy, we found that the sum of total interactions on each NTP, with the exception of ATP, approximately correlates with their stability enhancement effects (Table 3; Fig. 1E). Unlike GTP, the adenine base of ATP is bound with an anti-conformation, with both rings of adenine participating in the interactions. It is not currently clear whether the conformation of the base plays a role. We also tried to quantitate the affinity of each NTP to the enzyme using isothermal titration calorimetry (ITC), but the protein precipitates during the titration process due to its instability.
TABLE 3.
Summary of total interactions of each NTP with EV-D68-3Dpol and their stability enhancement effectsa
| Sample (PDB accession no.) | No. of hydrogen bonds | No. of salt bridges | Total no. of interactions | Distance to Asp234/Asp238b (Å) | Stability enhancementc |
|---|---|---|---|---|---|
| GTP-3Dpol-EV-D68 | 8 | 3 | 11 | 2.42 | ++++ |
| GTP-3Dpol-EV71 (3N6M) | 8 | 2 | 10 | 2.67 | ++++ |
| GTP-3Dpol-polio (1RA7) | 6 | 3 | 9 | 2.76 | ++++ |
| UTP-3Dpol-polio (2IM2) | 6 | 2 | 8 | 2.49 | +++ |
| ATP-3Dpol-polio (2ILY) | 6 | 3 | 9 | 2.99 | ++ |
| CTP-3Dpol-polio (2IM0) | 5 | 2 | 7 | 2.83 | + |
Cutoff distance, ≤3.35 Å.
Asp234 is according to EV-D68 numbering, and Asp238 is according to EV71 numbering.
Stability enhancement was measured by the unfolding temperature differences between NTP-bound EV-D68-3Dpol and apoenzyme. The number of plus signs indicates the extent of protection conferred by each NTP.
Therefore, both the NTPs and the N terminus are important to the stability of 3Dpol, but we detected notable differences in terms of their stabilizing natures, as characterized by various techniques. First, the N terminus connects the fingers and the thumb domains, which helps to maintain the general 3D structure and thus contributes to the overall stability of the protein. There were generally Tm differences of 4.0 to 5.0°C (±0.2°C) between the full-length and the truncated proteins, as measured by DLS and DSF. On the other hand, the NTPs bridge the fingers and the palm domains, and these interactions stabilize the core of the protein and contribute to its local stability. Consequently, the NTPs led to unfolding temperature differences of only 2.0 to 2.5 °C (±0.5°C), as shown by DLS, while they barely offer any stability enhancement, as detected by DSF (data not shown). Second, the limited proteolysis is meant to probe the flexible regions or the compactness of proteins, which may become more compact and rigid after the binding of the NTPs due to their strong interactions. This biochemical assay is more sensitive to local structure changes, and we could differentiate each NTP by this means (GTP > UTP > ATP > CTP). Furthermore, our studies revealed the approximate digestion sites within the protein.
In summary, we determined the structure of GTP-bound EV-D68-3Dpol, the first RdRP crystal structure of EV-D68-3Dpol. Our biochemical and biophysical characterization on 3Dpol demonstrated that the N terminus is critical to the overall stability of the protein, while the binding of the NTPs stabilizes the core of the protein. The combined structural analysis and experimental assays yield a clear view of the physiochemical properties of 3Dpol, which may be applied to RdRPs in general.
MATERIALS AND METHODS
Expression and purification of the WT and variants of EV-D68-3Dpol.
The gene encoding the EV-D68-3Dpol (BAK08578.1) was synthesized by The General Company (Shanghai, China) and cloned into pET-28a(+) with an additional GSSG-6×His tag at the C terminus. An extra ATG codon was added to the N terminus for translation initiation. However, an unwanted V17A mutation, which occurs in a nonconserved region, was produced in the cloning stage. It did not affect the overall structure of the polymerase as suggested by our structure, nor did it affect the polymerase activity (data not shown). EV-D68-3Dpol-ΔN30 was constructed by removing the 30 residues from the N terminus (Gly1 to Phe30). The four point mutations of EV-D68-3Dpol were obtained by using Fast Mutagenesis V2 kit (Vazyme, China), and they are all on the full-length gene.
The recombinant constructs were transformed into the E. coli strain BL21(DE3) for overexpression. Cells were cultured to an optical density at 600 nm (OD600) of 0.6 to 0.8 before induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated for an additional 20 h at 16°C. E. coli cells were harvested by centrifugation and disrupted by sonication in lysis buffer (100 mM K2HPO4, pH 8.0, 0.5 mM EDTA, pH 8.0, 60 μM ZnCl2, 20% glycerol, 3 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Polyethyleneimine (PEI; 0.25%) was added to the cell lysate, and the mixture was stirred at 4°C for 30 min to precipitate nucleic acids. The mixture was then centrifuged in a JA25.50 rotor at 17,000 rpm for 30 min, and the resulting supernatant was taken for further processing.
Ammonium sulfate powder was slowly added to the supernatant until 40% saturation was reached (280 g per liter), followed by a 30-min centrifugation in a JA25.50 rotor at 17,000 rpm to precipitate the target protein. The pellet from the ammonium sulfate precipitation was resuspended in 50 ml of lysis buffer per liter of cell culture (50 mM Tris-Hcl, pH 8.0, 300 mM NaCl, 20% glycerol, 3 mM β-mercaptoethanol, and 1 mM PMSF). Insoluble material was removed using a 0.45-μm-pore-size polyethersulfone (PES) membrane syringe filter (Millex, USA), and the filtrate was loaded onto 2 ml of preequilibrated Ni-nitrilotriacetic acid (NTA) beads (Qiagen, German). The Ni-NTA column was washed with 10 column volumes of lysis buffer plus 20 mM, 30 mM, and 40 mM imidazole in three stages to remove nonspecifically bound proteins.
The elution was performed with 200 mM imidazole present in the lysis buffer. The eluted fractions were pooled and then diluted using a buffer without salt (50 mM Tris, pH 8.0, 20% glycerol, and 3 mM β-mercaptoethanol) to a final salt concentration of 100 mM NaCl and loaded directly onto a HiTrap Q column (GE Healthcare). 3Dpol was eluted using a linear salt gradient from 100 mM NaCl to 500 mM NaCl. The peak fractions containing the target protein were pooled and concentrated to 9.0 mg/ml using Amicon Ultra concentrators (cutoff size of 30 kDa; Millipore) and loaded onto a Superdex 200 column (GE Healthcare) for further purification and characterization, with a buffer of 50 mM HEPES, pH 7.6, 300 mM NaCl, 3% glycerol, 50 mM MgCl2, and 10 mM dithiothreitol (DTT).
The purified protein was concentrated to 12.0 mg/ml, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. EV-D68-3Dpol-ΔN30 and four mutations were expressed in the same manner as the wild-type 3Dpol and purified by one-step Ni-affinity chromatography (12, 32).
Oligomerization characterization of EV-D68-3Dpol.
Gel filtration analysis was performed on a Superdex 200 (10/300) Increase column (GE Healthcare). A sample containing four standard protein markers was separated using the same column for calibration (Sigma, USA). The marker sizes are 150 kDa (alcohol dehydrogenase), 66 kDa (albumin bovine serum), 29 kDa (carbonic anhydrase), and 12.4 kDa (cytochrome c).
Crystallization and structure determination.
Purified 3Dpol was mixed with GTP at a final concentration of 11.2 mg/ml protein and 6.2 mM GTP and incubated on ice for >30 min before setting up drops. 3Dpol (0.6 μl) and reservoir solution (0.6 μl) were mixed and incubated over a 200-μl reservoir solution using the sitting-drop method in 96-well trays. Crystals were grown from 0.1 M Tris-HCl, pH 8.5, 20.1% isopropanol, and 20.1% polyethylene glycol (PEG) 3350 and mounted for data collections after 10 days of growth at 16°C.
Data collection was performed on an in-house Oxford Diffraction Xcalibur Nova diffractometer operating at 50 kV and 0.8 mA. The crystal-to-detector distance was set to 65 mm, exposure time was set to 60 s, and the data were recorded with a 165-mm Onyx charge-coupled-device (CCD) detector.
Data were processed using the CrysAlisPro software (version 1.171.33.49; Oxford Diffraction). The structure was solved by molecular replacement using the program Phaser (33) in the Phenix suite with the crystal structure of the EV71-3Dpol (PDB code 3N6M; sequence identity of 63%) (13) as the search model. For subsequent refinement, Refmac5 in CCP4 was used by setting jelly body restraints to 0.02 with four cycles of refinement (34). The refined model was further built manually according to the electron density map with COOT (35). The GTP molecule was added in the late stage of refinement. A final round of refinement was performed using Phenix.refine (36) and yielded a final R factor of 22.4% (Rfree, 26.7%). The final model was validated by MolProbity (37), and the structure figures were produced with PyMOL (www.pymol.org). The final data collection and refinement statistics are shown in Table 1.
Dynamic light scattering.
Dynamic light scattering measurements were performed on DynaPro Plate Reader II (Wyatt Technology, Goleta, CA) (38, 39). EV-D68-3Dpol (2.0 mg/ml) or the mutants were spun down at 14,000 rpm for 10 min, and 25 μl of each was loaded into 384-well plates. The NTP-bound samples were prepared by addition of NTPs to the protein to a final concentration of 10 mM, and the mixtures were incubated on ice for 30 min. An additional 20 μl of mineral oil was placed on the top of the protein to prevent evaporation during the heating process. The sample plate was centrifuged for 2 min at 2,000 rpm to remove any air bubbles. Measurements were taken using a temperature range of 25 to 50°C with a temperature increment of 0.66°C/min. Each well was measured five times, with a 5-s acquisition time. Each sample was repeated three times.
Thermoshift assay.
DSF analyses were performed on an Applied Biosystems real-time PCR instrument (CFX96; Bio-Rad, USA). Samples (at 1.0 mg/ml) were present in a buffer of 50 mM HEPES, pH 7.6, 300 mM NaCl, 3% glycerol, 50 mM MgCl2, and 10 mM DTT. SYPRO orange dye (100×; Thermo Scientific, USA) was mixed with the protein samples, and 10 μl of each sample was loaded into 96-well multiplate PCR plates (Bio-Rad, USA). Each sample was prepared in triplicate, sealed, and protected from light before being loaded onto the machine. The samples were heated at 0.2°C/min, from 25 to 90°C. The fluorescence intensity was measured every 0.2°C. All DSF figures were produced by Origin, version 8.0.
Proteolytic digestion analysis.
Twenty micrograms of EV-D68-3Dpol was partially digested with 0.24 μg of trypsin or chymotrypsin in a 40-μl reaction volume system. A sample of 8.0 μl was taken out at different time points (0, 5, 15, 30, and 45 min) and added to 2 μl of SDS-PAGE loading buffer. For the mutants, the reaction was conducted for up to 30 min. The aliquoted samples at each time point were immediately heated at 100°C for 10 min. Samples were analyzed by SDS-PAGE (15%) and stained by Coomassie blue. The NTP-bound samples were processed in the same manner. ImageJ was used to quantify the intensities of the target bands on SDS-PAGE gels, and the results were quantified by the ratios of the intensities of the remaining proteins to those of the undigested samples at each time point. Figures were produced by GraphPad Prism, version 6.
Accession number(s).
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 5XE0.
ACKNOWLEDGMENTS
We thank David Bushnell for his help throughout this study and in the manuscript-writing process.
Wei Xie conceived and designed the research, Chunnian Wang and Caiyan Wang performed the research, and Wei Xie, Qing Li, and Zhong Wang analyzed the data and wrote the paper.
This work was supported by the Fundamental Research Funds for the Central Universities (16lgjc76), the Science and Technology Program of Guangzhou (201504010025 and 201605030012), and Guangdong Innovative Research Team Program number 2011Y038.
We declare that we have no competing financial interests.
REFERENCES
- 1.Khan F. 2015. Enterovirus D68: acute respiratory illness and the 2014 outbreak. Emerg Med Clin North Am 33:e19– e32. doi: 10.1016/j.emc.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Aliabadi N, Messacar K, Pastula DM, Robinson CC, Leshem E, Sejvar JJ, Nix WA, Oberste MS, Feikin DR, Dominguez SR. 2016. Enterovirus D68 infection in children with acute flaccid myelitis, Colorado, USA, 2014. Emerg Infect Dis 22:1387–1394. doi: 10.3201/eid2208.151949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson R. 2014. Outbreaks of enterovirus D68 continue across the USA. Lancet Respir Med 2:791. doi: 10.1016/S2213-2600(14)70190-0. [DOI] [PubMed] [Google Scholar]
- 4.Holm-Hansen CC, Midgley SE, Fischer TK. 2016. Global emergence of enterovirus D68: a systematic review. Lancet Infect Dis 16:e64–75. doi: 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown E. 25 February 2014. Mysterious polio-like illnesses reported in some California children. Los Angeles Times, Los Angeles, CA: http://articles.latimes.com/2014/feb/23/local/la-me-ln-polio-like-paralysis-california-20140223. [Google Scholar]
- 6.Liu Y, Sheng J, Fokine A, Meng G, Shin WH, Long F, Kuhn RJ, Kihara D, Rossmann MG. 2015. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science 347:71–74. doi: 10.1126/science.1261962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen JL, Long AM, Schultz SC. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109–1122. doi: 10.1016/S0969-2126(97)00261-X. [DOI] [PubMed] [Google Scholar]
- 8.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem 279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 9.Love RA, Maegley KA, Yu X, Ferre RA, Lingardo LK, Diehl W, Parge HE, Dragovich PS, Fuhrman SA. 2004. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure 12:1533–1544. doi: 10.1016/j.str.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Thompson AA, Peersen OB. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J 23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson AA, Albertini RA, Peersen OB. 2007. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J Mol Biol 366:1459–1474. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campagnola G, Weygandt M, Scoggin K, Peersen O. 2008. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J Virol 82:9458–9464. doi: 10.1128/JVI.00647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Lou Z, Miao Y, Yu Y, Dong H, Peng W, Bartlam M, Li X, Rao Z. 2010. Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China. Protein Cell 1:491–500. doi: 10.1007/s13238-010-0061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng KK, Cherney MM, Vazquez AL, Machin A, Alonso JM, Parra F, James MN. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J Biol Chem 277:1381–1387. doi: 10.1074/jbc.M109261200. [DOI] [PubMed] [Google Scholar]
- 15.Zamyatkin DF, Parra F, Alonso JM, Harki DA, Peterson BR, Grochulski P, Ng KK. 2008. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem 283:7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 16.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417–1426. doi: 10.1016/S0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 17.Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol 6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 18.Choi KH, Gallei A, Becher P, Rossmann MG. 2006. The structure of bovine viral diarrhea virus RNA-dependent RNA polymerase and its amino-terminal domain. Structure 14:1107–1113. doi: 10.1016/j.str.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 19.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol 81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 21.Salgado PS, Makeyev EV, Butcher SJ, Bamford DH, Stuart DI, Grimes JM. 2004. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Tao Y, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synthesis in a cage–structural studies of reovirus polymerase lambda3. Cell 111:733–745. doi: 10.1016/S0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 23.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandman K, Grayling RA, Reeve JN. 1995. Improved N-terminal processing of recombinant proteins synthesized in Escherichia coli. Biotechnology 13:504–506. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Bassat A, Bauer K, Chang SY, Myambo K, Boosman A, Chang S. 1987. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol 169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopalan PT, Datta A, Pei D. 1997. Purification, characterization, and inhibition of peptide deformylase from Escherichia coli. Biochemistry 36:13910–13918. doi: 10.1021/bi971155v. [DOI] [PubMed] [Google Scholar]
- 27.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. 2009. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol 16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Lee JY, Nowotny M. 2006. Making and breaking Nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell 22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Smidansky ED, Maksimchuk KR, Lum D, Welch JL, Arnold JJ, Cameron CE, Boehr DD. 2012. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20:1519–1527. doi: 10.1016/j.str.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J 20:1153–1163. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards OC, Yu P, Neufeld KL, Ehrenfeld E. 1992. Nucleotide binding by the poliovirus RNA polymerase. J Biol Chem 267:17141–17146. [PubMed] [Google Scholar]
- 32.Gong P, Kortus MG, Nix JC, Davis RE, Peersen OB. 2013. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS One 8:e60272. doi: 10.1371/journal.pone.0060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gast K, Damaschun H, Misselwitz R, Muller-Frohne M, Zirwer D, Damaschun G. 1994. Compactness of protein molten globules: temperature-induced structural changes of the apomyoglobin folding intermediate. Eur Biophys J 23:297–305. doi: 10.1007/BF00213579. [DOI] [PubMed] [Google Scholar]
- 39.Morris AM, Watzky MA, Finke RG. 2009. Protein aggregation kinetics, mechanism, and curve-fitting: a review of the literature. Biochim Biophys Acta 1794:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]