ABSTRACT
The replication cycle of human cytomegalovirus (CMV) leads to drastic reorganization of domains in the host cell nucleus. However, the mechanisms involved and how these domains contribute to infection are not well understood. Our recent studies defining the CMV-induced nuclear proteome identified several viral proteins of unknown functions, including a protein encoded by the UL31 gene. We set out to define the role of UL31 in CMV replication. UL31 is predicted to encode a 74-kDa protein, referred to as pUL31, containing a bipartite nuclear localization signal, an intrinsically disordered region overlapping arginine-rich motifs, and a C-terminal dUTPase-like structure. We observed that pUL31 is expressed with true late kinetics and is localized to nucleolin-containing nuclear domains. However, pUL31 is excluded from the viral nuclear replication center. Nucleolin is a marker of nucleoli, which are membrane-less regions involved in regulating ribosome biosynthesis and cellular stress responses. Other CMV proteins associate with nucleoli, and we demonstrate that pUL31 specifically interacts with the viral protein, pUL76. Coexpression of both proteins altered pUL31 localization and nucleolar organization. During infection, pUL31 colocalizes with nucleolin but not the transcriptional activator, UBF. In the absence of pUL31, CMV fails to reorganize nucleolin and UBF and exhibits a replication defect at a low multiplicity of infection. Finally, we observed that pUL31 is necessary and sufficient to reduce pre-rRNA levels, and this was dependent on the dUTPase-like motif in pUL31. Our studies demonstrate that CMV pUL31 functions in regulating nucleolar biology and contributes to the reorganization of nucleoli during infection.
IMPORTANCE Nucleolar biology is important during CMV infection with the nucleolar protein, with nucleolin playing a role in maintaining the architecture of the viral nuclear replication center. However, the extent of CMV-mediated regulation of nucleolar biology is not well established. Proteins within nucleoli regulate ribosome biosynthesis and p53-dependent cellular stress responses that are capable of inducing cell cycle arrest and/or apoptosis, and they are proposed targets for cancer therapies. This study establishes that CMV protein pUL31 is necessary and sufficient to regulate nucleolar biology involving the reorganization of nucleolar proteins. Understanding these processes will help define approaches to stimulate cellular intrinsic stress responses that are capable of inhibiting CMV infection.
KEYWORDS: cytomegalovirus, nucleolus, rRNA expression, dUTPase, nucleolin, pUL31
INTRODUCTION
Human cytomegalovirus (CMV) is a member of the betaherpesvirus family, which also includes human herpesviruses 6A, 6B, and 7 (reviewed in reference 1). While often asymptomatic in healthy adults, CMV infection can cause severe disease in immunocompromised patients and is a leading infectious cause of birth defects. Anti-CMV pharmaceuticals, such as ganciclovir (GCV), have significantly improved the management of CMV-associated diseases. However, the use of these medications is limited due to toxicity and the emergence of drug-resistant strains (2). Human CMV consists of an approximately 235-kbp double-stranded DNA genome that is contained within an icosadeltahedral nucleocapsid and surrounded by an envelope. The genome encodes approximately 200 open reading frames (ORFs), with the potential to express over 750 viral proteins that are separated into different kinetic classes of expression (1, 3–8). The functions of many of these proteins remain unknown.
The viral replication cycle of CMV leads to drastic reorganization of the nucleus in infected cells (reviewed in reference 9). This is evident based on the profound enlargement of the nucleus, which doubles in size by 72 h postinfection (hpi), and its deformation late during infection (10, 11). The changes are partially the result of an accumulation of nuclear viral proteins exhibiting a wide range of activities. Our recent studies defining changes in the nuclear proteome identified 36 viral proteins associated with the nucleus by 24 hpi (10), and this includes proteins involved in genome replication (e.g., pUL54 DNA polymerase, pUL44 processivity factor, and pUL102 helicase-primase subunit), nucleotide metabolism (e.g., pUL45 RNase reductase, pUL114 uracil-DNA glycosylase, and pUL98 DNase), transcriptional regulation (e.g., TIP60 regulator pUL27, NuRD complex regulator pUL29, and STAT3 regulator pUL123), and capsid assembly (pUL46 capsid triplex subunit, pUL80 capsid maturation protease, and pUL86 major capsid protein) (1, 12–18). Several viral proteins were identified which lack known functions (e.g., US23, US24, UL25, and UL31) (10). In addition to altered protein content, infection also changes host cell nuclear architecture. The nucleus contains diverse membrane-less subdomains that compartmentalize cellular processes. Many of the changes are associated with the formation of a viral nuclear replication compartment. This process results in partitioning of the host genome (19, 20) and rearranging a large subset of cellular proteins (9). For instance, components of the cellular proteasome and the nucleolar marker, nucleolin, are relocalized to the periphery of the nuclear replication compartment (21, 22). Nucleolin is required for targeting pUL84 and the viral processivity factor, pUL44, to this compartment for efficient viral genome synthesis (23, 24). The nuclear architecture is also impacted by the creation of subnuclear domains that impede viral replication. These structures include promyelocytic leukemia protein nuclear bodies (PML-NBs). After entry into the nucleus, most CMV genomes localize with PML-NBs, which repress viral transcription (reviewed in reference 25). However, CMV proteins IE1 and pp71 disrupt PML-NBs, promoting viral replication. These examples support the conclusion that CMV-mediated rearrangement of functional nuclear domains is crucial for viral replication.
Prominent subdomains in the nucleus are nucleoli (reviewed in reference 26). Nucleoli are membrane-less regions of concentrated ribosomal DNA (rDNA) that function in ribosome biogenesis. In uninfected cells, nucleoli are made up of three subnucleolar compartments, known as the fibrillar center (FC), dense fibrillar component (DFC), and granular component (GC), with each functioning in a distinct phase of ribosome biogenesis. Transcription of rDNA into pre-rRNA occurs at the border of FC and DFC. The FC contains RNA polymerase I and transcription factors, such as UBF (upstream binding factor), while the multifunctional protein, nucleolin, is observed in both FC and DFC regions. The early processing steps of pre-rRNA occur in the DFC, and the final stages of ribosome rRNA-protein assembly occur in the GC. The structure of nucleoli is characterized by fluid organization that is maintained by multivalent protein-protein and protein-nucleic acid interactions (27, 28). Proteins with intrinsically disordered regions (IDRs) often mediate these interactions. For example, purified nucleolar proteins fibrillarin and nucleophosmin, both of which contain IDRs, can phase separate into distinct subnucleolar-like domains in vitro (27). Nucleoli also play essential roles in sensing and responding to cellular stresses. Stresses, such as viral infection and DNA damage, lead to changes in the organization and protein composition of nucleoli. This results in stress-induced activation of p53-dependent and -independent signaling pathways due to changes in nucleolar protein-protein interactions (26). Recently, chemical induction of this pathway was shown to be antiviral against both mouse and human CMV (29). However, the impact of CMV infection on nucleolar biology remains unclear.
In previous studies, we detected multiple peptides matching to the predicted ORF of UL31 (10). UL31 is in all published genomes of CMV, contains a conserved dUTPase-like motif, and is detected in ribosomal profile studies as CMV ORFL87W (3, 30). We set out determine the contribution of the protein pUL31 to CMV replication. We observed that pUL31 is expressed with true late kinetics and accumulates in nucleolar-like domains at late times during infection. Further, we demonstrate that pUL31 is both necessary and sufficient to regulate pre-rRNA levels and nucleolar organization, contributing to efficient CMV replication.
RESULTS
Characterization of CMV pUL31 protein expression.
CMV expresses a diverse repertoire of proteins involved in regulating cellular processes, with many of these processes occurring within the nucleus. Our previous studies defined CMV-mediated changes in the nuclear proteome and uncovered several viral nuclear proteins of unknown functions (10). This included three peptides that matched to a protein expressed from the predicted UL31 ORF. CMV UL31 is positioned antisense to the neighboring UL30 (early expression kinetics) and UL32 (true late expression kinetics) genes (Fig. 1A) (7, 8). Recent studies demonstrated that mRNAs containing UL32 use a polyadenylation signal between UL30 and UL31 with small noncoding RNAs expressed from UL31 (31). The UL31 gene is conserved in published genomes of CMV, including clinical and laboratory-adapted strains (4). Further, ribosomal profiling studies by Stern-Ginossar et al. (3) defined the UL31 ORF (ORFL87W) in the CMV Merlin strain starting at an ATG upstream of most annotated genomes. CMV UL31 codes for a 671-amino-acid protein, pUL31, with a predicted molecular mass of 74 kDa.
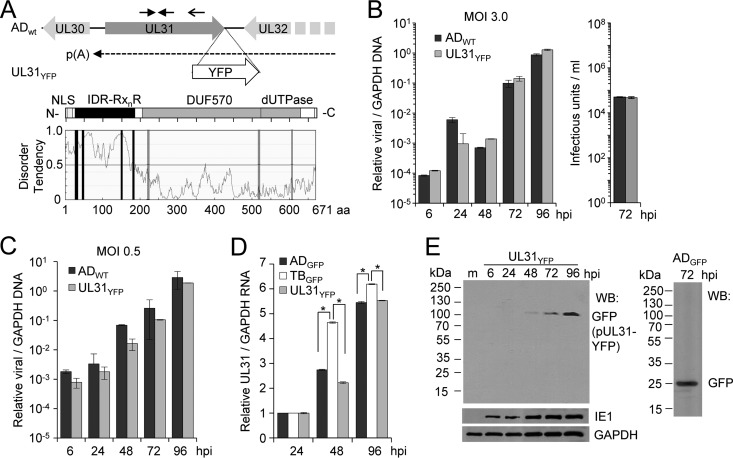
FIG 1.
Expression of CMV pUL31 during infection. (A) The UL30 and UL32 ORFs are positioned antisense to the UL31 ORF, with the UL32 mRNA using a polyadenylation signal (pA) between UL30 and UL31. A recombinant virus, UL31YFP, contains the yellow fluorescent protein (YFP) gene in frame with the UL31 ORF at the 3′ end in the CMV AD169 background. Solid arrows indicate locations of primers for gene-specific (primer; open arrow) quantitative RT-PCR analysis. The UL31 ORF is predicted to express a 671-amino-acid protein with an N-terminal bipartite nuclear localization signal (NLS), multiple arginine-rich R-motifs (RxnR; where x is any amino acid and n is ≤2) overlapping an intrinsic disordered region (IDR), and a domain of unknown function (DUF570) in beta-herpesviruses containing a dUTPase-like motif. (B) Fibroblasts were infected with ADWT or UL31YFP virus at an MOI of 3. Relative viral DNA levels were determined at the indicated times using quantitative PCR and primers against UL123 and GAPDH. Viral titers at 72 hpi were determined using a TCID50 assay following infections using ADWT or UL31YFP virus. Data are from three biological and two technical replicates, with error bars representing standard deviations from the means. (C) Viral DNA levels were determined as described for panel B using an MOI of 0.05. (D) MRC-5 fibroblasts were infected with AD169 (ADGFP), TB40/E (TBGFP), or UL31YFP virus at an MOI of 3.0. Total RNA was isolated at the indicated times, and UL31 and GAPDH levels were determined by quantitative RT-PCR using gene-specific primers. Data are from three biological and two technical replicates, with error bars representing standard deviations from the means. To calculate statistical significance, an ANOVA was used to compare the three conditions. An asterisk indicates a significant P value of less than 0.05. (E) Fibroblasts were mock infected (m) or infected with UL31YFP virus at an MOI of 3. Whole-cell lysates were collected at multiple times postinfection and analyzed by Western blotting using antibodies against the indicated proteins. As a control, ADGFP was analyzed at 72 hpi.
To begin investigating pUL31, we evaluated the primary amino acid sequence with the results summarized in Table 1 and Fig. 1A. CMV pUL31 contains a predicted amino-terminal bipartite nuclear localization signal (NLS), (K/R)(K/R)x10–12(K/R)3/5, where x represents any amino acid (32). This type of NLS uses the classical nuclear import machinery, importin-α and -β (reviewed in reference 40), and exists in the nucleolar protein, nucleolin. At the carboxyl terminus, the protein contains a motif conserved in the U10 proteins of beta-herpesviruses HHV6A, -6B, and -7 (DUF570; domain of unknown function), which exhibit structural features similar to those of monomeric dUTPases (30, 36). dUTPases are nucleotidohydrolases that hydrolyze dUTP to dUMP, and several CMV proteins have dUTPase-like features, including pUL72, pp71, pp65, and pUL84, yet do not exhibit dUTPase activity (30). Finally, the amino terminus of pUL31 is predicted to be an intrinsically disordered region (IDR) and contains several arginine-rich motifs (R-motifs, for RxnR, where x is any amino acid at an n value of ≤2) (41). These features have been observed in cellular proteins associated with nucleoli and are proposed to participate in the liquid-like phase separation that defines nucleolar compartments (27).
TABLE 1.
Distinct features identified in the CMV pUL31 primary amino acid sequence
| Motif | Amino acids | Predicted function | Search algorithm | Reference(s) |
|---|---|---|---|---|
| Bipartite NLSa | 2–24 | Classical nuclear import pathway | cNLS Mapper | 32, 33 |
| IDR | 1–182 | Conformational flexibility, phase separation with R-motif | DisEMBL, IUPred | 27, 34–35, 79 |
| R-motifb | 26–33, 44–47, 148–152, 181–186, 219–222, 518–521, 608–610 | Nucleolar localization, phase separation with IDR | 27, 41 | |
| DUF570 | 206–626 | Conserved in HHV-6A, -6B, and -7 U10 | Conserved Domain Database | 36 |
| Monomeric dUTPase-like fold | 521–669 | Nucleotide metabolism, PAMPc | Robetta, Phyre2 | 30, 37, 38, 39 |
(K/R)(K/R)X10–12(K/R)3/5 where X is any amino acid.
(RXn1R) where X is any amino acid and n1 is ≤2.
PAMP, pathogen-associated molecular patterns.
To examine CMV pUL31 during infection, we constructed a recombinant virus containing the YFP (yellow fluorescence protein) gene in frame with UL31 at the carboxy terminus in an AD169 background, and we refer to this bacterial artificial chromosome (BAC)-derived virus as the UL31YFP virus (Fig. 1A). Because the neighboring genes are important for CMV replication (42), we evaluated the replication kinetics of UL31YFP virus compared to those of the BAC-derived AD169 virus (ADWT). We infected MRC-5 fibroblasts at a multiplicity of infection (MOI) of 3 or 0.05 infectious units per cell (IU/cell) and quantified changes in viral DNA levels normalized to the cellular gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH). We observed similar increases in viral genomes between the two viruses as well as similar viral titers (Fig. 1B and C). These data demonstrate that introduction of the YFP tag does not disrupt CMV replication and likely preserves wild-type levels of UL30 and UL32 gene expression. To begin evaluating UL31, we quantified UL31 RNA levels during infection using the UL31YFP virus along with AD169 encoding green fluorescent protein (ADGFP) and BAC-derived TB40/E (TBGFP) viruses. We infected fibroblasts as described above and isolated total RNA from infected cells at multiple time points. Because the UL32 mRNA contains the antisense UL31 sequence, we used gene-specific primers for UL31 as well as GAPDH to synthesize cDNA (Fig. 1A). We observed increasing levels of UL31 RNA after 24 hpi, with slightly higher levels detected during TB40GFP infection compared to both ADGFP and UL31YFP infection (Fig. 1D). Finally, the UL31 gene is predicted to express a 74-kDa protein. We tested for pUL31 protein expression following infection of fibroblasts using the UL31YFP virus and Western blot (WB) analysis using an antibody against GFP cross-reacting with YFP. We detected a single band at approximately 100 kDa in size starting at 48 hpi, with increasing steady-state levels out to 96 hpi (Fig. 1E). This signal is consistent with pUL31 (74 kDa) containing the YFP tag (26 kDa) and follows the kinetics of UL31 RNA expression (Fig. 1D). As a control, we infected fibroblasts with a recombinant strain, ADGFP, expressing free GFP, which shows an approximately 26-kDa protein that is not seen upon UL31YFP infection (Fig. 1E). Our data demonstrate that the CMV UL31 gene, corresponding to ORFL87W (3), encodes an approximately 74-kDa protein at late times during infection.
CMV pUL31 is a nucleolar protein expressed with true late kinetics.
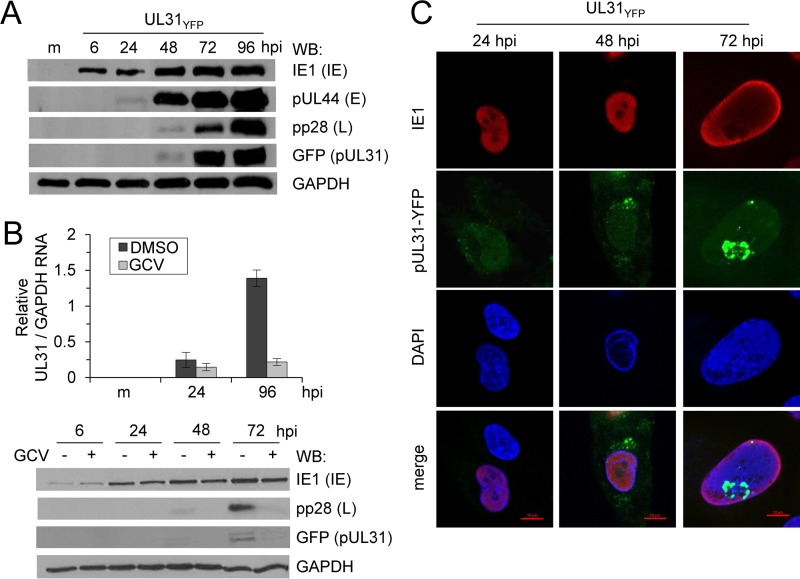
To determine pUL31 protein expression kinetics, fibroblasts were mock infected or infected with the UL31YFP virus, and cell lysates were collected between 6 and 96 hpi. We analyzed expression of three kinetic classes of CMV proteins using Western blotting. This included CMV IE1, pUL44, and pp28, representing proteins with immediate-early, early-late, and true late kinetics, respectively (Fig. 2A). Expression of pUL31 again was detected starting at 48 hpi and increased out to 96 hpi. Under these conditions, pUL31 expression kinetics were similar to those of the true late gene pp28 (Fig. 2A) (8). Expression of true late viral genes is dependent upon viral DNA synthesis. To determine whether pUL31 is a true late viral gene, fibroblasts were infected with UL31YFP virus in the presence or absence of ganciclovir (GCV), an inhibitor of the viral DNA polymerase (Fig. 2B). CMV IE1, whose expression is not dependent upon viral DNA synthesis, was minimally affected by GCV treatment (Fig. 2B). pUL31 protein expression at both 48 and 72 hpi and UL31 RNA expression at 96 hpi were inhibited by GCV, similar to what we observed for pp28 (Fig. 2B). These data demonstrate that pUL31 is expressed as a true late viral gene.
FIG 2.
CMV pUL31 is expressed as a true late gene product. (A) MRC-5 fibroblasts were infected with the UL31YFP virus at an MOI of 3. Whole-cell lysates were collected at multiple times postinfection and analyzed by Western blotting using antibodies against the indicated proteins. (B) Fibroblasts were infected using the UL31YFP virus at an MOI of 3 and treated with 10 μM ganciclovir (GCV) or vehicle control for the duration of the infection. Total RNA was isolated at the indicated times, and UL31 and GAPDH levels were determined by quantitative RT-PCR using gene-specific primers. Whole-cell lysates were collected at the indicated times and analyzed using Western blotting. (C) At 24, 48, and 72 hpi, UL31YFP-infected fibroblasts were fixed, permeabilized, and stained using DAPI and an antibody against CMV IE1.
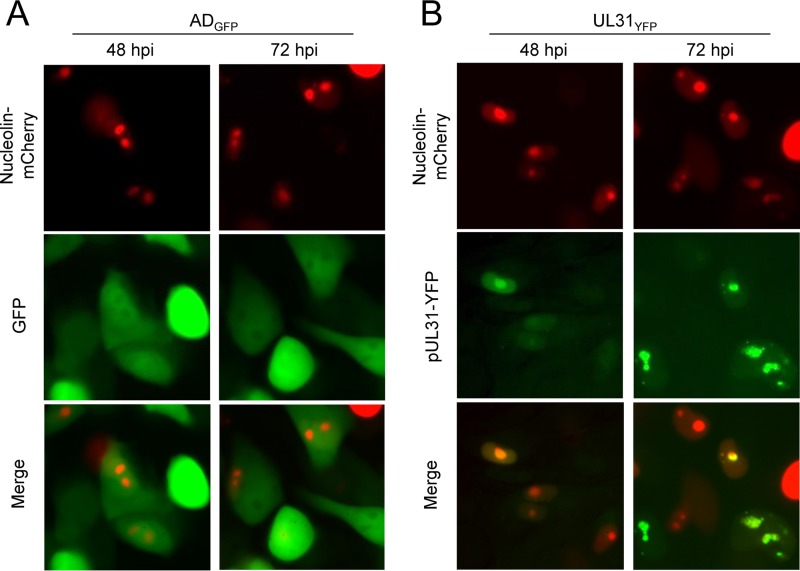
To investigate the subcellular localization of pUL31 during infection, fibroblasts were infected with the UL31YFP virus and fixed at 24, 48, and 72 hpi for analysis using immunofluorescence (IF) and confocal microscopy. Cells were stained for CMV IE1 as a control for infected cells (Fig. 2C). At 48 hpi, pUL31 expression, defined by YFP fluorescence, was localized juxtanuclear and throughout the nucleus of infected cells (Fig. 2C). By 72 hpi, pUL31 was visible in the nucleus and seemed to accumulate inside distinct subnuclear compartments (Fig. 2C). The juxtanuclear staining was not visible at 72 hpi (Fig. 2C). Areas of the nucleus that exhibit a reduced 4′,6-diamidino-2-phenylindole (DAPI) staining include nucleoli. To evaluate if pUL31 localized to nucleoli, we transfected U373 cells with the nucleolar protein nucleolin fused with the mCherry fluorescence tag. We infected nucleolin-mCherry-expressing cells with either ADGFP or UL31YFP virus and evaluated localization using live-cell imaging (Fig. 3). At 48 and 72 hpi for both infections, we observed nucleolin-mCherry fluorescence within subnucleolar domains, consistent with the known localization of nucleolin to nucleoli (Fig. 3A and B). Because CMV reorganizes cellular processes and intracellular domains, it remains to be determined whether nucleoli seen in CMV-infected cells represent nucleoli as defined in uninfected cells. Therefore, we have referred to the area as nucleolar-like domains. ADGFP-infected cells, which express free GFP, exhibited diffuse GFP fluorescence with reduced levels colocalizing with nucleoli-mCherry (Fig. 3A). In contrast, pUL31 colocalized with nucleolin-mCherry (Fig. 3B). Our studies demonstrate that the localization of pUL31 within infected cells is dynamic, being juxtanuclear initially upon expression, accumulating in the nucleus, and ending within nucleolin-containing domains at late times.
FIG 3.
pUL31-YFP localizes with the nucleolar marker nucleolin-mCherry in live cells during infection. U373 cells were transfected with a plasmid expressing nucleolin fused to the mCherry fluorescent protein. At 24 h posttransfection, cells were infected with either ADGFP (A) or UL31YFP (B) virus at an MOI of 3 and analyzed at 48 and 72 hpi using live cell imaging.
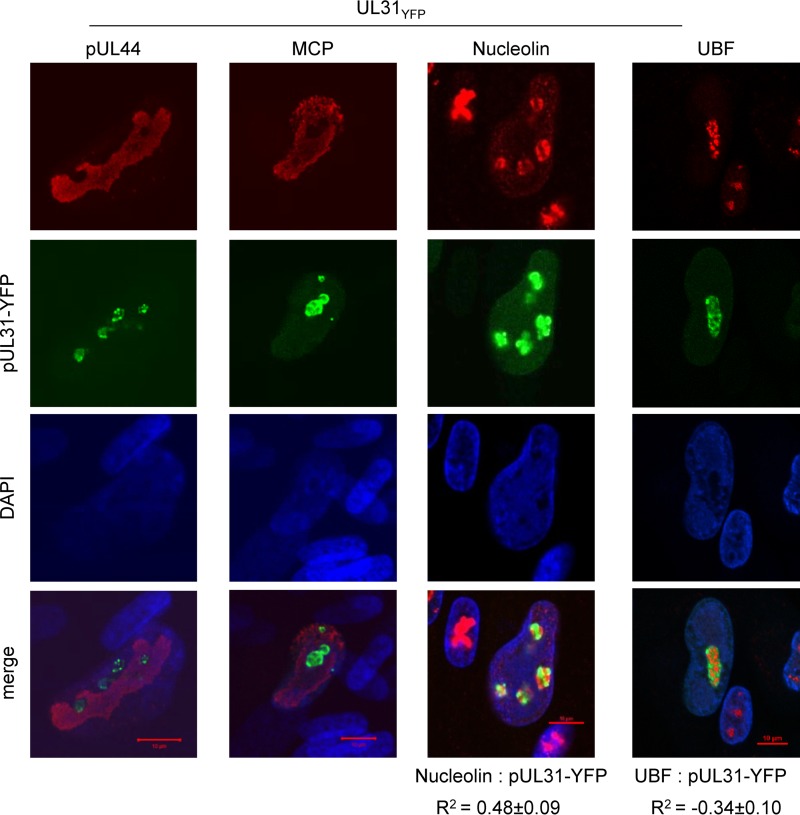
To more precisely define localization, we fixed and stained infected fibroblasts at 72 hpi using antibodies to CMV pUL44 and major capsid protein (MCP) as well as nucleolar markers, nucleolin, and the Pol I transcription factor, UBF (upstream binding factor). Both pUL44 and MCP are known to localize to the viral nuclear replication compartment during infection. Using confocal microscopy, we observed pUL44 and MCP within replication compartments (Fig. 4). In contrast, the majority of signal for pUL31 was excluded from this compartment and was concentrated in domains consistent with nucleolar localization (Fig. 4). We observed that pUL31 localized within domains containing both nucleolin and UBF. Nucleolin is a nucleolar marker that accumulates in both fibrillar center (FC) and dense fibrillar compartment (DFC), while UBF accumulates in the FC (26). We evaluated the localization of these factors with pUL31 using Pearson's correlation coefficient (R2) as a statistic for quantifying colocalization. We determined the average R2 value for pUL31 (YFP) and nucleolin to be 0.48 ± 0.09 (Fig. 4). This suggests that pUL31 and nucleolin exhibit a moderate degree of colocalization (43). In contrast, the R2 value defined by UBF for pUL31 (YFP) and UBF is −0.34 ± 0.10, suggesting the absence of colocalization (Fig. 4). These data demonstrate that pUL31 is expressed within nucleolus-like domains by 72 hpi while being excluded from the nuclear replication compartment. Within nucleolus-like domains, pUL31 overlaps nucleolin but not the transcription factor UBF.
FIG 4.
CMV pUL31 is excluded from the viral nuclear replication compartment and differentially localized to nucleolar markers. Fibroblasts were infected using the UL31YFP virus, fixed and permeabilized at 72 hpi, and stained using the indicated antibodies. Colocalization of proteins was calculated by defining a region of interest determined by DAPI staining and determining the average Pearson correlation coefficient (R2) between the indicated signals from 15 to 20 cells.
A UL31-deficient virus exhibits a replication defect at low multiplicities.
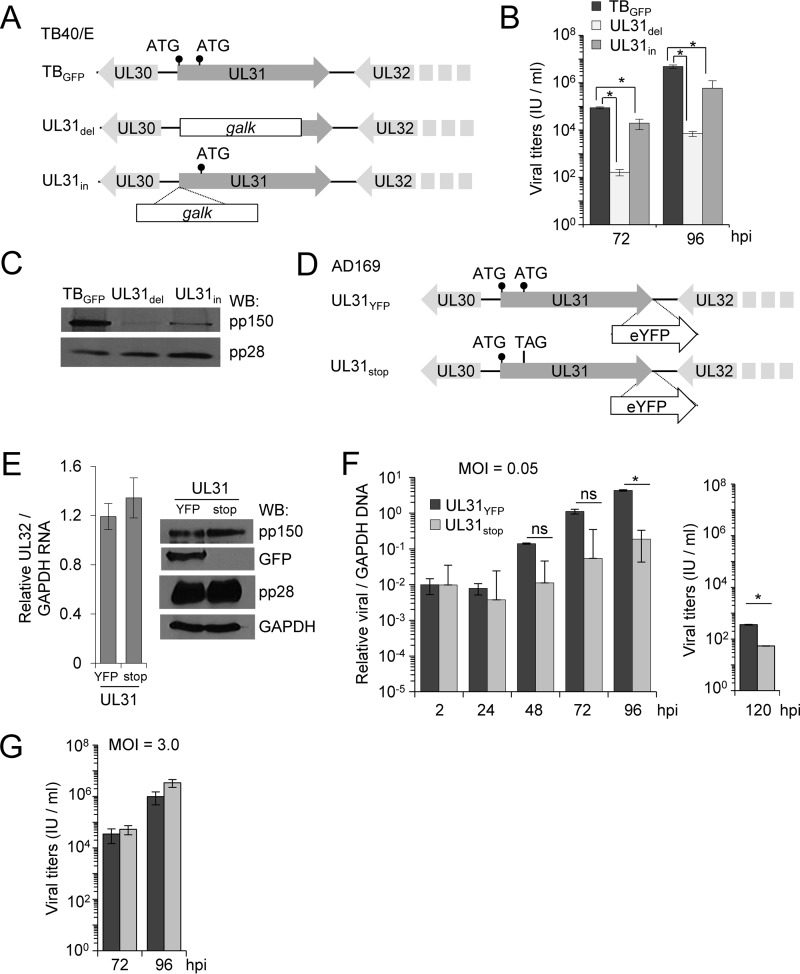
To study the impact of pUL31 on CMV replication, we generated UL31-deficient mutants in the TB40/E strain of CMV using BAC recombineering. For one mutant, we replaced the first approximately 1,200 bp of the UL31 gene with the bacterial galK gene and designated the resulting virus the UL31del virus (Fig. 5A). This mutant is similar to that constructed in CMV Towne UL31, which replicates poorly in fibroblasts (44). We also generated the UL31in virus, where the galK sequence was introduced into the start codon of UL31, replacing the start codon with galK while retaining the remaining UL31 sequence (Fig. 5A). These mutations were confirmed by sequence analysis and restriction enzyme digestion of the BAC DNA (data not shown). To investigate the impact of pUL31 on viral replication, we infected fibroblasts with BAC-derived TBGFP, UL31del, or UL31in at an MOI of 3 and measured viral release at 72 and 96 hpi. We quantified significant growth defects for both the UL31del and UL31in mutant viruses compared to the parental control (Fig. 5B). These data are consistent with observations made using the CMV Towne strain (44) but not those made using the AD169 strain (42). A transposon insertion in UL31 of AD169 exhibited wild-type-like plaque sizes, which prompted the authors to define UL31 as a nonessential gene (42). To investigate these differences, we evaluated expression of the neighboring essential UL32 gene product pp150 and a second late gene product, pp28, by Western blotting (31, 42). Infection with either UL31del or UL31in resulted in a decrease in pp150 expression compared to that of TBGFP, with a larger decrease seen during UL31del infection (Fig. 5C). Expression of the late viral protein pp28 remained unchanged. These data suggest that the defects in viral replication for UL31del and UL31in are the result of having disrupted pp150 expression, which may explain the differences between previous studies (42, 44). As an aside, our observations confirm the importance of the UL31 region to the expression of the neighboring UL32 gene.
FIG 5.
Recombinant CMV mutant lacking expression of pUL31-YFP exhibits an MOI-dependent replication defect. (A) UL31 was disrupted in CMV TB40/E (TBwt) by replacing 1,200 bp of the UL31 ORF with the galK gene (UL31del) or insertion of galK into the first ATG of UL31 (UL31in). The locations of the first two ATG sequences are indicated. (B) MRC-5 fibroblasts were infected using TBGFP, UL31del, or UL31in virus at an MOI of 3. Viral titers from culture supernatants at 72 and 96 hpi were quantified using a TCID50 assay. Data are from three biological replicates and two technical replicates, with error bars representing standard deviations from the means. (C) Whole-cell lysates were collected at 96 hpi and analyzed using Western blotting and the indicated antibodies. (D) A stop codon, TAG, was introduced into the second ATG of AD169 UL31YFP, resulting in UL31stop virus. (E) Fibroblasts were infected with UL31YFP or UL31stop virus at an MOI of 3. Total RNA was collected at 96 hpi and analyzed using qRT-PCR with primers to UL32 (pp150) and GAPDH. Data include three biological replicates and two technical replicates, with error bars representing standard deviations from the means. Whole-cell lysates were analyzed by Western blotting. (F) Fibroblasts were infected with UL31YFP or UL31stop virus at an MOI of 0.05 IU per cell. Whole-cell DNA was isolated at the indicated times, and relative viral genomes were determined by qPCR using primers for UL123 and GAPDH. Viral titers were determined from cell-free virus at 120 hpi. (G) Titers were also determined following infection at an MOI of 3. Data include three biological replicates and two technical replicates, with error bars representing standard deviations from the means.
To avoid disrupting pp150 expression, we constructed a third mutant virus, UL31stop, by introducing a TAG amber stop codon into the UL31 ORF (Fig. 5D). Since no antibody exists for UL31, the mutant was generated in the background of the UL31YFP virus so we could monitor the loss of YFP expression. To confirm the loss of pUL31-YFP, we infected fibroblasts with BAC-derived UL31YFP or UL31stop viruses at an MOI of 3 (Fig. 5E). Cell lysates were collected at 96 hpi and processed for quantitative reverse transcription-PCR (qRT-PCR) and Western blot analyses. We observed equivalent levels of UL32 RNA and protein expression between viruses (Fig. 5E). However, we failed to detect YFP expression during infection using the UL31stop virus, indicating that pUL31 is no longer expressed (Fig. 5E). We next evaluated the impact of pUL31 on viral replication. We infected fibroblasts using UL31YFP or UL31stop virus at an MOI of 0.05 or 3 IU/cell and then quantified changes in replication. At 120 hpi under low-MOI conditions, we quantified a 6-fold decrease in UL31stop viral genomes and an accompanying drop in the viral titers compared to the parental virus (Fig. 5F). Under high-MOI conditions, UL31stop virus replicated similarly to UL31YFP virus (Fig. 5G). Together, our data indicate that a UL31-deficient virus, exhibiting wild-type levels of pp150 expression, has a minor multiplicity-dependent replication defect in cultured cells.
Alteration of pre-rRNA levels occurs during pUL31-deficient CMV infection.
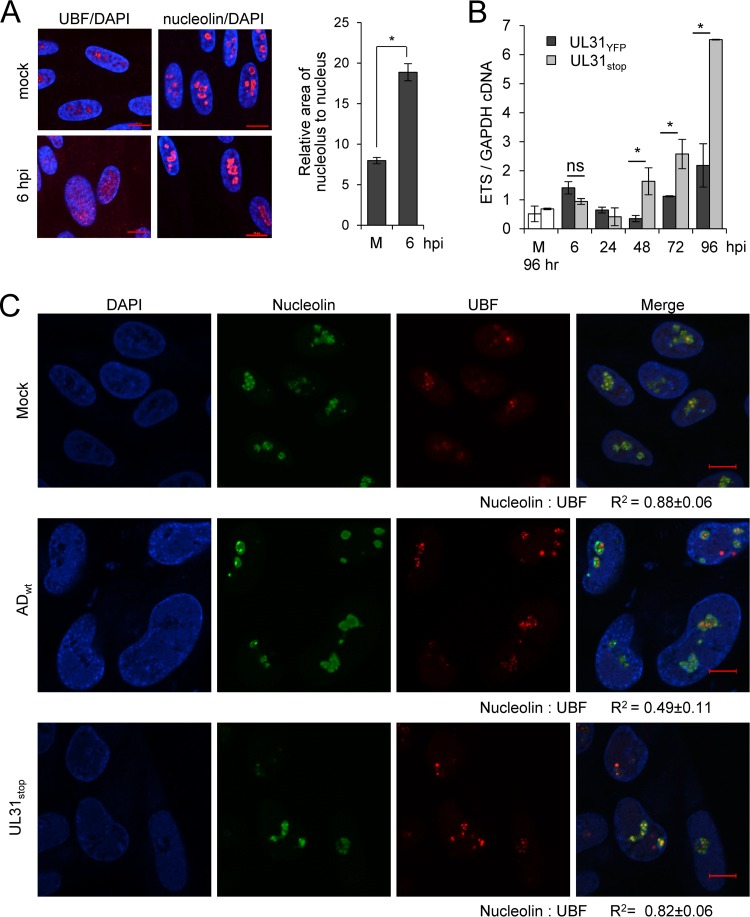
One well-defined function of the nucleoli is ribosomal biogenesis (26). rRNA is transcribed by RNA polymerase I and involves the transcription factor UBF as a 45S pre-rRNA containing internal and external transcribed spacers (ITS/ETS). This 45S pre-rRNA is processed into 28S, 18S, and 5.8S fragments while removing ITS/ETS. To determine whether CMV alters nucleoli and transcription of rDNA into pre-rRNA, we infected fibroblasts at an MOI of 3 and evaluated changes in nucleolar morphology and pre-rRNA levels. As early as 6 hpi, we detected changes in nucleolar organization by immunofluorescence analysis using antibodies to UBF and nucleolin (Fig. 6A). CMV infection resulted in relocalization of UBF from within nucleolar-like domains to diffuse nuclear staining. Further, the size of domains containing nucleolin was increased in infected cells (Fig. 6A). To measure changes in transcription of rDNA, we quantified pre-rRNA levels using quantitative RT-PCR and primers for the 5′ ETS region. We detected an average 2.8-fold transient increase in pre-rRNA at 6 hpi during UL31YFP virus infection compared to levels for uninfected cells (Fig. 6B). A similar change was detected during infection using TB40GFP virus (data not shown). Late during UL31YFP virus infection, we again observed increasing levels of pre-rRNA compared to those for mock infection (Fig. 6B). We completed these studies in parallel using the UL31-deficient UL31stop virus. Surprisingly, starting at 48 hpi, we found a significant increase in pre-rRNA levels during infection with UL31stop compared to the parental UL31YFP virus (Fig. 6B). The steady-state levels of processed 18 and 28S rRNAs remained unchanged between the UL31stop and UL31YFP infections (data not shown), suggesting that the observed change in ETS1 levels was not due to altered pre-rRNA processing. These data demonstrate that infection in the absence of pUL31 results in significantly elevated levels of host cell pre-rRNA.
FIG 6.
CMV pUL31 alters nucleolar organization and function during infection. (A) MRC-5 fibroblasts were mock treated or infected at an MOI of 3, fixed at 6 hpi, and stained using antibodies to UBF (red; left) or nucleolin (red; right) and DAPI (blue). Data represent the average change in area of nucleoli relative to nucleus from mock-infected (m) or infected cells at 6 hpi. Nucleolin and DAPI staining were used to define perimeters of nucleoli and the nucleus, respectively. The relative area of nucleolus to nucleus per cell (in pixels) was determined and averaged from 50 cells. (B) Fibroblasts were infected with UL31YFP or UL31stop virus or mock infected at an MOI of 3, and total RNA was isolated at the indicated times. Mock-infected samples were collected at 96 h. Pre-rRNA levels were determined using qRT-PCR and primers against the 5′ external transcribed spacer (ETS) of rRNA relative to GAPDH. Data are from three biological and two technical replicates, with error bars representing standard deviations from the means. (C) Fibroblasts were mock treated or infected with the parental UL31YFP or UL31stop virus at an MOI of 3. At 96 hpi, samples were fixed and stained with antibodies to nucleolin or UBF. Colocalization was calculated by defining a region of interest determined by nucleolin staining from 15 to 20 cells, and the average Pearson correlation coefficient (R2) between the indicated signals was determined.
Changes in the colocalization of nucleolin with UBF have been associated with altered pre-rRNA expression (45). To test whether pUL31 expression alters nucleolin-UBF colocalization, we mock treated or infected fibroblasts using ADWT or UL31stop virus and evaluated localization of these nucleolar proteins at 96 hpi. We determined the R2 value for colocalization of UBF and nucleolin. Upon ADWT infection, we observed a decrease in colocalization between nucleolin and UBF (R2 = 0.49) compared to both mock-treated and UL31stop-infected cells (R2 = 0.88 and 0.82, respectively) (Fig. 6C). These data suggest that pUL31-mediated changes in nuclear and/or nucleolar organization contribute to regulating pre-rRNA levels. However, future studies are required to define the underlying relationship.
pUL31 interacts with CMV pUL76 with coexpression altering localization.
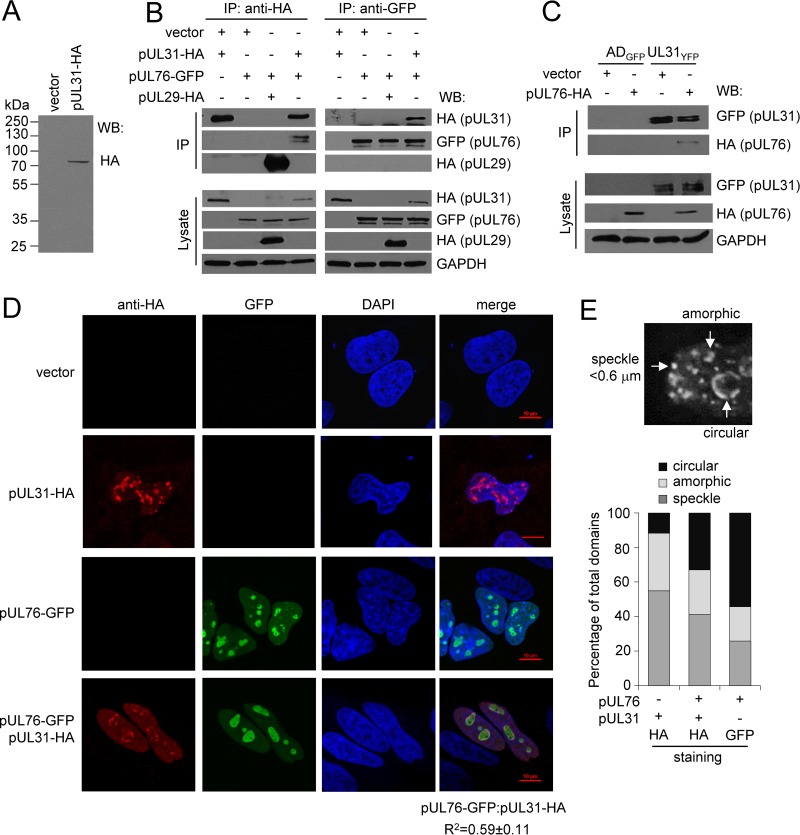
Several CMV proteins have been observed to localize to nucleolar-like domains or intranuclear aggregates, including pUL83, pUL27, pUL29, and pUL76 (13, 46–48). Preliminary studies to identify pUL31 binding partners using mass spectrometry detected peptides to pUL76 (data not shown). CMV pUL76 has been shown to associate with intranuclear aggregates and to alter gene expression involving DNA damage machinery (46, 49–52). Therefore, we investigated the possible association between pUL31 and pUL76. For these studies, we constructed a pUL31 expression vector containing an amino-terminal hemagglutinin (HA) epitope tag. Transfection of the plasmid into U373 resulted in expression of a single protein at the predicted size of pUL31, i.e., 74 kDa (Fig. 7A). To investigate interactions, we transfected U373 cells with pUL31-HA, pUL76-GFP, or empty vector control and performed an immunoprecipitation (IP) experiment using an anti-HA antibody to isolate pUL31-HA (Fig. 7B). As a control, we used CMV pUL29-HA, which also localizes to nucleolus-like domains (46). Protein interactions were analyzed by Western blotting using the indicated antibodies (Fig. 7B). The HA antibody efficiently immunoprecipitated both pUL31-HA and pUL29-HA (Fig. 7B); however, only pUL76-GFP was observed to interact with pUL31 (Fig. 7B). The reciprocal experiment gave the same result; pUL31-HA was found in pUL76-GFP isolated complexes, verifying the interaction (Fig. 7B). We next evaluated this interaction during infection. We transfected U373 cells with pUL76-HA or empty vector control and then infected the cells with ADGFP or UL31YFP virus (Fig. 7C). At 72 hpi, we immunoprecipitated UL31YFP and checked for the presence of pUL76-HA by Western blotting. We observed an interaction between pUL31-YFP and pUL76-HA using the UL31YFP virus but not ADGFP virus (Fig. 7C). These data demonstrate that pUL31 interacts with pUL76 in both the presence and absence of CMV infection.
FIG 7.
CMV pUL31 specifically associates with pUL76, altering pUL31 localization. (A) U373 cells were transfected with pUL31-HA or vector control. At 18 h posttransfection, proteins from whole-cell lysates were analyzed using Western blotting and an anti-HA antibody. (B) U373 cells were transfected with pUL31-HA, pUL76-GFP, or pUL29-HA as a control. At 18 h posttransfection, proteins from whole-cell lysates were immunoprecipitated using anti-HA or anti-GFP antibody and analyzed using Western blotting and antibodies to the indicated epitopes. (C) U373 cells were transfected with pUL76-HA or empty vector control. At 18 h posttransfection, cells were infected with UL31YFP or ADGFP virus at an MOI of 3. Whole-cell lysates were collected at 72 hpi. Proteins were immunoprecipitated with an anti-GFP antibody and analyzed using Western blotting. (D) U373 cells were transfected with pUL31-HA, pUL76-GFP, or both pUL31-HA and pUL76-GFP. Cells were fixed at 18 h posttransfection, stained using DAPI and an anti-HA antibody, and analyzed by confocal microscopy. Colocalization was calculated by defining a region of interest determined by DAPI staining from 15 to 20 cells and determining the average Pearson correlation coefficient (R2) between the indicated signals. (E) Multiple subnuclear domains were observed (circular, speckled, or amorphic), counted in a blinded fashion, and presented as a percentage of all domains observed using the indicated antibody.
We observed that pUL31 localizes to nucleolin-containing domains during infection, and other laboratories observed pUL76 within intranuclear aggregates. To determine if pUL31 and pUL76 colocalize in nucleoli, we analyzed the localization of pUL31 and pUL76 in transfected cells. We transfected U373 cells with empty vector, pUL31-HA, pUL76-GFP, or pUL31-HA and pUL76-GFP together. At 18 h posttransfection, cells were fixed and analyzed by immunofluorescence using an anti-HA antibody and GFP fluorescence. When expressed separately, pUL31-HA and pUL76-GFP localized to subnuclear domains (Fig. 7D). Upon coexpression, we observed a strong degree of colocalization between pUL31-HA and pUL76-GFP with an average R2 value of 0.59 ± 0.11 (Fig. 7D). In addition, expression of pUL76-GFP resulted in altered pUL31 localization compared pUL31 alone (Fig. 7D). We observed pUL31 alone localized to pleomorphic domains as circular, amorphic, and speckled subnuclear domains (Fig. 7E). However, in cells expressing both pUL31 and pUL76, we detected increased circular subnuclear domains containing pUL31-HA and a corresponding decrease in amorphic and speckled structures (Fig. 7E). These data demonstrate that pUL31 localizes to subnuclear domains and that pUL76 alters the localization of pUL31 to distinct subnuclear domains. Further, the data suggest that pUL76, like pUL31, associates with host cell nucleoli.
Expression of pUL31 and pUL76 is sufficient to suppress pre-rRNA levels.
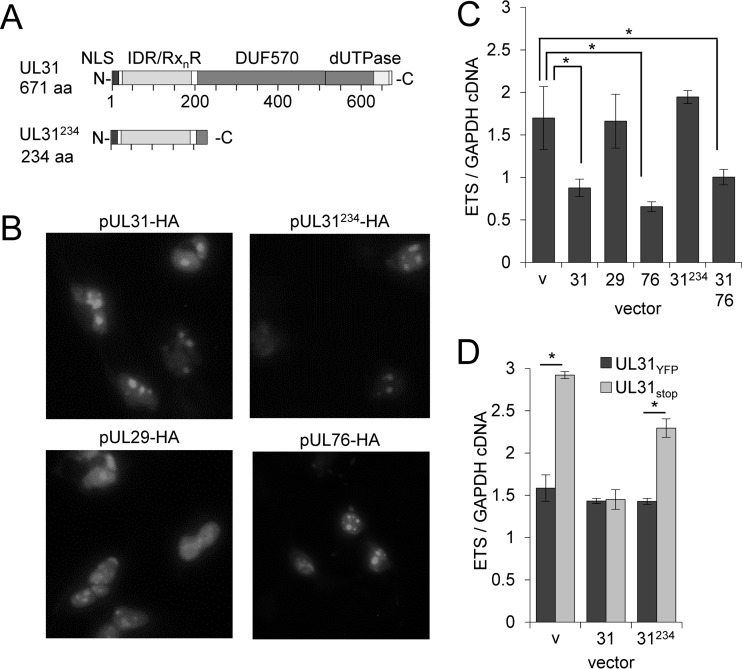
We were interested in determining whether pUL31 could influence pre-rRNA levels in the absence of infection. For these studies, we again transfected U373 cells with empty control or the pUL31-HA expression vector. We included expression vectors for pUL29 and pUL76 as well as a pUL31 carboxy-terminal truncation mutant in order to evaluate any contributions of the conserved dUTPase-like motif (Fig. 8A). Following transfection, each protein localized to nucleolar-like subnuclear structures (Fig. 8B). We determined changes in pre-rRNA levels at 18 h posttransfection using qRT-PCR and primers for the 5′ ETS. We observed an average decrease of 48% in pre-rRNA levels upon pUL31 expression (Fig. 8C). Expression of pUL76 resulted in a 61% decrease in pre-rRNA levels, while no changes were observed for pUL29 or pUL31234 (Fig. 8C). Coexpression of pUL31 with pUL76 resulted in a 41% reduction, similar to the decrease observed with pUL31 alone (Fig. 8C). These data demonstrate that pUL31 or pUL76 expression is sufficient to reduce pre-rRNA levels, and that the pUL31 dUTPase-like motif contributes to this activity.
FIG 8.
Expression of pUL31 is sufficient to reduce rRNA levels. (A) Domains identified within full-length UL31, including nuclear localization sequence (NLS), arginine-rich motifs (RxnR) within an intrinsic disorder region (IDR), and a conserved domain in betaherpesviruses (DUF570) containing a predicted dUTPase. The truncated UL31224 contains only NLS, RxnR, and IDR motifs. (B) U373 cells were transfected with pUL31-HA, pUL29-HA, pUL76-HA, or pUL31234-HA expression vectors. Cells were fixed and permeabilized at 18 h posttransfection and evaluated with immunofluorescence analysis using an anti-HA antibody. (C) U373 cells were transfected with the indicated vectors, including an empty control (v). Pre-rRNA levels relative to those of GAPDH were determined from total RNA isolated at 18 h posttransfection using qRT-PCR. Data are from three biological and two technical replicates, with error bars representing standard deviations from the means. (D) U373 cells were transfected with empty vector or with pUL31-HA or pUL31234-HA expression vector, and at 18 h posttransfection they were infected using UL31YFP or UL31stop virus at an MOI of 3. Total RNA was isolated at 96 hpi, and pre-rRNA levels were determined using qRT-PCR and primers against the 5′ external transcribed spacer (ETS) relative to GAPDH. Data are from three biological and two technical replicates, with error bars representing standard deviations from the means.
We next asked whether transient pUL31 expression could repress pre-rRNA levels during infection with UL31stop virus. To address the question, we transfected U373 cells with empty control, full-length pUL31, or truncated pUL31234 expression vectors (Fig. 8A). After 18 h, transfected cells were infected with either UL31YFP or UL31stop virus, and pre-rRNA levels at 96 hpi were quantified using primers to the 5′ ETS (Fig. 8D). As presented earlier (Fig. 6B), we observed increased pre-RNA levels after infection with the UL31stop virus compared to the level for UL31YFP (Fig. 8D). Similar levels of pre-rRNA were found in UL31YFP-infected cells transfected with pUL31 or a vector control (Fig. 8D), which indicates that pUL31 overexpression did not result in further inhibition. In contrast, pUL31 expression in trans prevented the increase in pre-rRNA levels seen during UL31stop infection (Fig. 8D). Expression of the carboxy-terminal truncation pUL31234 partially reversed the increase in pre-rRNA. Overall, our data demonstrate that pUL31 expression is sufficient to reduce pre-rRNA levels and suggest that the dUTPase-like domain of pUL31 contributes to the activity.
DISCUSSION
During CMV replication, profound changes occur to the host cell nucleus, including increased size, an altered proteome, and reorganized nuclear domains (9, 10, 53, 54). Our recent studies investigating the nuclear proteome identified numerous viral proteins associated with the nucleus at the onset of viral DNA synthesis (10). This included multiple peptides matching to the predicted ORF of UL31, which contains a conserved dUTPase-like motif (Fig. 1A and Table 1) (30). Here, we have investigated pUL31 and its contribution to CMV replication. We have demonstrated that pUL31 is approximately 74 kDa in size (Fig. 1E and 7A), with expression kinetics of a true late gene and member of the Tp5 class of proteins (Fig. 2B) (6, 8). Intriguingly, around the onset of expression, we detected perinuclear localization at 48 hpi, followed by accumulation within the nucleus and eventual accumulation within nucleolar-like subdomains by 72 hpi (Fig. 2C and 3B). Several viral proteins have been observed to associate with nucleoli, including pUL76 (46), and we demonstrate that pUL31 interacts with pUL76 in the presence (Fig. 7C) and absence of infection (Fig. 7B). Expression of pUL31 and pUL76 altered nuclear subdomain organization (Fig. 7D) and disrupted pre-rRNA expression levels (Fig. 8C). However, unlike pUL76, disruption of pUL31 resulted in a minor disruption to CMV replication in cultured cells (Fig. 5F).
Consistent with pUL31 localization, we detected a bipartite nuclear localization motif at the amino terminus (Fig. 1A and Table 1). Proteins containing this signal use the classical nuclear import machinery, importin-α/β, and we have previously demonstrated that CMV induces expression of a subset of importin-α adaptor proteins starting at 24 hpi to support replication (10). Current annotation of the UL31 ORF in published genomes identifies a downstream methionine as its starting codon, which lacks the NLS motif. Our studies indicate that expression during infection (Fig. 1E) as well as upon transfection of an expression vector (Fig. 7A) involves a methionine located 76 amino acids upstream and includes the predicted NLS. Use of the upstream start is also supported by ribosomal profiling studies done during CMV replication (3). As infection progresses, pUL31 eventually accumulates in domains overlapping the nucleolar protein, nucleolin (Fig. 3 and 4). In contrast, pUL31 is excluded from the nuclear replication compartment, defined by the pUL44 DNA processivity factor and pUL86 major capsid protein (Fig. 4). Nucleolin is a common marker for nucleoli, which are fluid membrane-less organelles within the nucleus. Further sequence analysis of pUL31 identified arginine motifs (R-motifs) within an intrinsically disordered region (IDR) (Fig. 1A and Table 1). This combination of features is commonly found in proteins localizing to nucleoli, including nucleolin (27). We observed that expression of a carboxy-terminal truncated version of pUL31 retaining these amino-terminal features remained associated with nucleoli (Fig. 8B). However, we were unable to express truncation mutants lacking the amino terminus (data not shown). Recent biochemical studies indicate that proteins containing similar R-motifs within IDRs can participate in liquid-liquid phase separation, which is hypothesized to regulate the dynamic nature and protein composition of nucleoli (27, 28). Our data demonstrate that pUL31 localizes to nucleolar-like domains late during infection, possessing motifs similar to those found in nucleolar proteins and possibly involved in nucleolus-associated phase separation.
Nucleoli function in ribosome biogenesis and act as a central regulator of cellular stress responses (55). To begin investigating the role of pUL31 in CMV replication, we constructed several UL31-deficient viruses. A previous study disrupting the UL31 ORF in CMV Towne strain demonstrated a replication defect (44). We introduced similar mutations which also resulted in replication defects (Fig. 5B). However, these changes inhibited expression of the neighboring antisense gene, UL32, which encodes the tegument protein, pp150 (Fig. 5C). Upon introducing a stop codon within the UL31 ORF and confirming wild-type levels of pp150 expression, we observed reduced replication kinetics of the UL31-deficient virus at a low MOI but not high MOI compared to the parental UL31YFP virus (Fig. 5F and G). We also evaluated the impact of pUL31 on rDNA transcription by quantifying pre-rRNA levels. As early as 6 hpi we found a transient increase in pre-rRNA levels (Fig. 6B), which coincided with CMV-mediated relocalization of the nucleolar proteins UBF and nucleolin (Fig. 6A). The levels increased again after 48 hpi, and this occurred using both AD169 (Fig. 6B) and TB40/E strains (data not shown). Other laboratories have shown that nucleolin participates in the formation of the CMV nuclear replication compartment after 24 hpi (22), and we postulate that this change contributes to the observed fluctuations in rRNA expression levels. In the absence of pUL31, however, we observed a significant increase in pre-rRNA above that detected in cells infected using the parental virus (Fig. 6B). Conversely, expressing pUL31 in the absence of infection decreased pre-rRNA levels (Fig. 8C) and suppressed the high levels observed using the UL31-deficient virus (Fig. 8D). These studies demonstrate that pUL31 is necessary and sufficient to regulate and suppress high levels of pre-rRNA expression. Determining the functional significance of regulating pre-rRNA to CMV infection and pathogenesis will be the focus of future studies.
Changes in pre-rRNA expression and processing alter nucleolar protein composition, resulting in p53-dependent and p53-independent nucleolar stress responses (reviewed in reference 56). These changes ultimately activate mechanisms that can lead to cell cycle arrest and apoptosis. The importance of regulating nucleolar stress responses is highlighted by recent studies demonstrating that a compound capable of inducing a nucleolar stress response can serve as a potent antiviral agent against CMV replication (29). As a nucleolar protein, it is conceivable that pUL31 controls pre-rRNA levels to protect from or regulate a cell stress response. We postulate that suppression of a nuclear stress response at late times is important during persistent infections and/or a slower replication cycle. pUL31 may regulate the response by altering UBF-nucleolin localization. We observed that colocalization of nucleolin and UBF is reduced upon infection compared to the level for mock-infected cells but not when infected using the UL31-deficient virus (Fig. 6C). In Huntington's disease, nucleolin is sequestered away from UBF and Pol I, resulting in reduced rDNA transcription and induction of a nuclear stress response (45). Similar to pUL31, several proteins that participate in nucleolar stress responses possess R-motifs within intrinsic disordered regions of the protein (27). Our studies suggest a role for the dUTPase-like structure in the observed activity of pUL31. This dUTPase-like domain exists in proteins from most herpesviruses, including CMV proteins pUL31, pUL72, pp65, pp71, and pUL84 (30). However, the CMV proteins lack additional domains required for dUTPase activity. Recently, herpesvirus proteins containing dUTPase-like features have been implicated as pathogen-associated molecular pattern (PAMP) proteins functioning to modulate immune responses (37). In contrast to regulating nucleolar stress responses, it is conceivable that pUL31 has an immunomodulatory role. Both pp65 and pp71 regulate intrinsic immune responses with pp65 binding the double-stranded DNA sensors and IFI16 restricting it to the nucleus and nucleoli (57–64). Perhaps the movement of pUL31 from a juxtanuclear location to nucleoli participates in a similar activity (Fig. 2C). Like pUL31, pp65-deficient viruses exhibit a minor replication defect in culture (57, 65). Intriguingly, we observed that a pUL31 mutant lacking the dUTPase-like domain still localized to nucleoli yet failed to suppress rRNA levels (Fig. 8C). Future studies will explore the role of pUL31 as an immune modulator and its relationship to nucleolar biology.
As noted earlier, several CMV proteins have been observed to associate with nucleoli, including pUL29 and pUL76 (46). The absence of a substantial growth defect for the UL31-deficient virus may be the result of redundant functions. For example, we have previously shown that pUL29 and pUL29/28 suppress p53-mediated gene expression (15, 16). Any role of pUL31 in regulating p53-dependent stress responses may be masked by pUL29/28 expression. We demonstrated that pUL76 interacts with pUL31 in the presence and absence of infection (Fig. 7B and C). While not studied comprehensively, we found that pUL76 expression decreases pre-rRNA levels similar to pUL31 in the absence of infection (Fig. 8C). These data support the notion of a redundancy in function between pUL31 and pUL76. The concept that CMV expresses a protein that decreases pre-rRNA levels in a cell that requires increased protein production to support the viral life cycle seems counterintuitive. An alternative role of pUL31 may be to simply help reorganize membrane-less domains within the nucleus, and the changes that we observed in pre-rRNA expression are an indirect consequence. We observed that pUL31 does not colocalize with UBF, a crucial player in the initiation of RNA polymerase 1-mediated transcription, but moderately colocalizes with nucleolin (Fig. 4). In addition to binding pUL31, expression of pUL76 changed the localization of pUL31 (Fig. 7D). pUL76 expression induces double-strand breaks and aberrant spindle formation in uninfected cells (49–51). Proper spindle formation is important during mitosis and regulated by nucleolar proteins. This further supports the notion that pUL76, along with pUL31, participates in reorganizing domains within the nucleus to support infection.
Our studies demonstrate that CMV pUL31 associates with cellular nucleolar-like domains late during infection and regulates aspects of nucleolar biology. The contribution of these changes to replication remains unclear. However, nucleoli are important regulators of cellular stress responses capable of inducing cell cycle arrest and/or apoptosis, and they are proposed targets for cancer therapies (66). It is our goal to understand how CMV proteins, including pUL31, regulate nuclear and nucleolar biology as a means to uncover unique therapeutic strategies to stimulate the cell's intrinsic ability to suppress infection.
MATERIALS AND METHODS
Biological reagents.
MRC-5 fibroblasts (ATCC) and U373 cells were propagated in Dulbecco's modified Eagle medium (DMEM) (ThermoFisher Scientific) containing 7% fetal bovine serum (FBS) (Atlanta Biologicals) and 1% penicillin-streptomycin (ThermoFisher Scientific). Cells were grown to approximately 70% confluence and serum starved using 0.5% FBS for 24 h to synchronize the population prior to infections and transfections unless otherwise noted.
The CMV strain AD169 (ADWT), AD169 encoding GFP (ADGFP), and strain TB40/E encoding GFP (TBGFP) were produced from the bacterial artificial chromosome (BAC) clones (67–69). The AD169 UL31YFP BAC clone (referred to as UL31YFP) was constructed by introducing the YFP gene at the 3′ end of UL31 using recombineering methods previously described for other CMV genes and primers specific to UL31 (5′-TGGGCGACATGCAATTGCCCGCGGACAACTTTCTCACGTCTCCCCATCCCCCGGAAGAAGATGGAAAAAG-3′ and 5′-AATGAAACCATCGGATAGTGACGTGTCGGGAAAGGAGGACGGACGGAGGGGTCGTGGAATGCCTTCG-3′, with CMV sequence underlined) (Integrated DNA Technologies) (57). For UL31stop BAC (UL31stop), a stop codon, TGA, was introduced into the second predicted start codon, ATG, using the UL31YFP BAC. This was done in order to monitor the loss of pUL31 protein expression by YFP. To construct the BAC clone, the galK gene was amplified by PCR using primers flanking the second ATG codon of UL31 (5′-GGCGACGACACGGCGACGGTCAACAGGGTCACAAGCGTGGGTTTGTCCCCTCAGCACTGTCCTGCTCCTT-3′ and 5′-CGCCGGAGGAAACGGGGACCGGCAACGACGGCGGTGGCGGCGACCAGATTCCTGTTGACAATTAATCATCGGCA-3′, with CMV sequence underlined). The resulting PCR product was electroporated into Escherichia coli SW105 cells containing AD169 UL31YFP BAC clone and grown under chloramphenicol and galactose selection (70). The galK gene was replaced by introducing a double-stranded oligonucleotide containing UL31 sequence with an amber mutation, TAG (5′-CCGGAGGAAACGGGGACCGGCAACGACGGCGGTGGCGGCGACCAGATTTAGTGGGGACAAACCCACGCTTGTGACCCTGTTGACCGTCGCCGTGTCGTCG-3′, with the nucleotide substitution underlined) and chloramphenicol and 2-deoxygalactose counterselection. Additional UL31 mutations were constructed using the TB40/E BAC by replacing the majority of the UL31 gene (UL31del) or disrupting the gene (UL31in) with the insertion of galK using sequence-specific primers (upstream primer 5′-AGTCGGCTACATGCGCCCTGGGTCTGACGCTCCAAAGCGTACGCAGTCTGTCAGCACTGTCCTGCTCCTT-3′ with UL31in, 5′-CCGGGCCCCGCGTTCAAGACGGCGTGCCGTGACGCTCGATGGGTCCGCTTCCTGTTGACAATTAATCATCGGCA-3′, or with UL31del, 5′-GCCTCGTCGTCGTCCATCGTGCCGCGGAACTGCGCGAGGTAGCGCGTAATCCTGTTGACAATTAATCATCGGCA-3′; CMV sequences are underlined). All clones were confirmed by sequencing across the junctions of specific changes and evaluated by restriction enzyme digestion compared to the parental BAC clone. Viral stocks were prepared by transfecting BAC DNA into MRC-5 fibroblasts with a UL82 expression vector (71) using electroporation at 260 mV for 23 ms with 4-mm-gap cuvettes and a Gene Pulser Xcell electroporation system (Bio-Rad Laboratories). Culture medium was then collected and pelleted through a sorbitol cushion (20% sorbitol, 50 mM Tris-HCl, pH 7.2, 1 mM MgCl2) at 55,000 × g for 1 h in a Sorvall WX-90 ultracentrifuge and SureSpin 630 rotor (ThermoFisher Scientific) (72). Titers of viral stocks were determined using a limiting dilution assay (50% tissue culture infectious dose [TCID50]) and MRC-5 cells in 96-well dishes. We determined CMV IE1-positive cells at 2 weeks postinfection and refer to the resulting titers as infectious units (IU) per milliliter.
Infections were completed using a multiplicity of infection (MOI) of 3 IU/cell, unless otherwise noted, with the number of cells counted using a hemocytometer prior to infection. After 2 hpi, cells were washed with phosphate-buffered saline (PBS) and new medium was added. In drug treatment experiments, compounds were added at 0 hpi and replaced every 24 h. Cells were treated with 10 μM ganciclovir (GCV; Sigma-Aldrich) or dimethyl sulfoxide (DMSO) as a vehicle control.
Plasmids were transfected using Fugene 6 reagent (Promega Corporation) by following the manufacturer's protocol. The pCGN-pUL29 and pCGN-pp71 plasmids have been previously described (71, 73). The nucleolin-mCherry plasmid was generously provided by Keiichi Nakayama (Kyushu University) (74), the pUL76-GFP plasmid was generously provided by Shang-Kwei Wang (Kaohsiung Medical University) (48), and the pUL76-HA vector was generously provided by Robert Parkhouse (Instituto Gulbenkian de Ciência) (75). The pUL31 expression plasmid, pcDNA-pUL31HA, was constructed using the pcDNA3 vector. The UL31 gene was amplified by PCR using Platinum Taq DNA polymerase high fidelity (ThermoFisher Scientific) and an upstream primer encoding the HA epitope sequence (5′-TCCTCGAGGCCACCATGTACCCATACGATGTTCCAGATTACGCTAAGCGGACCCATCGAGCGTC-3′, with HA sequence underlined) and a downstream primer (5′-ATATCTAGATCAGGGATGGGGAGACGTGAG-3′) containing XhoI and XbaI restriction enzyme sites. The resulting product was introduced into the equivalent sites in pcDNA3. To construct the pUL31234-HA expression vector, we PCR amplified the first third of UL31 using the template pcDNA-pUL31HA and an upstream primer (5′-TCACGGCACGCCGTCTTGAACGCAGGGCCC-3′) with a downstream primer containing a stop codon and sequences to pcDNA3 (5′-CTCTAGCATTTAGGTGACACTATAGAATAGGGCCCTCAGGCCGTCAGGTGCGAGGC-3′, with UL31 sequence underlined). The product was inserted into pcDNA-pUL31HA using the restriction enzyme ApaI. Expression plasmids were verified using sequence analysis of the inserted gene.
The following antibodies were used in these studies for Western blot (WB), immunoprecipitation (IP), or immunofluorescence (IF) analysis with the amount of antibody or dilution used indicated: mouse anti-GAPDH (clone 0411; WB, 1:10,000; Santa Cruz Biotechnology), mouse anti-HA (clone HA-7; WB, 1:1,000; IP, 5 μg; Sigma-Aldrich), mouse anti-GFP (WB, 1:1,000; IP, 5 μg; IF, 1:100; Santa Cruz Biotechnology), mouse anti-UBF (clone F-9; IF, 1:100; Santa Cruz Biotechnology), mouse anti-nucleolin (clone C-23; IF, 1:100; Santa Cruz Biotechnology), preconjugated anti-nucleolin Alex Fluor 488 (IF, 1:100; Abcam), and mouse anti-pUL44 (clone 10D8; WB, 1:1,000; IF, 1:100; Virusys). The CMV antibodies mouse anti-pUL123 (clone 1B12; WB, 1:1,000; IF, 1:100) and mouse anti-pp28 (clone 10B4; WB, 1:1,000) were generously provided by Tom Shenk (Princeton University) (76, 77). Mouse anti-MCP/UL86 (clone MAb 28-4; IF, 1:5) and mouse anti-pp150 (clone MAb 36-14; WB, 1:500) were generously provided by William Britt (University of Alabama, Birmingham) (78). Goat anti-mouse immunoglobulin (IgG) conjugated with horseradish peroxidase (HRP) secondary antibody was used for Western blot analysis at 1:10,000 (Jackson ImmunoResearch). Donkey anti-mouse and anti-rabbit IgG conjugated with Alexa Fluor 488 and 568 secondary antibodies were used for IF analysis at 1:500 (Thermo Fisher Scientific).
Analysis of protein and nucleic acid.
Steady-state protein levels were measured by Western blotting. Cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40) and lysed by sonication. Protein samples then were resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a Protran nitrocellulose membrane (Sigma-Aldrich) by semidry transfer using a Trans-Blot turbo transfer system (Bio-Rad). The membrane was blocked in 5% milk in PBS-T (PBS, 0.05% Tween 20) for 1 h, incubated in primary antibody diluted in 5% milk in PBS-T for 2 h at room temperature or 4°C overnight, and then incubated in secondary antibody conjugated to HRP in 5% milk for 1 h at room temperature. HRP-conjugated antibodies were detected using enhanced chemiluminescence reagents (ECL) (GE Healthcare) and film. For immunoprecipitation experiments, cells were resuspended in lysis buffer containing cOmplete protease inhibitor cocktail (Roche) and lysed by sonication. Cell lysates were precleared for 30 min at 4°C with Protein G Dynabeads (ThermoFisher Scientific). Protein G Dynabeads were washed in lysis buffer, bound to the antibody for 30 min at room temperature, washed again, and incubated with precleared lysates for 3 h at 4°C with rotation. Protein-bound beads were washed five times in lysis buffer, resuspended in Laemmli sample buffer, and analyzed using Western blotting as described above.
Cellular and viral DNA and RNA contents were determined using quantitative PCR (qPCR) analysis. Cells were collected, resuspended in lysis buffer (400 mM NaCl, 10 mM Tris, pH 8.0, 10 mM EDTA, 0.1 mg/ml proteinase K, 0.2% SDS), and incubated at 37°C overnight. DNA was extracted using phenol-chloroform and precipitated using ethanol. qPCR was completed using primers for CMV UL123 (5′-GCCTTCCCTAAGACCACCAAT-3′ and 5′-ATTTTCTGGGCATAAGCCATAATC-3′) and cellular GAPDH (5′-ACCCACTCCTCCACCTTTGAC-3′ and 5′-CTGTTGCTGTAGCCAAATTCCGT-3′). Quantification was performed using FastStart Universal SYBR green master mix (Roche) and the QuantStudio 6 Flex real-time PCR system (ThermoFisher Scientific). Relative quantities of DNA were determined using an arbitrary standard curve within each experiment for each primer set and normalized to GAPDH levels. For studies involving RNA, we isolated total RNA from cells using TRIzol reagent (ThermoFisher Scientific) by following the manufacturer's instructions. Approximately 2 μg of RNA was treated with DNA-free DNA removal kit (ThermoFisher Scientific) and used to synthesize cDNA with random hexamers and Superscript III reverse transcriptase (ThermoFisher Scientific). Studies on UL31 expression required cDNA synthesis using a gene-specific primer, 5′-TGCCGCATGGTTTCCTCG-3′. qPCR was performed as described above using primers against UL31 (5′-TGTCCATCACCGAGCAGTACAACA-3′ and 5′-TGGTGGAGCAGAGCAGAAAGAAGA-3′), UL32 (5′-CATCTTTACGCCCATCAAGAAAC-3′ and 5′-TCTGACTGCCCGAGGATAA-3′), GAPDH (5′-ACCCACTCCTCCACCTTTGAC-3′ and 5′CTGTTGCTGTAGCCAAATTCCGT-3′), and pre-rRNA external transcribed spacer 1 (5′-GAACGGTGGTGTGTCGTTC-3′ and 5′-GCGTCTCGTCTCGTCTCACT-3′). Relative quantities of cDNA were determined using an arbitrary standard curve within each experiment for each primer set, and viral cDNA was normalized to GAPDH cDNA.
Immunofluorescence studies.
For immunofluorescence, cells were plated in 6-well dishes containing glass coverslips at approximately 60% confluence. After the indicated perturbation, cells were analyzed by either live-cell microscopy using a Nikon Eclipse TS100 inverted microscope or fixed for immunofluorescence analysis using 4% paraformaldehyde for 20 min at room temperature. Fixed cells were permeabilized in 0.1% Triton X-100 for 30 min, blocked using 3% bovine serum albumin (BSA) in PBS-T for 30 min, and then incubated using the indicated primary antibody diluted in 3% BSA in PBS-T for 2 h at room temperature or overnight at 4°C. Samples were washed three times with PBS-T, incubated using the appropriate Alexa-Fluor-conjugated secondary antibody for 1 h at room temperature, and washed. Coverslips were placed on glass slides in Prolong Gold with DAPI antifade reagent (ThermoFisher Scientific). Images were acquired at 60× oil magnification on a Nikon A1 spectral confocal microscope (Nikon). Colocalization was determined by manually defining a region of interest (ROI) based upon either DAPI or nucleolar staining, as indicated in the figure legends, and colocalization coefficients were calculated using NIS Elements analysis software (Nikon).
Statistical analysis.
The data are representative of 3 or more biological replicates. Reported values are given as the means from all replicates ± standard deviations. To calculate statistical significance, the Student t test was used to compare two samples, and analysis of variance (ANOVA) was used when more than two samples were being analyzed. An asterisk indicates a significant P value of less than 0.05.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the NIAID of the NIH under award number R01AI083281 to S.S.T. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Thomas Shenk and William Britt for providing antibodies against CMV proteins and Shang-Kwei Wang, Keiichi Nakayama, and Robert Parkhouse for the pEGFP-UL76, nucleolin-mCherry, and pUL76-HA plasmids, respectively. We thank Carol Williams and Patrick Gonyo for reagents and advice on nucleolar biology, Blake Hill for advice on protein analysis, and Glen Slocum for assistance with microscopy. Finally, we thank members of our department, the BBC, and past laboratory members John Savaryn, Justin Reitsma, Tarin Bigley, and Dominique Carter for their support, assistance, and insightful suggestions.
REFERENCES
- 1.Mocarski E, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Chou S. 2015. Approach to drug-resistant cytomegalovirus in transplant recipients. Curr Opin Infect Dis 28:293–299. doi: 10.1097/QCO.0000000000000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS. 2012. Decoding human cytomegalovirus. Science 338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. 2003. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A 100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang EC, Aicheler R, Murrell I, Wilkinson GW, Lehner PJ, Gygi SP. 2014. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J Gen Virol 85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 8.Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan JS, Bittner A, Frueh K, Jackson MR, Peterson PA, Erlander MG, Ghazal P. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol 73:5757–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strang BL. 2015. Viral and cellular subnuclear structures in human cytomegalovirus-infected cells. J Gen Virol 96:239–253. doi: 10.1099/vir.0.071084-0. [DOI] [PubMed] [Google Scholar]
- 10.Carter DM, Westdorp K, Noon KR, Terhune SS. 2015. Proteomic identification of nuclear processes manipulated by cytomegalovirus early during infection. Proteomics 15:1995–2005. doi: 10.1002/pmic.201400599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchkovich NJ, Maguire TG, Alwine JC. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J Virol 84:7005–7017. doi: 10.1128/JVI.00719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigley TM, Reitsma JM, Terhune SS. 2015. Antagonistic relationship between human cytomegalovirus pUL27 and pUL97 activities during infection. J Virol 89:10230–10246. doi: 10.1128/JVI.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakki M, Drummond C, Houser B, Marousek G, Chou S. 2011. Resistance to maribavir is associated with the exclusion of pUL27 from nucleoli during human cytomegalovirus infection. Antiviral Res 92:313–318. doi: 10.1016/j.antiviral.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reitsma JM, Savaryn JP, Faust K, Sato H, Halligan BD, Terhune SS. 2011. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27-dependent degradation of Tip60 acetyltransferase and cell-cycle arrest. Cell Host Microbe 9:103–114. doi: 10.1016/j.chom.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Savaryn JP, Reitsma JM, Bigley TM, Halligan BD, Qian Z, Yu D, Terhune SS. 2013. Human cytomegalovirus pUL29/28 and pUL38 repression of p53-regulated p21CIP1 and caspase 1 promoters during infection. J Virol 87:2463–2474. doi: 10.1128/JVI.01926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terhune SS, Moorman NJ, Cristea IM, Savaryn JP, Cuevas-Bennett C, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS Pathog 6:e1000965. doi: 10.1371/journal.ppat.1000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reitsma JM, Sato H, Nevels M, Terhune SS, Paulus C. 2013. Human cytomegalovirus IE1 protein disrupts interleukin-6 signaling by sequestering STAT3 in the nucleus. J Virol 87:10763–10776. doi: 10.1128/JVI.01197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reitsma JM, Terhune SS. 2013. Inhibition of cellular STAT3 synergizes with the cytomegalovirus kinase inhibitor maribavir to disrupt infection. Antiviral Res 100:321–327. doi: 10.1016/j.antiviral.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Dowd JM, Zavala AG, Brown CJ, Mori T, Fortunato EA. 2012. HCMV-infected cells maintain efficient nucleotide excision repair of the viral genome while abrogating repair of the host genome. PLoS Pathog 8:e1003038. doi: 10.1371/journal.ppat.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strang BL, Boulant S, Chang L, Knipe DM, Kirchhausen T, Coen DM. 2012. Human cytomegalovirus UL44 concentrates at the periphery of replication compartments, the site of viral DNA synthesis. J Virol 86:2089–2095. doi: 10.1128/JVI.06720-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran K, Mahr JA, Spector DH. 2010. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J Virol 84:3079–3093. doi: 10.1128/JVI.02236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strang BL, Boulant S, Kirchhausen T, Coen DM. 2012. Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. mBio 3:e00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strang BL, Boulant S, Coen DM. 2010. Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication. J Virol 84:1771–1784. doi: 10.1128/JVI.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender BJ, Coen DM, Strang BL. 2014. Dynamic and nucleolin-dependent localization of human cytomegalovirus UL84 to the periphery of viral replication compartments and nucleoli. J Virol 88:11738–11747. doi: 10.1128/JVI.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherer M, Stamminger T. 2016. Emerging role of PML nuclear bodies in innate immune signaling. J Virol 90:5850–5854. doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James A, Wang Y, Raje H, Rosby R, DiMario P. 2014. Nucleolar stress with and without p53. Nucleus (Austin) 5:402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. 2016. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5:e13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay R, Roy S, Venkatadri R, Su YP, Ye W, Barnaeva E, Mathews Griner L, Southall N, Hu X, Wang AQ, Xu X, Dulcey AE, Marugan JJ, Ferrer M, Arav-Boger R. 2016. Efficacy and mechanism of action of low dose emetine against human cytomegalovirus. PLoS Pathog 12:e1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davison AJ, Stow ND. 2005. New genes from old: redeployment of dUTPase by herpesviruses. J Virol 79:12880–12892. doi: 10.1128/JVI.79.20.12880-12892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, Gao S, Wang L, Wang N, Li M, Zheng B, Qi Y, Sun Z, Liu W, Ruan Q. 2013. Analysis and mapping of a 3′ coterminal transcription unit derived from human cytomegalovirus open reading frames UL30-UL32. Virol J 10:65. doi: 10.1186/1743-422X-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robbins J, Dilworth SM, Laskey RA, Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623. doi: 10.1016/0092-8674(91)90245-T. [DOI] [PubMed] [Google Scholar]
- 33.Kosugi S, Hasebe M, Tomita M, Yanagawa H. 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Dosztanyi Z, Csizmok V, Tompa P, Simon I. 2005. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol 347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 36.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams MV, Cox B, Ariza ME. 2016. Herpesviruses dUTPases: a new family of pathogen-associated molecular pattern (PAMP) proteins with implications for human disease. Pathogens 6:E2. doi: 10.3390/pathogens6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. 2009. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77(Suppl 9):S89–S99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto Y, Yamada K, Yoneda Y. 2016. Importin alpha: a key molecule in nuclear transport and non-transport functions. J Biochem 160:69–75. doi: 10.1093/jb/mvw036. [DOI] [PubMed] [Google Scholar]
- 41.Mitrea DM, Grace CR, Buljan M, Yun MK, Pytel NJ, Satumba J, Nourse A, Park CG, Madan Babu M, White SW, Kriwacki RW. 2014. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc Natl Acad Sci U S A 111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinchuk V, Wu Y, Grossenbacher-Zinchuk O. 2013. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci Rep 3:1365. doi: 10.1038/srep01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsoi H, Chan HY. 2013. Expression of expanded CAG transcripts triggers nucleolar stress in Huntington's disease. Cerebellum 12:310–312. doi: 10.1007/s12311-012-0447-6. [DOI] [PubMed] [Google Scholar]
- 46.Salsman J, Zimmerman N, Chen T, Domagala M, Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog 4:e1000100. doi: 10.1371/journal.ppat.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arcangeletti MC, De Conto F, Ferraglia F, Pinardi F, Gatti R, Orlandini G, Calderaro A, Motta F, Medici MC, Martinelli M, Valcavi P, Razin SV, Chezzi C, Dettori G. 2003. Human cytomegalovirus proteins PP65 and IEP72 are targeted to distinct compartments in nuclei and nuclear matrices of infected human embryo fibroblasts. J Cell Biochem 90:1056–1067. doi: 10.1002/jcb.10655. [DOI] [PubMed] [Google Scholar]
- 48.Lin SR, Jiang MJ, Wang HH, Hu CH, Hsu MS, Hsi E, Duh CY, Wang SK. 2013. Human cytomegalovirus UL76 elicits novel aggresome formation via interaction with S5a of the ubiquitin proteasome system. J Virol 87:11562–11578. doi: 10.1128/JVI.01568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa H, Nascimento R, Sinclair J, Parkhouse RM. 2013. Human cytomegalovirus gene UL76 induces IL-8 expression through activation of the DNA damage response. PLoS Pathog 9:e1003609. doi: 10.1371/journal.ppat.1003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siew VK, Duh CY, Wang SK. 2009. Human cytomegalovirus UL76 induces chromosome aberrations. J Biomed Sci 16:107. doi: 10.1186/1423-0127-16-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang SK, Duh CY, Wu CW. 2004. Human cytomegalovirus UL76 encodes a novel virion-associated protein that is able to inhibit viral replication. J Virol 78:9750–9762. doi: 10.1128/JVI.78.18.9750-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang SK, Duh CY, Chang TT. 2000. Cloning and identification of regulatory gene UL76 of human cytomegalovirus. J Gen Virol 81:2407–2416. doi: 10.1099/0022-1317-81-10-2407. [DOI] [PubMed] [Google Scholar]
- 53.Alwine JC. 2012. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog 8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clippinger AJ, Alwine JC. 2012. Dynein mediates the localization and activation of mTOR in normal and human cytomegalovirus-infected cells. Genes Dev 26:2015–2026. doi: 10.1101/gad.196147.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.James A, Wang Y, Raje H, Rosby R, DiMario P. 2014. Nucleolar stress with and without p53. Nucleus (Austin) 5:402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol 84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Browne EP, Shenk T. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A 100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abate DA, Watanabe S, Mocarski ES. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol 78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmolke S, Drescher P, Jahn G, Plachter B. 1995. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J Virol 69:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J Virol 79:15494–15502. doi: 10.1128/JVI.79.24.15494-15502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li T, Diner BA, Chen J, Cristea IM. 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A 109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog 8:e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmolke S, Kern HF, Drescher P, Jahn G, Plachter B. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J Virol 69:5959–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. 2014. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta 1842:802–816. doi: 10.1016/j.bbadis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 67.Yu D, Smith GA, Enquist LW, Shenk T. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol 76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 69.Umashankar M, Petrucelli A, Cicchini L, Caposio P, Kreklywich CN, Rak M, Bughio F, Goldman DC, Hamlin KL, Nelson JA, Fleming WH, Streblow DN, Goodrum F. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog 7:e1002444. doi: 10.1371/journal.ppat.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baldick CJ Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Britt WJ. 1984. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology 135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell DP, Savaryn JP, Moorman NJ, Shenk T, Terhune SS. 2009. Human cytomegalovirus UL28 and UL29 open reading frames encode a spliced mRNA and stimulate accumulation of immediate-early RNAs. J Virol 83:10187–10197. doi: 10.1128/JVI.00396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kita Y, Nishiyama M, Nakayama KI. 2012. Identification of CHD7S as a novel splicing variant of CHD7 with functions similar and antagonistic to those of the full-length CHD7L. Genes Cells 17:536–547. doi: 10.1111/j.1365-2443.2012.01606.x. [DOI] [PubMed] [Google Scholar]
- 75.Nascimento R, Dias JD, Parkhouse RM. 2009. The conserved UL24 family of human alpha, beta and gamma herpesviruses induces cell cycle arrest and inactivation of the cyclinB/cdc2 complex. Arch Virol 154:1143–1149. doi: 10.1007/s00705-009-0420-y. [DOI] [PubMed] [Google Scholar]
- 76.Zhu H, Shen Y, Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol 69:7960–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva MC, Yu QC, Enquist L, Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol 77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/JVI.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dosztanyi Z, Csizmok V, Tompa P, Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]