ABSTRACT
Human bocavirus 1 (HBoV1) is an autonomous parvovirus that infects well-differentiated primary human airway epithelia (HAE) in vitro. In human embryonic kidney HEK293 cells, the transfection of a duplex HBoV1 genome initiates viral DNA replication and produces progeny virions that are infectious in HAE. HBoV1 takes advantage of signaling pathways in the DNA damage response for efficient genome amplification in both well-differentiated (nondividing) HAE and dividing HEK293 cells. On the other hand, adeno-associated virus 2 (AAV2) is a helper-dependent dependoparvovirus, and productive AAV2 replication requires coinfection with a helper virus (e.g., adenovirus or herpesvirus) or treatment with genotoxic agents. Here, we report that HBoV1 is a novel helper virus for AAV2 replication. Coinfection by HBoV1 and AAV2 rescued AAV2 replication in HAE cells. The helper function of HBoV1 for AAV2 is not limited to HAE cells but also includes HEK293 and HeLa cells. Importantly, the helper function of HBoV1 for AAV2 relies on neither HBoV1 replication nor the DNA damage response. Following transfection of HEK293 cells, the minimal requirements for the replication of the AAV2 duplex DNA genome and the production of progeny virions included the HBoV1 NP1 and NS4 proteins and a newly identified viral long noncoding RNA (BocaSR). However, following infection of HEK293 and HeLa cells with AAV2 virions, HBoV1 NS2 (but not NS4), NP1, and BocaSR were required for AAV2 DNA replication and progeny virion formation. These new methods for packaging the AAV2 genome may be useful for generating recombinant AAV-packaging cell lines and the directed evolution of AAV capsids.
IMPORTANCE We first report that an autonomous parvovirus, HBoV1, helps the replication of a dependoparvovirus, AAV2, in differentiated human airway epithelia. We identified the minimal sets of HBoV1 genes required to facilitate the replication of the AAV2 duplex genome and for AAV2 infection. Notably, together with the expression of the NP1 and BocaSR genes, HBoV1 NS2 is required for the productive infection of HEK293 and HeLa cells by AAV2, whereas NS4 is sufficient for viral DNA replication of an AAV2 duplex genome. The identification of HBoV1 as a helper virus for AAV2 replication has implications for the improvement of recombinant AAV production in HEK293 cells and cell types that do not express the adenovirus E1 gene as well as for the rescue of wild-type AAV genomes from tissues during directed evolution in the absence of wild-type adenovirus. A further understanding of the mechanism underlying HBoV1 helper-dependent AAV2 replication may also provide insights into its functions in HBoV1 replication.
KEYWORDS: DNA replication, adeno-associated virus, human bocavirus, parvovirus
INTRODUCTION
Human bocavirus 1 (HBoV1) causes acute respiratory illness in young children, and the main clinical symptoms are bronchiolitis, pneumonia, asthma, and the common cold (1). HBoV1 is an autonomous parvovirus newly identified in 2005, belonging to the genus Bocaparvovirus of the family Parvoviridae (2). It replicates in polarized/well-differentiated human airway epithelia cultured at the air-liquid interface (HAE-ALI) in vitro. The HBoV1 duplex DNA genome can also replicate in human embryonic kidney HEK293 cells following transfection and produces progeny virions, which are infectious in HAE-ALI (3, 4).
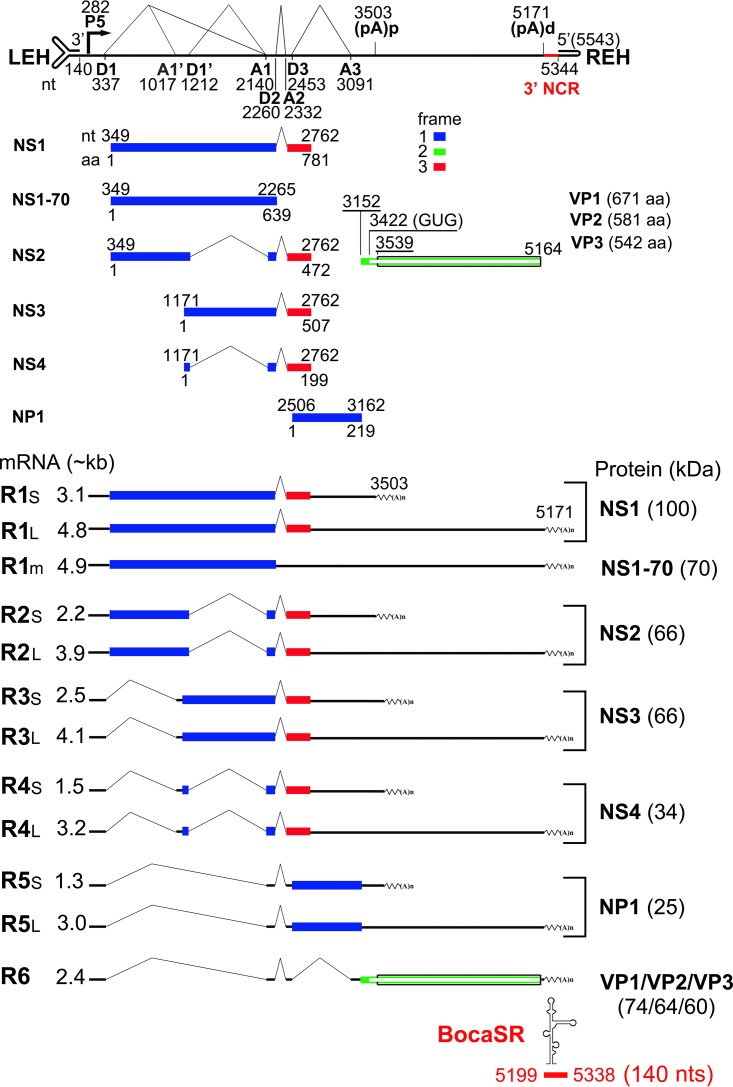
HBoV1 is known to express five nonstructural proteins, namely, NS1, NS2, NS3, NS4, and NP1, by mRNA transcripts generated through alternative splicing and the polyadenylation of a single viral pre-mRNA (Fig. 1) (5). The NS1 to NS4 (NS1-4) proteins are encoded in different regions of the same open reading frame (ORF). NS1 consists of an origin-binding/endonuclease domain (OBD), a helicase domain, and a putative transactivation domain (TAD) in the N terminus, middle, and C terminus, respectively. NS1 binds to the HBoV1 replication origin and presumably nicks single-stranded DNA (ssDNA) of the origin during rolling-hairpin replication (6). NS2 harbors the OBD and TAD, NS3 contains the helicase domain and TAD, and a large portion of NS4 is the TAD. Mutagenesis analysis reveals that NS2, NS3, and NS4 are not essential for HBoV1 replication of the duplex replicative-form (RF) genome in HEK293 cells. However, NS2 is required for HBoV1 infection of HAE-ALI (5). A unique feature of bocaparvoviruses is the presence of a middle ORF in the center of the genome, which encodes NP1 (7, 8). NP1 is a multifunctional protein that is required for efficient virus replication (9). It regulates alternative splicing and polyadenylation of the viral pre-mRNA to control viral capsid protein expression (10, 11). HBoV1 expresses three viral structural (capsid) proteins, termed VP1, VP2, and VP3 (11). Importantly, HBoV1 expresses a novel bocavirus-transcribed small noncoding RNA, termed BocaSR, from the 3′ noncoding region of the viral duplex RF genome (Fig. 1) (12). BocaSR is essential for viral DNA replication both in HEK293 cells and in HAE-ALI. Mechanistically, BocaSR not only regulates NS1, NS2, NS3, and NP1 expression but also colocalizes with viral DNA replication centers, and thus, it may play a direct role in viral DNA replication.
FIG 1.
Genetic map of HBoV1 (1, 5, 8, 11, 12, 60). The HBoV1 ssDNA genome is shown in the negative sense, along with its transcriptional and posttranscriptional units, including the P5 promoter, 5′ splice donor sites (D1, D1′, D2, and D3), 3′ splice acceptor sites (A1, A1′, A2, and A3), the proximal polyadenylation site [(pA)p], the distal polyadenylation site [(pA)d], and the 3′ noncoding region (NCR). The left-end hairpin (LEH) and right-end hairpin (REH) structures of the genome are shown. Six groups of major HBoV1 mRNA transcripts are shown with either long-form mRNA (RXL) that reads through the (pA)p site or short-form mRNA (RXS) that is polyadenylated at the (pA)p site. R1 mRNA has a minor species (R1m) that is unspliced at the central small intron (D3-A3) and encodes a minor protein, NS1-70. Major ORFs are depicted as colored boxes, with the nucleotides (nt) and amino acids (aa) at the start and stop codons indicated. Proteins expressed from each mRNA are indicated beside their respective mRNA transcripts (molecular mass in kilodaltons). A bocaviral noncoding small RNA (BocaSR) is transcribed from the 3′ noncoding region spanning nucleotides 5199 to 5338, as indicated.
Unlike other autonomous parvoviruses, whose replication is cell cycle dependent (13), HBoV1 replicates in mitotically quiescent HAE cells by taking advantage of the DNA damage response (DDR) and DNA repair signaling (14). DNA repair polymerases (Pols) of the Y family, Pol η and Pol κ, are involved in HBoV1 genome amplification. In HEK293 cells, cellular DDR signaling, Pol η, and Pol κ were also involved in HBoV1 DNA replication (15). In addition, we have known that HBoV1 utilizes the replication origin at the right-end hairpin for the terminal resolution of its duplex genome (16), and NS1, NP1, and BocaSR are essential for viral DNA replication in HEK293 cells (5, 12).
Adeno-associated virus 2 (AAV2), on the other hand, is a member of the genus Dependoparvovirus of the Parvoviridae family (2). In the absence of helper virus coinfection, AAV2 establishes a latent infection (17). The AAV2 genome commonly integrates into human chromosomes at a unique locus on human chromosome 19, called AAVS1 (18). Productive AAV2 infection is dependent on helper functions that can be supplied by helper viruses, e.g., adenovirus (Ad), herpesvirus, papillomavirus, and vaccinia virus, or genotoxic agents in mammalian cells (19–21). During lytic infection, AAV2 replicates its genome through a rolling-hairpin-dependent strand displacement mechanism (17). The viral ssDNA genome is packaged into preassembled capsids in the nucleus (22). AAV2 infection has not been associated with any human disease identified so far (23). Recombinant AAV (rAAV) infects both dividing and nondividing cells (24). These unique features make rAAV an ideal vector for human gene delivery.
Ad E1a, E1b55K, E2a, E4orf6, and virus-associated RNA I (VAI RNA) have been identified as the Ad helper genes for AAV2 replication in HEK293 cells, which has allowed the development of the Ad-free rAAV production system (25). E1a gene products trans-activate Ad late promoters and also relieve the repression of the AAV2 P5 promoter (26, 27). E2a is an ssDNA-binding protein and localizes with AAV2 replication centers (28). The E1b55K and E4orf6 proteins collaborate to accelerate AAV2 replication and second-strand synthesis (20, 29, 30). E4orf6 has also been demonstrated to have a role in degrading Mre11, which is important for AAV2 replication (31, 32). VAI RNA promotes the synthesis of AAV2 proteins by inhibiting the protein kinase R (PKR)-eukaryotic translation initiation factor 2α (eIF2α) pathway (33).
As Ad and HBoV1 share a natural host cell/tissue, the human airway tract (3, 34–36), we wondered whether HBoV1 could help AAV2 infection in human airway epithelia. In this study, we not only proved that HBoV1 is a novel helper virus for AAV2 infection of human airway epithelia but also demonstrated that HBoV1 gene expression supported AAV2 DNA replication at a level similar to that with Ad in HEK293 cells. Furthermore, we identified that the expression of the HBoV1 NS4, NP1, and BocaSR genes is essential for the replication of the duplex AAV2 genome in transfected cells. However, genome replication following AAV2 infection of HEK293 and HeLa cells required NS2 but not NS4.
RESULTS
Coinfection by HBoV1 and AAV2 rescues productive infection by AAV2 in well-differentiated HAE.
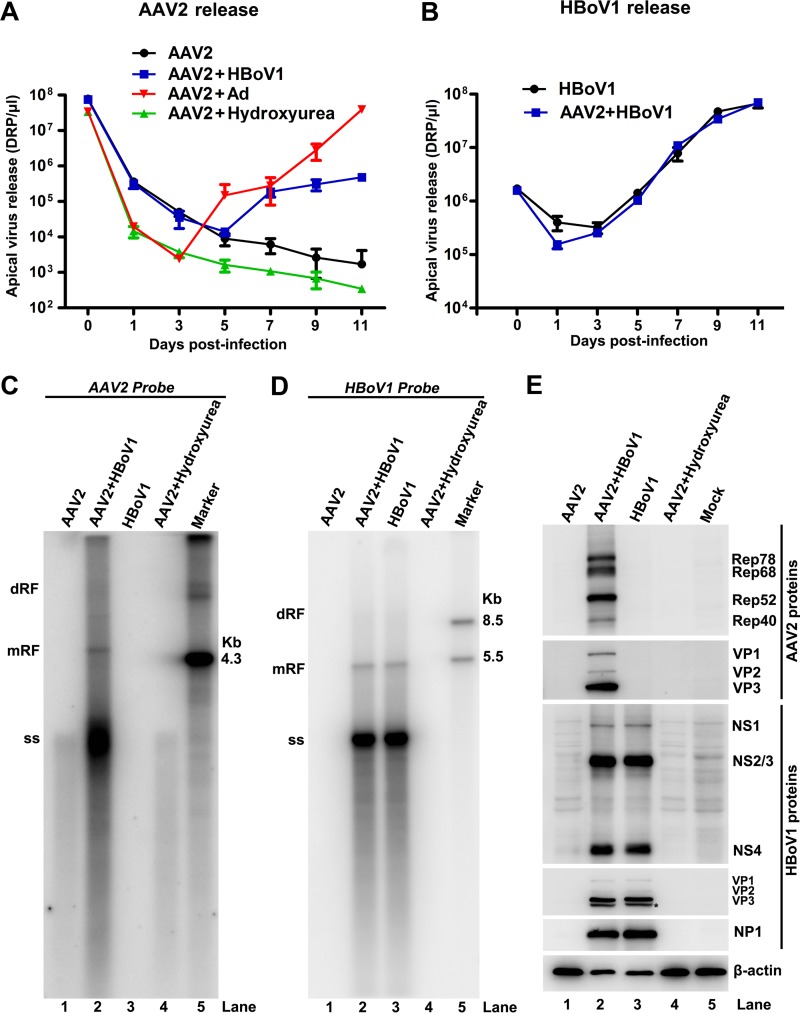
Since AAV2 has been reported to replicate in well-differentiated mitotically quiescent cells (37), similarly to HBoV1 (14), and both Ad and HBoV1 infect HAE-ALI (3, 38), we hypothesized that HBoV1 would facilitate AAV2 replication in nondividing HAE cells. To test this hypothesis, HAE cells of HAE-ALI were infected with AAV2, in the presence or absence of an HBoV1 or Ad helper. Apically released viruses were collected every other day and quantified. As expected, infection with AAV2 alone did not lead to the replication of virus in HAE cells, as shown by the continual drop in the amount of detectable AAV2 (DNase-resistant particles [DRP] per microliter) in apical washes following infection (Fig. 2A). Notably, HBoV1 coinfection rescued AAV2 replication. The amounts of apically released AAV2 virions were increased after day 5 postinfection (Fig. 2A), consistent with the lytic spread and replication of HBoV1 within the culture (Fig. 2B). As a positive control, Ad rescued AAV2 infection much more efficiently (∼1 to 2 logs higher in virus titer) and 2 days earlier than did HBoV1 (Fig. 2A). This observation suggests that HBoV1 is a modest helper virus for AAV2 replication in comparison to Ad. However, treatment of HAE-ALI cultures with a DNA damage-inducing reagent (hydroxyurea) failed to rescue AAV2 replication (Fig. 2A). At 11 days postinfection, infected cells were collected for analysis of viral DNA and proteins. Typical AAV2 DNA replicative forms, the dimer replicative form (dRF), monoreplicative form (mRF), and ssDNA, were observed only in cells coinfected with HBoV1 (Fig. 2C, lane 2 versus lanes 1 and 4); however, these forms were observed at levels ∼2 logs lower than those observed following Ad coinfection, despite similar ratios of viral ssDNA to mRF DNA (data not shown). Consistent with virus replication, AAV2 nonstructural (Rep) and structural (VP) proteins were expressed only in cells coinfected with HBoV1 and AAV2 (Fig. 2E, lane 2).
FIG 2.
HBoV1 rescues AAV2 replication in HAE-ALI cultures. (A) Virus release kinetics of AAV2. HAE-ALI cultures were infected with AAV2 in the presence of HBoV1 or Ad5 coinfection or treated with hydroxyurea at a final concentration of 2 mM. The released apical virions were collected every other day and measured by real-time PCR. Virus titers are shown as DRP per microliter. (B) HBoV1 virus release kinetics. HAE-ALI cultures were infected with HBoV1 alone or coinfected with AAV2. The amounts of HBoV1 virions released were measured by real-time PCR and are shown as DRP per microliter. (C and D) Southern blot analysis of AAV2 and HBoV1 DNA replication. Eleven days after infection or treatment with hydroxyurea, as indicated, 90% of the collected cells were lysed for Hirt DNA extraction. Hirt DNA samples were analyzed by Southern blotting with a 32P-labeled AAV2 probe (C) or a 32P-labeled HBoV1 probe (D). The dimer replicative form (dRF), monomer replicative form (mRF), and ssDNA (ss) are indicated. An AAV2 DNA at 4.3 kb (C) and HBoV1 DNA fragments at 5.5 kb and 8.5 kb (D) were used as size markers. (E) Western blot analysis of AAV2 and HBoV1 proteins. Eleven days after infection or treatment with hydroxyurea, as indicated, 10% of the collected cells were lysed and immunoblotted with anti-AAV2 Rep, anti-AAV2 VP, anti-HBoV1 NS1C, anti-HBoV1 VP3, anti-HBoV1 NP1, and anti-β-actin antibodies. Proteins detected are indicated next to the images. The asterisk indicates a possible cleaved VP3 protein.
Previously, it was reported that during AAV2-Ad coinfection, Ad replication was inhibited (39). In this study, we observed HBoV1 DNA replication in cells coinfected with HBoV1 and AAV2. As shown in Fig. 2B for apical virus release, in Fig. 2D for HBoV1 replicative DNA forms, and in Fig. 2E for HBoV1 proteins, there were no large differences with or without AAV2 coinfection.
Collectively, these results demonstrated that HBoV1 is a helper virus for productive AAV2 infection in well-differentiated HAE cells; however, AAV2 infection does not affect HBoV1 infection.
The HBoV1 NP1, BocaSR, and NS4 genes function as essential helper components for replication of the AAV2 duplex genome in HEK293 cells.
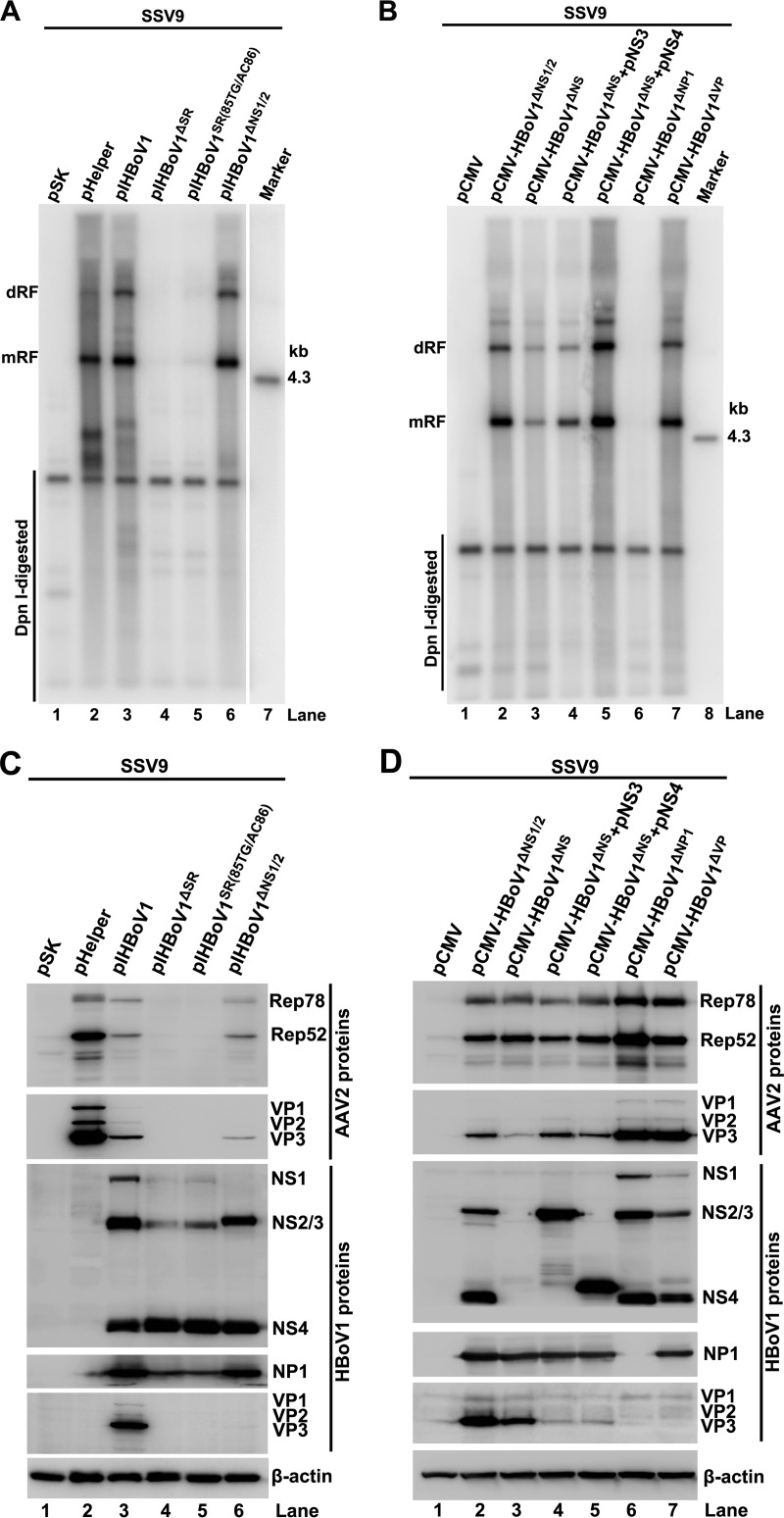
We next investigated whether this helper function depends on HBoV1 replication or only on viral gene expression. Due to the inefficiency of DNA transfection of HAE cells in polarized HAE-ALI, we tested AAV2 DNA replication in HEK293 cells by cotransfecting a duplex AAV2 genome (SSV9, an infectious clone of AAV2) (40) and various HBoV1 gene-expressing plasmids. Cotransfection of SSV9 with plasmid Ad pHelper, which expresses necessary adenovirus helper genes, i.e., E2a, E4, and virus-associated (VA) RNAs, served as a positive control. The results showed that cotransfection with pIHBoV1, an HBoV1 duplex DNA genome (an infectious clone) (3), rescued AAV2 DNA replication as efficiently as did cotransfection with Ad pHelper in HEK293 cells (Fig. 3A, lane 3 versus lane 2), although pIHBoV1 gave rise to lower expression levels of the AAV2 Rep and VP genes than did Ad pHelper (Fig. 3C, lane 3 versus lane 2). Mutagenesis analysis revealed that BocaSR was essential for AAV2 DNA replication. The helper function of HBoV1 for AAV2 was significantly reduced when BocaSR was deleted (pIHBoV1ΔSR) or mutated [pIHBoV1SR(85TG/AC86)] (Fig. 3A, lanes 4 and 5).
FIG 3.
Identification of minimal HBoV1 helper genes for AAV2 DNA replication in HEK293 cells. HEK293 cells were transfected with an AAV2 infectious clone (SSV9) and Ad pHelper or pIHBoV1-based mutants (A and C) or with SSV9 and pCMV-HBoV1-based mutants (B and D), as indicated. (A and B) Southern blot analysis. At 48 h posttransfection, 90% of the collected cells were lysed for Hirt DNA extraction. Hirt DNA samples were digested with DpnI and analyzed by Southern blotting with a 32P-labeled AAV2 probe. dRF and mRF DNAs, the 4.3-kb AAV2 marker, and DpnI-digested (input) DNA are indicated. (C and D) Western blot analysis. At 48 h posttransfection, 10% of the collected cells were lysed and immunoblotted with anti-AAV2 Rep, anti-AAV2 VP, anti-HBoV1 NS1C, anti-HBoV1 VP3, anti-HBoV1 NP1, and anti-β-actin antibodies. Proteins detected are indicated next to the images.
We next dissected the HBoV1 genes essential for facilitating AAV2 DNA replication. First, pIHBoV1ΔNS1/2, a replication-deficient mutant (3), was chosen to examine the role of HBoV1 NS1/NS2 in this function. The prevention of NS1 and NS2 expression did not abrogate the helper function of HBoV1 for AAV2 DNA replication (Fig. 3A, lane 3 versus lane 6) and AAV2 protein expression (Fig. 3C, lane 3 versus lane 6). Thus, these results demonstrated that the helper function of HBoV1 for AAV2 does not rely on HBoV1 DNA replication but rather relies on HBoV1 gene expression. However, HBoV1 NS1 and NS2 were not essential for helping AAV2 DNA replication in HEK293 cells following transfection of the AAV2 duplex genome.
We next evaluated the contribution of the NS3 and NS4 proteins to helping AAV2 DNA replication by transfecting an NS1-4-null plasmid (pCMV-HBoV1ΔNS) (Fig. 3D, lane 3). AAV2 DNA replication was significantly reduced when the expression of the NS3 and NS4 proteins was further prevented (Fig. 3B, lane 2 versus lane 3), suggesting that either the NS3 or the NS4 protein was involved in helping AAV2 DNA replication. To clearly define which NS protein was involved, pCMV-HBoV1ΔNS was replenished with the NS3 and NS4 genes. Southern blot analysis revealed that NS4, but not NS3, fully restored the function of pCMV-HBoV1ΔNS for supporting AAV2 DNA replication (Fig. 3B, lane 3 versus lane 5). Therefore, NS4 was an essential HBoV1 helper gene for supporting AAV2 DNA replication.
Further mutagenesis studies revealed that NP1 was also essential for supporting AAV2 DNA replication. There was little AAV2 RF DNA detected when NP1 was not expressed (Fig. 3B, lane 6). Furthermore, we found that HBoV1 VP proteins had negligible functions in AAV2 DNA replication. AAV2 DNA replicated at similar efficiencies in the presence and absence of HBoV1 capsid protein expression (Fig. 3B, lane 2 versus lane 7).
We have narrowed down the HBoV1 genes NP1, BocaSR, and NS4 as being essential helper genes for AAV2 DNA replication in HEK293 cells. We next extended the study to address whether these three genes were sufficient to facilitate AAV2 replication individually or together in an orchestrated manner.
HBoV1 NP1, NS4, and BocaSR compose the minimal set of HBoV1 helper genes for DNA replication of the AAV2 duplex genome in transfected HEK293 cells.
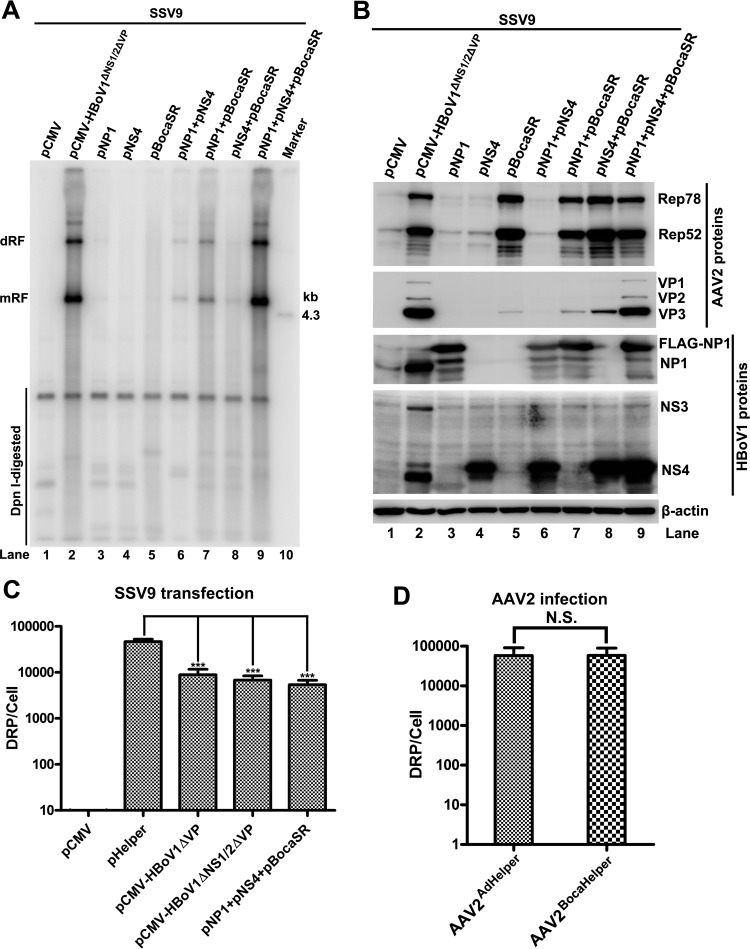
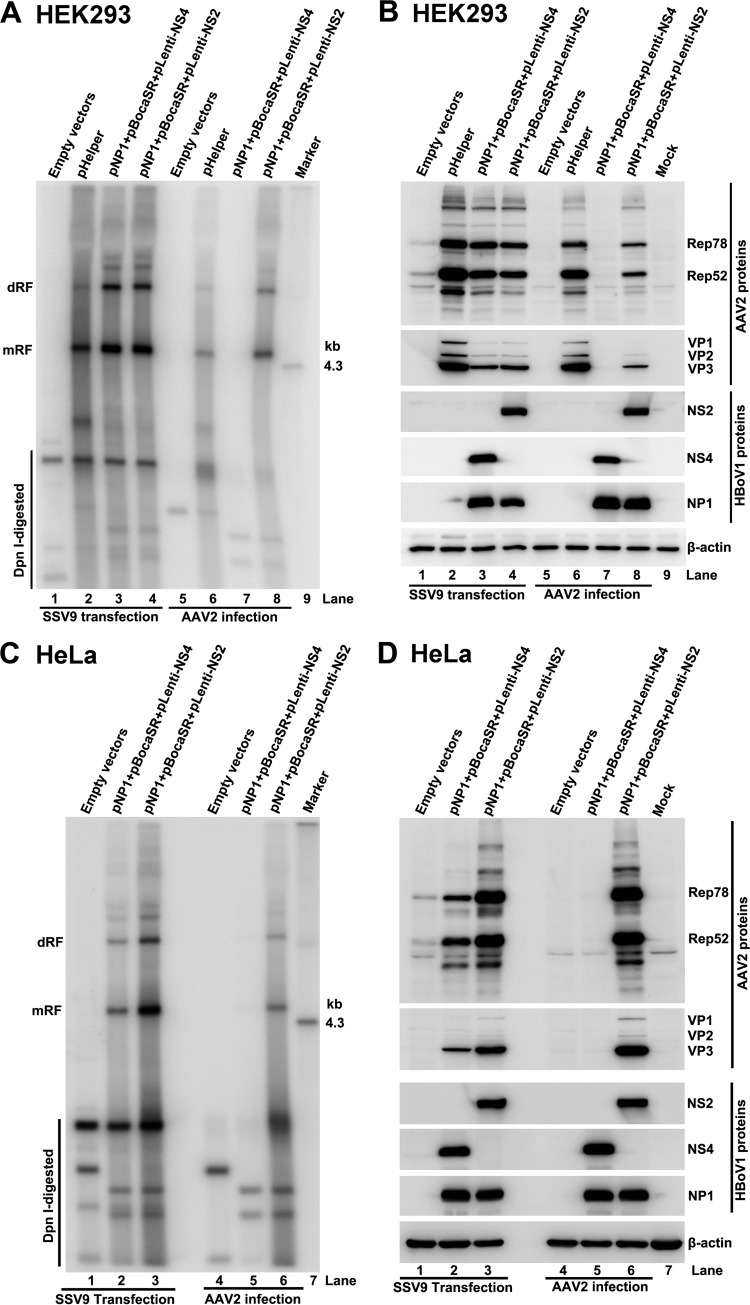
We tested the effects of the three HBoV1 helper genes alone and in various combinations on AAV2 DNA replication and protein expression. The results showed that the expression of individual genes (NP1, NS4, or BocaSR) did not facilitate AAV2 DNA replication (Fig. 4A, lanes 3 to 5). The simultaneous expression of any two genes only slightly stimulated AAV2 DNA replication (Fig. 4A, lanes 6 to 8) but less efficiently than with the maximum DNA replication that was achieved when all three genes were expressed (Fig. 4A, lane 9 versus lane 2). As for protein expression, BocaSR, but not NP1 and NS4, stimulated AAV2 Rep78 and Rep52 expression (Fig. 4B, lane 5 versus lanes 3 and 4). Notably, BocaSR-induced Rep expression was not sufficient to initiate AAV2 DNA replication (Fig. 4A and B, lane 5).
FIG 4.
AAV2 DNA replication, protein expression, and virus production in HEK293 cells cotransfected with different combinations of HBoV1 helper genes. HEK293 cells were transfected with an AAV2 infectious clone (SSV9) and various combinations of HBoV1 helper genes, as indicated. (A) Southern blot analysis. At 48 h posttransfection, 90% of the transfected cells were harvested for Hirt DNA extraction. Hirt DNA samples was examined for viral DNA replication by Southern blotting with a 32P-labeled AAV2 probe. dRF and mRF DNAs, DpnI-digested DNA, and the 4.3-kb AAV2 marker are indicated. (B) Western blot analysis. At 48 h posttransfection, 10% of the transfected cells were collected, lysed, and immunoblotted with anti-AAV2 Rep, anti-AAV2 VP, anti-HBoV1 NS1C, anti-HBoV1 NP1, and anti-β-actin antibodies. Proteins detected are indicated. (C) Production of AAV2. Transfected cells were collected, lysed, and quantified for DRP by real-time PCR. The virus production levels are shown as DRP per cell. Error bars show standard deviations, which were obtained from three independent experiments. Statistical analysis was performed by using the Student t test. ***, P < 0.0001. (D) Infectivity of the progeny viruses. HEK293 cells were infected with Ad pHelper-produced AAV2 (AAV2AdHelper) or with HBoV1 Helper-produced AAV2 (AAV2BocaHelper) at an MOI of 300 DRP/cell, followed by transfection with Ad pHelper. At 48 h posttransfection, the cells were collected for analysis of virus production, as determined by real-time PCR. Error bars show standard deviations, which were obtained from three independent experiments. Statistical analysis was performed by using the Student t test. N.S., no significance.
The helper function of Ad pHelper was also compared to that of HBoV1 Helper (a set of three plasmids, pNP1, pNS4, and pBocaSR) for virus production, which was measured by real-time PCR and normalized to the number of DRP per cell. Ad pHelper supported AAV2 production most efficiently, with an average virus titer of 4.6 × 104 DRP/cell, whereas the level of HBoV1 Helper-supported AAV2 production was 5- to 8-fold lower than that with Ad pHelper (Fig. 4C). We reasoned that the lower level of progeny production might be the result of the low expression levels of AAV2 capsid proteins facilitated by HBoV1 Helper (Fig. 3C, lane 2 versus lanes 3 and 6). Progeny virus infectivity was also examined, as shown in Fig. 4D. AAV2 produced with HBoV1 Helper (pNP1, pNS4, and pBocaSR) infected HEK293 cells as efficiently as did that produced with Ad pHelper.
Thus, we identified that HBoV1 NP1, NS4, and BocaSR are a minimal set of helper genes whose expression supports AAV2 DNA replication and progeny virus production from the transfection of the AAV2 duplex genome in HEK293 cells.
The minimal set of HBoV1 helper genes supports AAV2 DNA replication in HeLa cells, independent of the expression of any Ad genes.
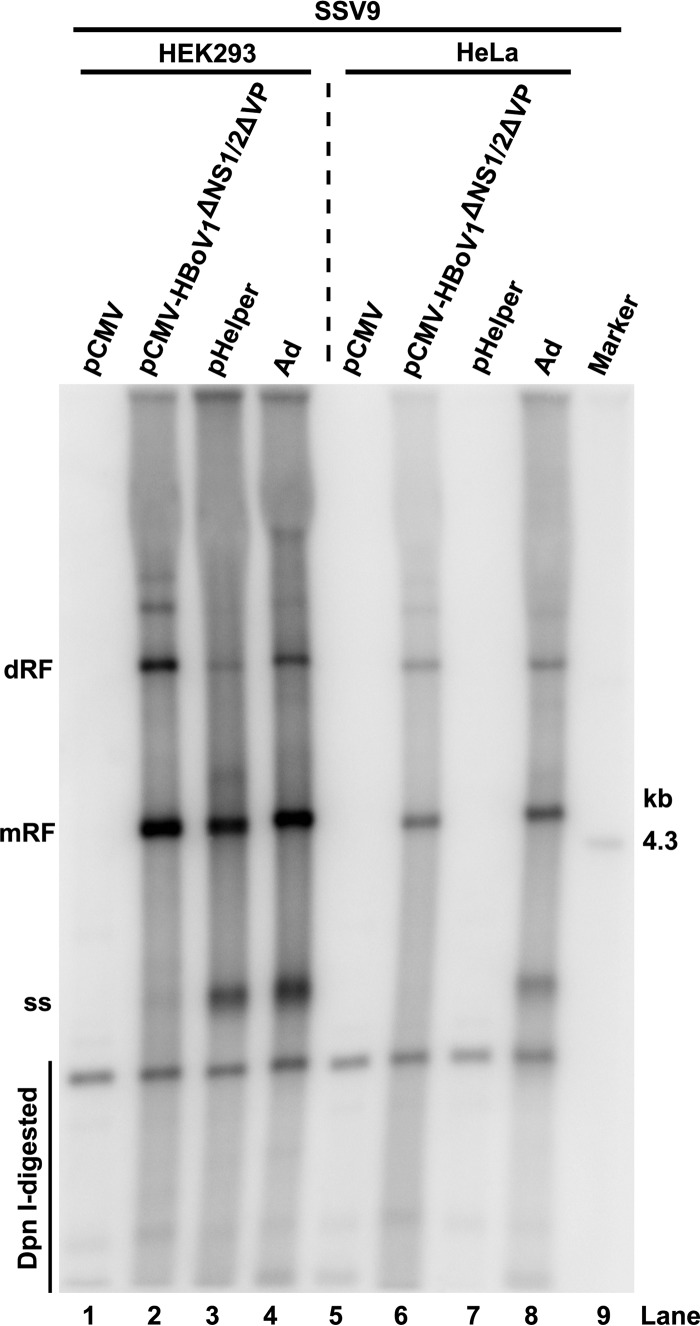
Considering that HEK293 cells express the Ad E1 gene, we next tested the helper function of HBoV1 in supporting AAV2 DNA replication in HeLa cells. In AAV2 duplex genome-transfected HEK293 and HeLa cells, we compared cotransfections with HBoV1 helper genes to those with Ad pHelper, and Ad infection served as a positive control. HBoV1 helper genes, provided by pCMV-HBoV1ΔNS1/2ΔVP, which expressed HBoV1 NP1, NS4, BocaSR, as well as NS3 (Fig. 4B, lane 2), stimulated AAV2 DNA replication as efficiently as did Ad pHelper or Ad infection (at a multiplicity of infection [MOI] of 1) in HEK293 cells (Fig. 5, lane 2 versus lanes 3 and 4). As expected, transfection of Ad pHelper did not result in any RF AAV2 DNA in HeLa cells (Fig. 5, lane 7), because the Ad E1 gene is not expressed in HeLa cells. Importantly, HBoV1 helper genes (NP1, NS3, NS4, and BocaSR) supported AAV2 DNA replication as efficiently as Ad infection in HeLa cells (Fig. 5, lane 6 versus lane 8), although the overall level of AAV2 RF DNA was lower in HeLa cells than in HEK293 cells (Fig. 5). The virus produced from HeLa cells was infectious and had a yield of ∼800 DRP/cell. Later, we further proved that the expression of the NP1, NS4, and BocaSR genes was sufficient to rescue AAV2 DNA replication in HeLa cells (see Fig. 7C, lane 2).
FIG 5.
Comparison of AAV2 duplex genome replication supported by Ad pHelper to that supported by HBoV1 helper genes in HEK293 and HeLa cells. HEK293 and HeLa cells were cotransfected with SSV9 and the Ad pHelper or HBoV1 helper plasmid or were infected with Ad5. At 48 h posttransfection, the cells were collected and lysed for Hirt DNA extraction. Hirt DNA samples were subjected to Southern blotting with a 32P-labeled AAV2 probe. dRF, mRF, ssDNA, the 4.3-kb AAV2 marker, and DpnI-digested DNA are indicated.
FIG 7.
NS2 fully replaces the function of NS4 for AAV2 DNA replication following transfection of the AAV2 duplex genome as well as following AAV2 infection. HEK293 cells (A and B) or HeLa cells (C and D) were cotransfected with SSV9 and Ad pHelper or different combinations of HBoV1 helper genes or were first infected with AAV2 and then transfected with pHelper or HBoV1 helper genes. Forty-eight hours after transfection with SSV9 or 72 h after infection with AAV2, 90% of the collected cells were lysed for Hirt DNA extraction, and 10% of the collected cells were lysed for protein analysis. (A and C) Hirt DNA samples were subjected to Southern blotting with a 32P-labeled AAV2 probe. dRF, mRF, DpnI-digested DNA, and a DNA size marker are indicated. (B and D) Cell lysates for protein analysis were subjected to Western blotting. AAV2 proteins (Rep78, Rep52, VP1, VP2, and VP3) were probed with anti-Rep and anti-VP antibodies. HBoV1 proteins were probed with anti-NS1C and anti-NP1 antibodies. β-Actin was probed as a loading control.
Together, the above-described results suggested that the HBoV1 helper genes NP1, NS4, and BocaSR facilitate AAV2 DNA replication in both HeLa and HEK293 cells without a requirement for Ad E1 gene expression.
The HBoV1 NS2 protein is required for AAV2 genome replication in HEK293 and HeLa cells following infection with AAV2 virions.
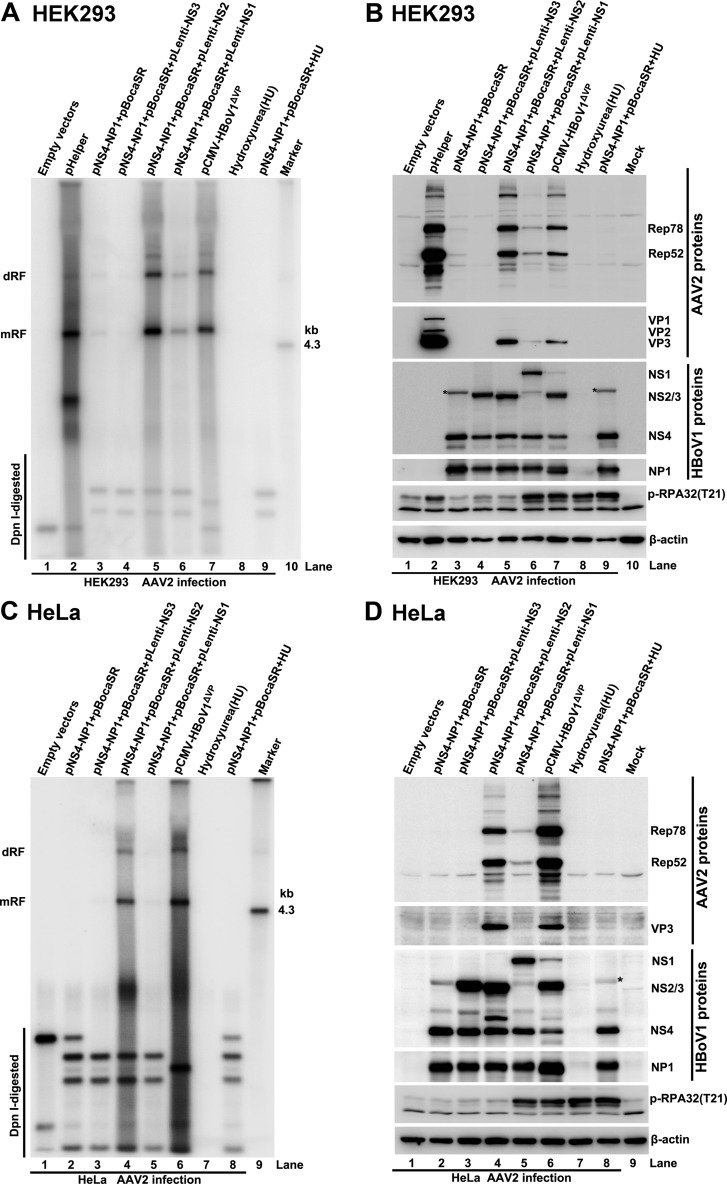
We next examined the minimal set of HBoV1 helper genes that support productive AAV2 infection in HEK293 and HeLa cells. Instead of transfection with infectious plasmid SSV9, HEK293 or HeLa cells were infected with AAV2, and analyses of DNA replication and protein expression levels for both AAV2 and HBoV1 were conducted following the transfection of HBoV1 helper genes. Our results showed that HBoV1 helper genes (NP1, NS4, and BocaSR) could not efficiently complement AAV2 DNA replication in HEK293 cells, compared with that with Ad pHelper (Fig. 6A, lane 3 versus lane 2). The addition of the NS3 gene by transfection of the pLenti-NS3 plasmid (Fig. 6B, lane 4) had no amendment effects (Fig. 6A, lane 4). Importantly, the additional expression of the NS2 protein (Fig. 6B, lane 5) from transfection of pLenti-NS2 or pCMV-HBoV1ΔVP, which expresses all HBoV1 nonstructural proteins, significantly enhanced AAV2 DNA replication (Fig. 6A, lanes 5 and 7 versus lane 3) and protein expression (Fig. 6B, lanes 5 and 7 versus lane 3). It was remarkable that the level of AAV2 DNA replication facilitated by the additional expression of the NS2 protein was similar to that supported by Ad pHelper (Fig. 6A, lanes 5 and 7 versus lane 2).
FIG 6.
The HBoV1 NS2 gene together with the HBoV1 NP1, NS4, and BocaSR genes are required for AAV2 infection. HEK293 cells (A and B) or HeLa cells (C and D) were preinfected with AAV2. At 12 h postinfection, the infected cells were transfected with Ad pHelper or different combinations of HBoV1 helper genes or were treated with hydroxyurea (at 2 mM) to rescue AAV2 DNA replication. At 72 h postinfection, 90% of the cells were used for Hirt DNA extraction, and 10% of the cells were lysed for protein analysis. (A and C) The Hirt DNA samples were subjected to Southern blotting using a 32P-labeled AAV2 probe. The AAV2 replicative DNA forms dRF and mRF, DpnI-digested DNA, and the 4.3-kb AAV2 DNA marker of the same load are indicated. (B and D) The protein samples were subjected to Western blotting. AAV2 proteins (Rep78, Rep52, VP1, VP2, and VP3) were probed with anti-Rep and anti-VP antibodies. The HBoV1 proteins (NS1, NS2, NS3, NS4, and NP1) were probed with anti-HBoV1 NS1C and anti-NP1 antibodies. Phosphorylation of RPA32 was probed with anti-PRA(pT21). β-Actin was probed as a loading control. Asterisks indicate the uncleaved NS4-P2A-NP1 fusion protein.
The additional expression of NS1 only slightly increased AAV2 DNA replication and protein expression in HEK293 cells (Fig. 6A and B, lanes 6). As NS1 induces a DDR in HEK293 cells (Fig. 6B, lane 6) (15), we used hydroxyurea to induce a DDR, as shown by the increased expression of phosphorylated RPA32 (p-RPA32) (Fig. 6B, lanes 8 and 9). However, hydroxyurea treatment did not obviously increase AAV2 DNA replication (Fig. 6A, lanes 8 and 9), which echoes the no-helper function in hydroxyurea-treated HAE cells. Thus, these results suggested that the HBoV1 helper supported AAV2 replication independently of DDR signaling.
We next performed the same experiments with HeLa cells, and similar results were obtained. As shown in Fig. 6C (lanes 4 and 6 versus lanes 2, 3, and 5) and D (lanes 4 and 6 versus lanes 2, 3, and 5), the coexpression of NS2 stimulated AAV2 DNA replication and protein expression following AAV2 infection in the presence of the NP1, NS4, and BocaSR genes. We noted that the overall level of AAV2 DNA replication in HeLa cells was lower than that in HEK293 cells during infection (Fig. 6A versus C). As noted below, an extended exposure time was required to visualize AAV2 DNA replication for the Southern blot shown in Fig. 6C.
Taken together, our results demonstrated that the HBoV1 NS2 protein plays an essential role in AAV2 DNA replication during AAV2 infection, in addition to NP1, NS4, and BocaSR.
NS2 fully substitutes for NS4 in supporting AAV2 DNA replication following both transfection of the AAV2 duplex genome and AAV2 infection.
The majority of the NS4 coding sequence is the TAD of NS1, and the four nonstructural proteins on the left side of the HBoV1 genome, NS1, NS2, NS3, and NS4, share this TAD in the C terminus (Fig. 1) (5). We hypothesized that the addition of a helper function from NS4 expression was redundant in the presence of NS2 during AAV2 infection. To this end, HEK293 cells were transfected with an AAV2 duplex genome and various combinations of HBoV1 helper genes, and in parallel, HEK293 cells were infected with AAV2 and transfected with HBoV1 helper genes. Analysis of AAV2 DNA replication showed that the combination of NS2, NP1, and BocaSR supported AAV2 DNA replication as efficiently as did the combination of the NS4, NP1, and BocaSR genes (Fig. 7A, lane 4 versus lane 3), suggesting that the function of NS4 can be fully replaced by NS2 for AAV2 DNA replication following transfection, although the function of the OBD of NS2 is not required. As we expected, the combination of NP1 and BocaSR with NS2, but not with the NS4 gene, efficiently supported viral DNA replication in AAV2-infected HEK293 cells (Fig. 7A, lane 8 versus lane 7), which was at a level similar to that supported by Ad pHelper (Fig. 7A, lane 8 versus lane 6). However, the expression levels of AAV2 proteins in the groups of HBoV1 helper genes were lower than those in the Ad pHelper group, in particular, the VP proteins (Fig. 7B, lane 4 versus lane 2 and lane 8 versus lane 6).
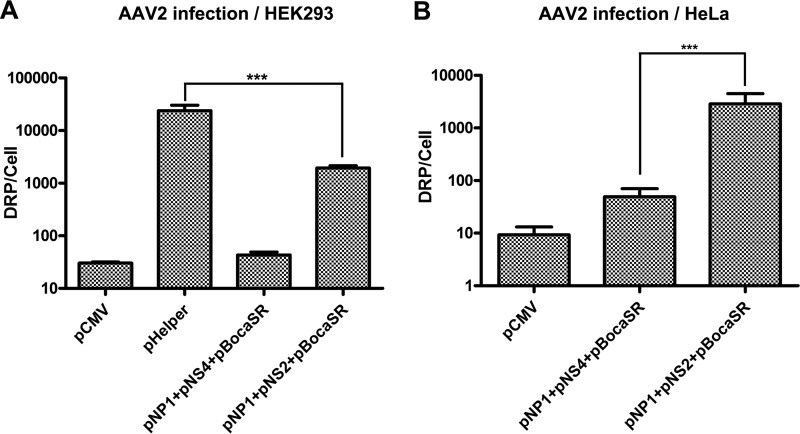
In parallel, the same experiments were conducted with HeLa cells, and similar results were observed. As shown in Fig. 7C (lane 3 versus lane 2) and D (lane 3 versus lane 2), NS2 fully replaced NS4 in supporting AAV2 DNA replication following transfection. However, following AAV2 infection, NS2, but not NS4, in combination with NP1 and BocaSR, rescued AAV2 DNA replication and protein expression efficiently (Fig. 7C and D, lane 5 versus lane 6). These results demonstrated that a combination of helper genes (NP1, NS2, and BocaSR) supported efficient AAV2 DNA replication not only following transfection but also during AAV2 infection. However, the number of progeny virions produced by the gene combination of NP1, NS2, and BocaSR was lower (∼1 log) than that produced by Ad pHelper in HEK293 cells (Fig. 8A), which is likely due to the insufficient expression of AAV2 capsid proteins (Fig. 7B, lane 8). In HeLa cells, the level of virus production was similar to that in HEK293 cells in the presence of NP1, NS2, and BocaSR as well as in the presence of NP1, NS4, and BocaSR (Fig. 8B).
FIG 8.
AAV2 infection is productive in HEK293 and HeLa cells transfected with HBoV1 helper genes. HEK293 (A) or HeLa (B) cells were infected with AAV2 (MOI = 1,000 DRP/cell), followed by transfection with Ad pHelper, HBoV1 helper gene sets, or control DNA (pCMV), as indicated. At 48 h posttransfection, the cells were collected for analysis of virus production. AAV2 production in HEK293 cells (A) or HeLa cells (B) was measured by real-time PCR as DRP per cell. Error bars show standard deviations, which were obtained from three independent experiments. Statistical analysis was performed by using the Student t test. ***, P < 0.0001.
Taken together, our results confirmed that the combination of the HBoV1 NP1, NS2, and BocaSR genes is the minimal and full helper for AAV2 DNA replication and also is the minimal helper for AAV2 infection.
DISCUSSION
In this study, we showed that HBoV1 can serve as a novel helper virus for AAV2. This is the first example of an autonomous parvovirus facilitating the replication of a dependoparvovirus. HBoV1 triggered AAV2 replication during coinfection in HAE-ALI. The minimal set of HBoV1 products required to support the replication of the AAV2 genome is composed of NP1, NS4, and BocaSR (HBoV1 Helper I). In contrast, the HBoV1 products required to support the production of AAV2 virions are NP1, NS2, and BocaSR (HBoV1 Helper II). Importantly, we now provide an alternative and simple system to replicate the AAV2 genome in non-Ad E1-expressing cells. The HBoV1/AAV2 helper system can be used to pinpoint the functions of HBoV1 genes by comparison with the Ad/AAV2 system, as many of the functions of Ad genes are already known.
Helper functions.
Productive AAV2 infection requires a helper virus or treatment with a genotoxic reagent in proliferating cells (20). Different helper viruses or genotoxic reagents help to facilitate productive AAV2 infection by various mechanisms. The minimal set of herpes simplex virus 1 (HSV-1) (human herpesvirus 1) genes required for AAV2 replication are those encoding the helicase/primase complex (UL5, UL8, UL52) and the DNA-binding protein ICP8 (41). It was suggested previously that in the HSV-1 helper system, AAV2 DNA replication utilizes the HSV-1-encoded viral DNA polymerase (42) rather than cellular DNA polymerases, which are used when an Ad helper is present (43). It was recently shown by the Xiao laboratory (44) that vaccinia virus can also serve as a helper for AAV replication (21); however, in this case, it is not a full helper because it does not activate AAV promoters. Thus, vaccinia virus can drive AAV replication and packaging only when all AAV promoters are already activated (44). In our study, we observed that HBoV1 is a less efficient helper for AAV2 than Ad following infection in HAE-ALI; there were 2 logs fewer AAV2 virions released from AAV-HBoV1-coinfected HAE-ALI than from AAV2/Ad-coinfected HAE-ALI (Fig. 2A). As HBoV1 infected nearly 80% of the HAE cells of the HAE-ALI, as detected by NS1 expression (data not shown), we speculate that HBoV1 is a modest helper for AAV2 replication. However, unlike HBoV1, AAV2 poorly transduces the apical surface of HAE-ALI due to an intracellular block of nuclear translocation (45). Thus, the inefficient AAV2 helper function of HBoV1 could be influenced by the inability of HBoV1 to enhance the nuclear translocation of AAV2, unlike Ad, which enhances AAV2 transduction by promoting nuclear translocation (46). Thus, it is possible that HBoV1 could more efficiently support the replication of alternative AAV serotypes (i.e., AAV1) that transduce HAE-ALI at a greater efficiency (47).
As helper viruses for AAV2 replication, there are similarities in the helper functions of Ad and HBoV1. First, both of these helper viruses infect differentiated HAE cells (3, 38). Second, AAV2 replication in the presence of Ad or HBoV1 utilizes cellular DNA polymerases (48). HBoV1 does not express a DNA replication polymerase, and its replication relies largely on cellular repair polymerases of the Y family (14, 15). Third, both Ad and HBoV1 helper systems use virally encoded noncoding RNAs, VA RNA and BocaSR, respectively, which share 50% sequence identity and high structural similarity (12).
While DNA damage-inducing agents were previously claimed to help AAV2 replication (49), we did not observe a significant increase in AAV2 DNA replication when cells were treated with hydroxyurea, which is likely because hydroxyurea-stimulated AAV2 replication was weak, compared with the Ad or HBoV1 helper (49). Apparently, HBoV1-facilitated AAV2 replication is independent of DNA damage response signaling, as the HBoV1 DDR inducer NS1 protein was not required for AAV2 replication (15).
Helper function in viral DNA replication versus virus production.
The HBoV1 helper produced 10-fold less virus than did Ad pHelper (Fig. 4C and 8A), although both the HBoV1 and Ad helpers supported equal levels of AAV2 replicative-form DNA (Fig. 3 and 7). This is likely due to the low expression levels of AAV2 capsid proteins in the presence of the HBoV1 helper (Fig. 3 and 7). It was reported previously that AAV2 VP-encoding mRNAs activate PKR and shut down the host cell translation machinery (33), while VAI RNA acts to overcome the AAV VP mRNA-induced activation of PKR (50). Unfortunately, in HBoV1-mediated helper functions, BocaSR does not inhibit PKR (12). However, it is also possible that HBoV1 helper gene products transactivate the AAV2 P40 promoter that transcribes VP-encoding mRNAs, or facilitate the splicing of these mRNAs, less efficiently than Ad helper gene products (51). Besides AAV2 VP expression, the small Rep proteins (Rep52 and Rep40) (22) are critical for the packaging of the AAV2 genome, and assembly-activating protein (AAP) (52) is critical for capsid assembly. We noticed that the expression level of Rep52 was lower with HBoV1 Helper than with Ad pHelper (Fig. 3C). Thus, the low expression levels of small Rep proteins and/or AAP may also account for the low level of production of AAV2 from the HBoV1 helper system. Further analysis of the limiting steps and mechanisms underlying AAV2 packaging in the HBoV1 helper system is warranted.
Function of NS2 versus NS4.
Although NS4 supported AAV2 DNA replication well in the transfection of an AAV2 duplex genome, NS2, but not NS4, is required for productive AAV2 replication following infection. Consistent with this observation, during HBoV1 infection of HAE-ALI, NS2 is also essential, and it is not required for DNA replication of the duplex HBoV1 genome and the production of infectious virions in HEK293 cells (5). In fact, bocaparvovirus NS2 is unique among all parvoviruses in that it contains a DNA-binding (endonuclease) domain and a C-terminal putative TAD (5, 53). It may function as a transcription transactivator, since the Ad E1a protein activates the AAV2 P5 promoter to express Rep proteins (26, 27), and this is a critical step in AAV2 replication. This assumption is supported by the fact that in HeLa cells, which lack the expression of Ad E1A, the expression of the minimal set of HBoV1 helper genes fully supports AAV2 DNA replication as efficiently as Ad infection. As NS4 can fully replace NS2 for the replication of the duplex AAV2 genome, we speculate that NS4, which largely is the TAD, also functions as a transactivator, like the HSV transcription transactivator VP16 (54) but without a DNA-binding domain. Following AAV2 infection, the AAV2 ssDNA genome must be converted to double-stranded DNA (dsDNA) replicative-form DNA that is a transcription-capable template for Rep expression, which is already available when the duplex AAV2 genome is provided. We believe that this step should not be a major limiting factor when only NS4, NP1, and BocaSR are provided, since conventional single-stranded rAAV2 vectors still transduce HEK293 cells and HeLa cells efficiently, although the so-called self-complementary rAAV vector, which bypasses the ssDNA-to-dsDNA conversion, delivers transgene expression at a higher level and with an earlier onset (55). Of note, both NS1 and NS2 contain the OBD of NS1 (Fig. 1); however, we currently do not know whether NS2 binds to the HBoV1 replication origin. The interaction between NS1 and HBoV1 origin DNA was weak, as assayed by an in vitro binding assay (16), suggesting that the DNA-binding activity of NS1 or NS2 (if there is any) requires additional viral or cellular factors. Why NS2 is needed and how it functions in helping AAV2 infection, beyond NS4, are major remaining questions.
Virus coevolution?
Ad, HBoV1, and AAV2 can infect the same tissue, human airway epithelia, and both Ad and HBoV1 can facilitate AAV2 replication during coinfection. Thus, one can imagine that these viruses may share some similarities by coevolution in human airway epithelia. To support the AAV2 lytic cycle, Ad E1a, E1b55k, E2a, E4orf6, and VAI RNA compose the minimal set of Ad helper genes, while NP1, NS2, and BocaSR compose the minimal set of HBoV1 genes. VAI RNA can partially replace the function of BocaSR in HBoV1 replication (12), implying that BocaSR may have evolved from VA RNA. From an evolutionary point of view, the NP1 and NS2 genes are unique genes in bocaparvoviruses, and BocaSR is unique in HBoVs; coincidently, these three genes are also the minimal set of helper genes for AAV2 infection. Therefore, it is reasonable to speculate that the NP1, NS2, and BocaSR genes not only make HBoV1 an autonomously replicating parvovirus but also may facilitate AAV2 replication in human airway epithelia.
In summary, we identified HBoV1 as a helper virus for AAV2 replication, and only three HBoV1 genes, NP1, NS2, and BocaSR, are essential for productive AAV2 infection. This is the first report that an autonomous parvovirus facilitates the replication of a dependoparvovirus. It will be critical to understand the mechanism underlying the helper functions of individual HBoV1 genes in AAV2 replication, which will provide a deep understanding of how AAV2 DNA replicates in cells and will help in the development of a better system to produce rAAV vectors using these novel helper genes.
MATERIALS AND METHODS
Ethics.
The use of primary human airway epithelial cells was approved by the Institutional Review Board (IRB) of the University of Iowa (IRB approval no. 9507432).
Cell lines and primary cultures.
HEK293 cells (ATCC CRL-1573) and HeLa cells (ATCC CRM-CCL-2) were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone, catalog no. SH30022.01; GE Healthcare Life Sciences, Logan, UT) supplemented with 10% fetal bovine serum (catalog no. F0926; Sigma, St. Louis, MO) at 37°C under a 5% CO2 atmosphere. Well-differentiated primary HAE-ALI (transwell inserts, catalog no. 3470; Corning, Corning, NY) were obtained from the Cell Culture Core of the Center for Gene Therapy, University of Iowa, as previously described (3). HAE-ALI cultures with a transepithelial electrical resistance (TEER) of >1,000 Ω · cm2 were selected for use in this study.
Plasmid construction. (i) pIHBoV1-based constructs.
HBoV1 infectious clone plasmid pIHBoV1 and its mutant, pIHBoV1ΔNS1/2, which does not express the NS1 and NS2 proteins, were previously described (3). pIHBoV1ΔSR, which does not express BocaSR RNA, and pIHBoV1SR(85TG/AC86), which expresses a BocaSR point mutant, were previously described (12).
(ii) pCMV-HBoV1NSCap-based constructs.
The pCMV-HBoV1 plasmid, which expresses all HBoV1 proteins under the control of the cytomegalovirus (CMV) promoter, was described previously (11). pCMV-HBoV1ΔNS1/2, which does not express the NS1 and NS2 proteins, and pCMV-HBoV1ΔNP1, which does not express the NP1 protein, were constructed by introducing an early stop codon in the ORFs of NS1 and NP1, respectively. pCMV-HBoV1ΔNS abolishes the expression of the NS1-4 proteins by replacing the D1-A1 intron with the IV intron of the Epo gene (56). pCMV-HBoV1ΔVP and HBoV1ΔNS1/2ΔVP were constructed by the introduction of a large deletion in the VP ORF using MfeI digestion followed by self-ligation of pCMV-HBoV1 and pCMV-HBoV1ΔNS1/2, respectively.
(iii) pcDNA3.1-based constructs.
pcDNA3.1-FLAG-NP1 (pNP1), which expresses N-terminally FLAG-tagged HBoV1 NP1, was constructed by inserting a codon-optimized HBoV1 NP1 ORF into the pcDNA3.1-FLAG vector (57) using EcoRI and XhoI sites. pcDNA3.1-Myc-NS3 (pNS3) and pcDNA3.1-Myc-NS4 (pNS4) were constructed by the insertion of N-terminally Myc-tagged codon-optimized HBoV1 NS3 and NS4, respectively, into the pcDNA3.1-Myc vector (57) using EcoRI and XhoI sites. pcDNA3.1-Myc-NS4-P2A-NP1 (pNS4-NP1) was constructed by the insertion of a coding sequence of NS4-P2A (a porcine teschovirus 1 peptide)-NP1 into the pcDNA3.1-Myc vector using EcoRI and XhoI sites.
(iv) pGEM-3Z-based constructs.
pGEM-3Z-BocaSR (pBocaSR), which expresses the HBoV1 noncoding RNA BocaSR, was previously described (12).
(v) pLenti-CMV-based constructs.
pLenti-CMV-HBoV1-NS1, -NS2, -NS3, and -NS4, which express the codon-optimized HBoV1 NS1, NS2, NS3, and NS4 proteins, respectively, were previously described (15).
(vi) Adenovirus plasmid pHelper.
pHelper, which contains the Ad5 E2, E4, and VA genes, was purchased from Agilent Technologies (Santa Clara, CA).
(vii) AAV2 duplex genome-containing plasmid (infectious clone) SSV9.
SSV9 (pSub201), which contains a full-length AAV2 genome and is an infectious clone of AAV2, was a gift from R. J. Samulski at the University of North Carolina, Chapel Hill (40, 51).
Plasmid DNA transfection.
HEK293 cells and HeLa cells seeded into 60-mm dishes were transfected by using the LipoD293 or GenJet transfection reagent (SignaGen Laboratories, Gaithersburg, MD) according to the manufacturer's instructions. The total amounts of plasmid DNA were kept constant (4 μg per 60-mm dish) in each group by supplementation with an empty vector to balance the transfection efficiency.
Viruses and infection.
We produced HBoV1 and AAV2 as previously described (3). Briefly, HEK293 cells were transfected with pIHBoV1, or SSV9 plus pHelper or HBoV1 helper plasmids, using PEI Max (Polysciences, Warrington, PA). The cells were collected at 72 h posttransfection, lysed, and treated with excess DNase I. The clarified cell lysates were subjected to cesium chloride gradient ultracentrifugation. Virus-enriched fractions were collected, dialyzed against phosphate-buffered saline (PBS) (pH 7.4), and quantified by real-time PCR. The titers were determined as DRP per microliters. The final virus stocks were kept in a −80°C freezer. Ad5 (dI309) was propagated in HEK293 cells (51) and purified by cesium chloride density gradient centrifugation according to a previously reported protocol (58). The Ad5 titer was determined as PFU.
HAE-ALI cultures were infected with HBoV1 at an MOI of 100 DRP/cell, with AAV2 at an MOI of 3,000 DRP/cell, and with Ad5 at an MOI of 0.5 PFU/cell. All viruses were diluted in 50 μl of culture medium and inoculated at the apical side for 4 h. HEK293 and HeLa cells were infected with AAV2 at an MOI of 100 DRP/cell or as otherwise specified in the figure legends. The virus (AAV2) was diluted in DMEM and incubated with the cells at 37°C for 4 h, and the cells were then washed once with PBS and fed fresh medium, followed by transfection at 12 h postinfection.
Quantification of virus production.
HEK293 cells seeded into a 6-well plate were transfected with SSV9 or infected with AAV2 in the presence of Ad pHelper or HBoV1 helper genes. The cells were lysed with fresh sodium deoxycholate lysis buffer (25 mM Tris [pH 8.0], 150 mM NaCl, 2 mM MgCl2, 0.5% sodium deoxycholate). The cell lysate was treated with Benzonase nuclease (250 U/ml) for 2 h at 37°C to digest free nucleic acids. The reaction was stopped by the addition of EDTA to 10 mM, and the mixture was further digested with protease K for viral DNA extraction using a DNeasy blood and tissue kit (Qiagen, Germantown, MD), according to the manufacturer's instructions. Viral genome numbers were determined by real-time PCR as DNase (nuclease) digestion-resistant particles and normalized to each cell.
Real-time PCR.
For AAV2, forward primer 5′-TCT GCA GCT CCC ACT CGA T-3′, reverse primer 5′-TTT GCT TCC TTC ATC ACA CAG TAC T-3′, and probe 5′-FAM (6-carboxyfluorescein)-TCC ACG CTG-ZEN (internal Zen quencher)-ACC GTG TCC CG-3IABkFQ (3′ Iowa Black FQ)-3′, purchased from IDT (Coralville, IA), were used. The primers and probe for HBoV1 genome quantification were previously described (3).
Western blotting.
Cells were directly lysed in 2× Laemmli sample buffer and boiled at 95°C for 5 min. The cell lysates were then loaded and separated on a 10% SDS-PAGE gel. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (catalog no. IPVH00010; Millipore, Bedford, MA), which was blocked and probed with primary and secondary antibodies sequentially. Signals were visualized by enhanced chemiluminescence, and images were acquired by using an ImageQuant LAS 4000 biomolecular imager (GE Healthcare Life Sciences, Pittsburgh, PA).
Antibodies used.
Antibodies against the HBoV1 NS1C, NP1, and VP proteins were previously described (8). The following antibodies were purchased: anti-AAV2 Rep (clone 303.9, catalog no. 03-65169) and anti-AAV2 VP (clone A69, catalog no. 03-61057), from American Research Products Inc. (Waltham, MA); anti-FLAG tag (catalog no. F1804) and anti-β-actin (catalog no. A5441), from Sigma (St. Louis, MO), anti-Myc tag (clone 9E10, catalog no. sc-40), from Santa Cruz Biotechnology; and anti-RPA32(pT21) [clone EPR2846(2), catalog no. ab109394], from Abcam (Cambridge, MA).
Hirt DNA extraction and Southern blotting. (i) Low-molecular-weight (Hirt) DNA extraction.
Cells were washed once with PBS and lysed with Hirt lysis buffer (10 mM Tris [pH 8.0], 10 mM EDTA, 0.6% SDS) for 15 min. Cell lysates were transferred into Eppendorf tubes, adjusted to a final NaCl concentration of 1.5 M, and incubated on ice overnight before being cleared by centrifugation at 17,000 × g for 20 min. The supernatants were collected and treated with protease K at a final concentration of 1 mg/ml for 1 h. Hirt DNA was purified with a DNA extraction kit (Qiagen) according to the manufacturer's instructions. For Hirt DNA extracted from transfected cells, DpnI was used to digest input DNA before Southern blotting was performed.
(ii) Southern blotting.
Southern blotting was performed according to our previously reported methods (59). Briefly, the Hirt DNA samples were digested with DpnI, resolved on a 1% agarose gel, blotted onto a nitrocellulose membrane, and probed with a [32P]dCTP-labeled probe. The template for the AAV2 probe was the XbaI/NdeI-digested 4.3 kb of AAV2, which contains the AAV2 Rep- and VP-encoding sequence. The HBoV1 probe was made as previously described (8) Hybridization signals were captured by using a storage phosphor screen and visualized on a Typhoon FLA 9000 biomolecular imager (GE Healthcare).
ACKNOWLEDGMENTS
We thank members of the Qiu laboratory for valuable discussions.
This study was supported by the following grants: PHS grants AI070723, AI105543, and AI112803 from the NIAID, NIH, to J.Q. and award YAN15XX0 from the Cystic Fibrosis Foundation to Z.Y. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Söderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Söderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng X, Yan Z, Luo Y, Xu J, Cheng Y, Li Y, Engelhardt J, Qiu J. 2013. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J Virol 87:4097–4102. doi: 10.1128/JVI.03132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen W, Deng X, Zou W, Cheng F, Engelhardt JF, Yan Z, Qiu J. 2015. Identification and functional analysis of novel nonstructural proteins of human bocavirus 1. J Virol 89:10097–10109. doi: 10.1128/JVI.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore SF, Tattersall P. 2005. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p 171–181. In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR (ed), The parvoviruses. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 7.Lederman M, Patton JT, Stout ER, Bates RC. 1984. Virally coded noncapsid protein associated with bovine parvovirus infection. J Virol 49:315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. 2010. Characterization of the gene expression profile of human bocavirus. Virology 403:145–154. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol 83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasina OO, Dong Y, Pintel DJ. 2015. NP1 protein of the bocaparvovirus minute virus of canines controls access to the viral capsid genes via its role in RNA processing. J Virol 90:1718–1728. doi: 10.1128/JVI.02618-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou W, Cheng F, Shen W, Engelhardt JF, Yan Z, Qiu J. 2016. Nonstructural protein NP1 of human bocavirus 1 plays a critical role in the expression of viral capsid proteins. J Virol 90:4658–4669. doi: 10.1128/JVI.02964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Shen W, Cheng F, Deng X, Engelhardt JF, Yan Z, Qiu J. 2017. Parvovirus expresses a small noncoding RNA that plays an essential role in virus replication. J Virol 91:e02375-16. doi: 10.1128/JVI.02375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berns KI, Parrish CR. 2015. Parvoviridae, p 1768–1791. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 14.Deng X, Yan Z, Cheng F, Engelhardt JF, Qiu J. 2016. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog 12:e1005399. doi: 10.1371/journal.ppat.1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng X, Xu P, Zou W, Shen W, Peng J, Liu K, Engelhardt JF, Yan Z, Qiu J. 2017. DNA damage signaling is required for replication of human bocavirus 1 DNA in dividing HEK293 cells. J Virol 91:e01831-16. doi: 10.1128/JVI.01831-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen W, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2016. Analysis of the cis and trans requirements for DNA replication at the right-end hairpin of the human bocavirus 1 genome. J Virol 90:7761–7777. doi: 10.1128/JVI.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berns KI. 1990. Parvovirus replication. Microbiol Rev 54:316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotin RM, Linden RM, Berns KI. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J 11:5071–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzman MD, Linden RM. 2011. Adeno-associated virus biology. Methods Mol Biol 807:1–23. doi: 10.1007/978-1-61779-370-7_1. [DOI] [PubMed] [Google Scholar]
- 20.Ward P. 2006. Replication of adeno-associated virus DNA, p 189–211. In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR (ed), The parvoviruses. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 21.Schlehofer JR, Ehrbar M, zur Hausen H. 1986. Vaccinia virus, herpes simplex virus, and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology 152:110–117. doi: 10.1016/0042-6822(86)90376-4. [DOI] [PubMed] [Google Scholar]
- 22.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava A, Carter BJ. 2017. AAV infection: protection from cancer. Hum Gene Ther 28:323–327. doi: 10.1089/hum.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flotte TR, Afione SA, Zeitlin PL. 1994. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am J Respir Cell Mol Biol 11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 25.Xiao X, Li J, Samulski RJ. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 72:2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Seto E, Chang LS, Shenk T. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 27.Chang LS, Shi Y, Shenk T. 1989. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J Virol 63:3479–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weitzman MD, Fisher KJ, Wilson JM. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol 70:1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samulski RJ, Shenk T. 1988. Adenovirus E1B 55-Mr polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J Virol 62:206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF, Wilson JM. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol 70:520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz RA, Palacios JA, Cassell GD, Adam S, Giacca M, Weitzman MD. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol 81:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrari FK, Samulski T, Shenk T, Samulski RJ. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol 70:3227–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayak R, Pintel DJ. 2007. Adeno-associated viruses can induce phosphorylation of eIF2alpha via PKR activation, which can be overcome by helper adenovirus type 5 virus-associated RNA. J Virol 81:11908–11916. doi: 10.1128/JVI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becroft DM. 1967. Histopathology of fatal adenovirus infection of the respiratory tract in young children. J Clin Pathol 20:561–569. doi: 10.1136/jcp.20.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prince GA, Porter DD, Jenson AB, Horswood RL, Chanock RM, Ginsberg HS. 1993. Pathogenesis of adenovirus type 5 pneumonia in cotton rats (Sigmodon hispidus). J Virol 67:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers C, Mane M, Kokorina N, Alam S, Hermonat PL. 2000. Ubiquitous human adeno-associated virus type 2 autonomously replicates in differentiating keratinocytes of a normal skin model. Virology 272:338–346. doi: 10.1006/viro.2000.0385. [DOI] [PubMed] [Google Scholar]
- 38.Kotha PL, Sharma P, Kolawole AO, Yan R, Alghamri MS, Brockman TL, Gomez-Cambronero J, Excoffon KJ. 2015. Adenovirus entry from the apical surface of polarized epithelia is facilitated by the host innate immune response. PLoS Pathog 11:e1004696. doi: 10.1371/journal.ppat.1004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timpe JM, Verrill KC, Trempe JP. 2006. Effects of adeno-associated virus on adenovirus replication and gene expression during coinfection. J Virol 80:7807–7815. doi: 10.1128/JVI.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samulski RJ, Chang LS, Shenk T. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol 63:3822–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weindler FW, Heilbronn R. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J Virol 65:2476–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alazard-Dany N, Nicolas A, Ploquin A, Strasser R, Greco A, Epstein AL, Fraefel C, Salvetti A. 2009. Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events. PLoS Pathog 5:e1000340. doi: 10.1371/journal.ppat.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni TH, McDonald WF, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol 72:2777–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore AR, Dong B, Chen L, Xiao W. 2015. Vaccinia virus as a subhelper for AAV replication and packaging. Mol Ther Methods Clin Dev 2:15044. doi: 10.1038/mtm.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Invest 105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao W, Warrington KH Jr, Hearing P, Hughes J, Muzyczka N. 2002. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J Virol 76:11505–11517. doi: 10.1128/JVI.76.22.11505-11517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan Z, Lei-Butters DC, Liu X, Zhang Y, Zhang L, Luo M, Zak R, Engelhardt JF. 2006. Unique biologic properties of recombinant AAV1 transduction in polarized human airway epithelia. J Biol Chem 281:29684–29692. doi: 10.1074/jbc.M604099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nash K, Chen W, McDonald WF, Zhou X, Muzyczka N. 2007. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J Virol 81:5777–5787. doi: 10.1128/JVI.02651-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yalkinoglu AO, Heilbronn R, Burkle A, Schlehofer JR, zur Hausen H. 1988. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res 48:3123–3129. [PubMed] [Google Scholar]
- 50.Vachon VK, Conn GL. 2016. Adenovirus VA RNA: an essential pro-viral non-coding RNA. Virus Res 212:39–52. doi: 10.1016/j.virusres.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 51.Qiu J, Pintel DJ. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol Cell Biol 22:3639–3652. doi: 10.1128/MCB.22.11.3639-3652.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonntag F, Schmidt K, Kleinschmidt JA. 2010. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fasina OO, Stupps S, Figueroa-Cuilan W, Pintel DJ. 2017. Minute virus of canines NP1 protein governs the expression of a subset of essential nonstructural proteins via its role in RNA processing. J Virol 91:e00260-17. doi: 10.1128/JVI.00260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Triezenberg SJ, LaMarco KL, McKnight SL. 1988. Evidence of DNA:protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev 2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 55.McCarty DM. 2008. Self-complementary AAV vectors; advances and applications. Mol Ther 16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 56.Yan Z, Zhang Y, Duan D, Engelhardt JF. 2000. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc Natl Acad Sci U S A 97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Mir MA. 2015. Andes virus nucleocapsid protein interrupts protein kinase R dimerization to counteract host interference in viral protein synthesis. J Virol 89:1628–1639. doi: 10.1128/JVI.02347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. 2007. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 59.Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus B19 can replicate in nonpermissive cells with the help of adenovirus genes and produces infectious virus. J Virol 83:9541–9553. doi: 10.1128/JVI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol 83:7739–7748. doi: 10.1128/JVI.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]