ABSTRACT
In this study, we sought to identify novel antimicrobial peptides (AMPs) in Python bivittatus through bioinformatic analyses of publicly available genome information and experimental validation. In our analysis of the python genome, we identified 29 AMP-related candidate sequences. Of these, we selected five cathelicidin-like sequences and subjected them to further in silico analyses. The results showed that these sequences likely have antimicrobial activity. The sequences were named Pb-CATH1 to Pb-CATH5 according to their sequence similarity to previously reported snake cathelicidins. We predicted their molecular structure and then chemically synthesized the mature peptide for three putative cathelicidins and subjected them to biological activity tests. Interestingly, all three peptides showed potent antimicrobial effects against Gram-negative bacteria but very weak activity against Gram-positive bacteria. Remarkably, ΔPb-CATH4 showed potent activity against antibiotic-resistant clinical isolates and also was observed to possess very low hemolytic activity and cytotoxicity. ΔPb-CATH4 also showed considerable serum stability. Electron microscopic analysis indicated that ΔPb-CATH4 exerts its effects via toroidal pore preformation. Structural comparison of the cathelicidins identified in this study to previously reported ones revealed that these Pb-CATHs are representatives of a new group of reptilian cathelicidins lacking the acidic connecting domain. Furthermore, Pb-CATH4 possesses a completely different mature peptide sequence from those of previously described reptilian cathelicidins. These new AMPs may be candidates for the development of alternatives to or complements of antibiotics to control multidrug-resistant pathogens.
KEYWORDS: antimicrobial peptides, cathelicidin, genome analysis, Python bivittatus, reptile
INTRODUCTION
Endogenous antimicrobial peptides (AMPs) are small charged peptides that function in the innate immune response, the first-line, nonspecific defense against pathogens and foreign substances (1). AMPs are found in diverse organisms, including animals, plants, insects, and microorganisms (2). The first AMP was discovered in the mucus of frog skin in the 1960s (3). Since then, more than 5,500 antimicrobial peptides with broad-spectrum antibacterial, antiviral, antifungal, antiparasitic, and anticancer activities have been reported (4, 5).
Most of the AMPs found in eukaryotic organisms can be classified into several families according to their structural characteristics. Defensins are found in diverse animal species and are characterized by three pairs of cysteine disulfide bonds that stabilize the peptide structure. Cathelicidins are also found in various vertebrates and are characterized by a conserved cathelin-like domain (CLD). Mature peptides linked to the C terminus of the CLD have different lengths, charges, and secondary structures, which determine their diverse functions.
Determining the molecular structures of AMPs and their association with the lipid bilayers of cell membranes is thought to be crucial for understanding their mechanisms of action (6, 7). Interactions between antimicrobial peptides and lipid membranes can be investigated by nuclear magnetic resonance and circular dichroism spectroscopy (8, 9). There are large differences in the membrane structures of bacteria and mammals, enabling AMPs to distinguish between these cells and thus providing specificity (10).
The use of high-throughput sequencing technologies, such as next-generation sequencing (11) and genomewide analyses, combined with AMP databases (12) and bioinformatic tools allows for the rapid identification of novel AMPs in genome sequences (13, 14). Genome information from several reptilian species has recently become available, providing interesting organisms for the investigation of the role of AMPs in species adapted to microbially challenging environments. Here, we attempted to analyze the AMP repertoire in the newly published python genome (15).
We first performed a genomewide identification of the AMPs in the Python bivittatus genome and then further analyzed five putative cathelicidins. We chemically synthesized the mature peptide sequences of three cathelicidins based on their simulated structural characteristics and evaluated their biological activities. Interestingly, despite their potent antimicrobial activities against clinically isolated antibiotic resistance pathogens, these peptides showed low hemolytic activity and cytotoxicity. These newly reported AMPs are likely to be important for the innate immunity of the host, P. bivittatus, and might be useful for the development of clinically applicable antibacterial substances.
RESULTS
Identification of 29 AMP-like proteins in a genomewide in silico analysis of the python genome.
The sequences of 2,488 nonredundant AMPs obtained from UniProtKB/Swiss-Prot were analyzed using the AMPA and APD3 databases. Ninety of these sequences were from reptiles, including 2 beta-defensins, 10 cathelicidins, 17 crotamine-myotoxins, 24 flavin monoamine oxidases (Fig. 1), 6 phospholipase A2s, 30 snake waprins, and 1 pelovaterin (see Table S1 in the supplemental material). In a BLAST search of these reptile AMP-like sequences against the genome of Python bivittatus, 29 AMP-like proteins from 5 families, including 12 flavin monoamine oxidases (Fig. 1), 8 phospholipase A2s, 3 snake waprins, 5 cathelicidins, and 1 crotamine-myotoxin, were identified (E value < 0.001; see Table S2 in the supplemental material). Interestingly, no classical β-defensins were identified in the python genome.
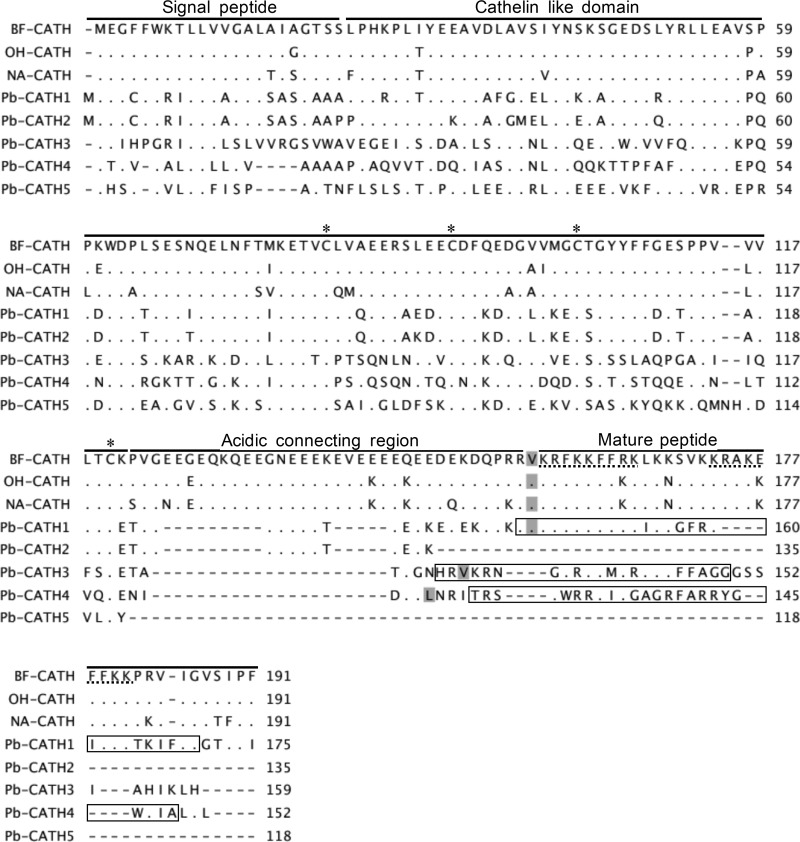
FIG 1.
Multiple-sequence alignment constructed using the amino acid sequences of representative cathelicidins of snakes together with putative python cathelicidins. The gaps are indicated by dashes (-), and amino acids that are identical to the reference (top) are indicated by dots (.). The regions corresponding to characteristic regions of snake cathelicidins are indicated by separated lines. The conserved cysteines are indicated by an asterisk (*). The previously reported repetitive cryptic nonapeptide (31) important for the antimicrobial activity is underlined with dots. The predicted cleavage sites that yield mature peptides are shaded. The regions corresponding to the synthesized peptides for Pb-CATH1, -3, and -4 are contained in boxes. Accession numbers for the sequences are as follows: NA-CATH (EU622892), BF-CATH (EU622893), OH-CATH (EU622894), Pb-CATH1 (XP_007443270.1), Pb-CATH2 (XP_007445262.1), Pb-CATH3 (XP_007442672.1), Pb-CATH4 (XP_007445036.1), and Pb-CATH5 (XP_015746367.1).
Genetic and evolutionary characteristics of the putative cathelicidin-related proteins in python.
Of the 29 AMP candidates in the python genome, the amino acid sequences of the five putative cathelicidins were subjected to further characterization and experimental validation. Comparison of the amino acid sequences of the five python cathelicidins to previously reported cathelicidins from three other snake species, Bungarus fasciatus, Ophiophagus hannah, and Naja atra, revealed high levels of conservation in the CLD region (Fig. 1). Additionally, four cysteines were present in the CLD of the cathelicidins. Because there is no established nomenclature available for these molecules, we have tentatively named them Pb-CATH1–5 according to their sequence similarity.
As expected, sequences near the carboxy terminus, corresponding to the mature peptide of python cathelicidin, were less well conserved than those near the amino terminus, corresponding to the CLD (Fig. 1). The CLD in Pb-CATH1 had the highest sequence homology (60%) to the CLDs in cathelicidins from other snake species. In contrast, Pb-CATH4 CLD had the lowest sequence identity (28%).
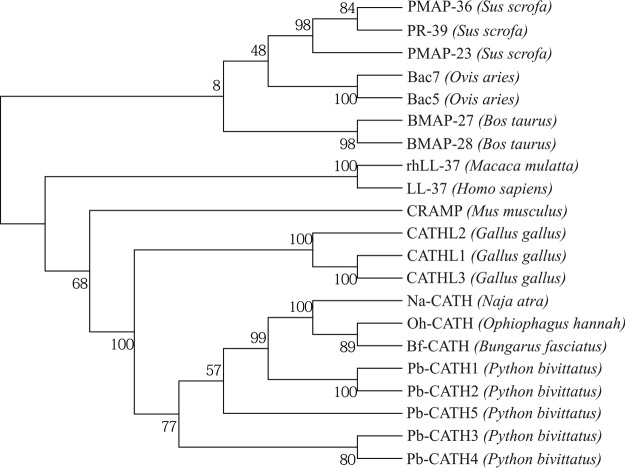
To investigate the evolutionary relationships among the Pb-CATHs and other cathelicidins from diverse species, we constructed a phylogenetic tree of 21 CLD sequences from 12 species, including 10 mammals, 6 snakes, and 3 birds (Fig. 2). The results were consistent with the evolutionary relationships among the species, and the reptile and avian CLDs were clustered together and separated from the mammalian CLDs. The relationships among the CLDs in the Pb-CATHs were also consistent with the results shown in Fig. 1, which used the full-length amino acid sequences.
FIG 2.
Phylogenetic relationships of 21 cathelicidins using the cathelin-like domain from 11 vertebrate species. A total of 102 amino acids were used for the analysis. The species names are indicated within parentheses. The numbers on the branches represent bootstrap values to support the branch (1,000 replications). Accession numbers of the sequences are as follows: LL-37 (NP_004336.3), PR-39 (NP_999615.1), PMAP-23 (NP_001123448.1), PMAP-36 (NP_001123437.1), BMAP-27 (NP_777257.1), BMAP-28 (NP_776935.1), Fowlicidin-1 (NP_001001605.1), Fowlicidin-2 (NP_001020001.2), Fowlicidin-3 (NP_001298106.1), rhLL-37 (NP_001028681.1), Bac7 (NP_001009301.1), Bac5 (NP_001009787.1), CRAMP (NP_034051.2), Na-CATH (EU622892), Bf-CATH (EU622893), Oh-CATH (EU622894), Pb-CATH1 (XP_007443270.1), Pb-CATH2 (XP_007445262.1), Pb-CATH3 (XP_007442672.1), Pb-CATH4 (XP_007445036.1), and Pb-CATH5 (XP_015746367.1).
In silico biochemical characteristics of cathelicidin-related proteins in Python bivittatus.
The antimicrobial domains of the five python cathelicidins were determined by analysis using the AMPA tool (Table 1). The results showed that Pb-CATH1, -3, and -4 contain putative antimicrobial domains longer than 20 amino acids. Pb-CATH2 and -5 do not encode mature peptides due to the presence of a stop codon or partial stop (data not shown). To evaluate their secondary structure, the PSIPRED protein sequence analysis workbench was used. The results showed that the antimicrobial domains form an α-helical structure, which is a common characteristic of AMPs (see Fig. S1 in the supplemental material). The hydrophobicity and net charges of the antimicrobial activity-containing regions were 36 to 40 and +9 to +15, respectively (Table 2). The maximum sequence similarities of ΔPb-CATH1, ΔPb-CATH4, and Pb-CATH3 to known antimicrobial peptides were 48.6%, 37.9%, and 38.5%, respectively. Taken together, our in silico analyses showed that the regions conferring antimicrobial activity in Pb-CATH1, -3, and -4 are consistent with the characteristics common to AMPs.
TABLE 1.
Predicted antimicrobial domains of the identified cathelicidins in Python bivittatus
| Peptide | Position (aa) | Sequence of the expected antimicrobial domain | Length (aa) |
|---|---|---|---|
| Pb-CATH1 | 142–172 | KRVKRFKKFFRKIKKGFRKIFKKTKIFIGGT | 31 |
| Pb-CATH3 | 128–149 | HRVKRNGFRKFMRRLKKFFAGG | 22 |
| Pb-CATH4 | 126–150 | TRSRWRRFIRGAGRFARRYGWRIAL | 25 |
TABLE 2.
Sequences and characteristics of synthesized peptides derived from the identified cathelicidins in Python bivittatus
| Peptide | Sequence | Length (aa) | Ha | z (+)b | Similarity (%)c |
|---|---|---|---|---|---|
| ΔPb-CATH1 | RVKRFKKFFRKIKKGFRKIFKKTKIFIG | 28 | 36 | 15 | 48.6 |
| Pb-CATH3 | HRVKRNGFRKFMRRLKKFFAGG | 22 | 36 | 9 | 38.5 |
| ΔPb-CATH4 | TRSRWRRFIRGAGRFARRYGWRIA | 24 | 40 | 9 | 37.9 |
H, hydrophobicity.
z, charge.
Sequence similarity to known AMPs.
Confirmation of the antimicrobial activity of python cathelicidins.
The antimicrobial activities of chemically synthesized python cathelicidins were evaluated against a panel of reference bacteria. Peptides corresponding to the antimicrobial activity-conferring regions of Pb-CATH1, -3, and -4, (28, 22, and 24 amino acids [aa], respectively) were successfully prepared by chemical synthesis (data not shown). MIC assays were performed using the synthesized peptides against six bacterial strains, Escherichia coli, Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, Staphylococcus aureus, Bacillus cereus, and Streptococcus iniae (Table 3). Ampicillin and gentamicin sulfate were used as controls of antimicrobial activity. MIC is considered the lowest concentration of antimicrobial substances required to completely prevent the visible growth of microorganism. All three peptides showed potent antimicrobial activities against E. coli, P. aeruginosa, and S. Typhimurium, which are Gram-negative strains. Of the tested peptides, ΔPb-CATH4 showed the strongest antimicrobial activity against the Gram-negative bacteria, with MICs of 0.5 to 3.0 μg/ml. However, no antimicrobial effects were observed against the Gram-positive strain S. aureus, even at a concentration of 128 μg/ml. Consistent with the results for S. aureus, ΔPb-CATH1 and Pb-CATH3 showed very weak or no antimicrobial activity against two other Gram-positive bacteria, B. cereus and S. iniae (Table 3). Because there are no predetermined values to evaluate the antimicrobial activity of these AMPs to B. cereus and S. iniae, we applied the general criteria of ≥32 μg/ml as the activity resistance value according to CLSI (Clinical and Laboratory Standards Institute) guidelines (16). ΔPb-CATH4 showed antimicrobial activity against B. cereus at 10 μg/ml, in contrast to two other Gram-positive strains, S. aureus and S. iniae, against which no activity was shown at 32 μg/ml (see Table S3 in the supplemental material). These results indicate that Pb-CATHs in this study show stronger activity toward Gram-negative bacteria than Gram-positive strains. To demonstrate that AMPs are able to kill the antibiotic resistance strains, ΔPb-CATH4 was tested against the clinically isolated antibiotic resistance strains E. coli and Klebsiella pneumoniae, which showed complete resistance particularly toward ampicillin and to a lesser extent with gentamicin. Remarkably, ΔPb-CATH4 showed very high potency toward these pathogens at 4.7 and 1.18 μg/ml, respectively.
TABLE 3.
Antimicrobial activity of Python bivittatus cathelicidin-derived peptides against bacterial strains
| Strains | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Pb-CATH1 | Pb-CATH3 | Pb-CATH4 | Ampicillin | Gentamicin | |
| Gram-negative bacteria | |||||
| E. coli ATCC 25922 | 2 | 3 | 1 | 1 | 0.5 |
| P. aeruginosa ATCC 27853 | 3 | 8 | 3 | >128 | 0.25 |
| S. typhimurium ATCC 14028 | 1.5 | 2 | 0.5 | 1 | 0.375 |
| Gram-positive bacteria | |||||
| S. aureus ATCC 29213 | >128 | >128 | >128 | 0.1 | 0.1 |
| B. cereus ATCC 10876 | 46 | 32 | 10 | 16 | 0.25 |
| Clinical isolates | |||||
| E. coli | NDa | ND | 4.7 | >64 | >64 |
| K. pneumoniae | ND | ND | 1.18 | 32 | 16 |
ND, not determined.
Evaluation of the serum stability of python cathelicidins.
The stability of antimicrobials in serum is important for clinical utility. Therefore, to test the effect of serum on the antimicrobial activity of Pb-CATHs, various concentrations of ΔPb-CATH4 were incubated with 50% (vol/vol) serum for 0.5 to 1 h, and their antimicrobial activity was measured. The results showed that the antimicrobial effects of ΔPb-CATH4 were maintained after 1 h of incubation in serum, although the efficacy decreased over time. At 4 μg/ml, ΔPb-CATH4 completely suppressed the growth of E. coli after 100 min of incubation (Fig. 3).
FIG 3.
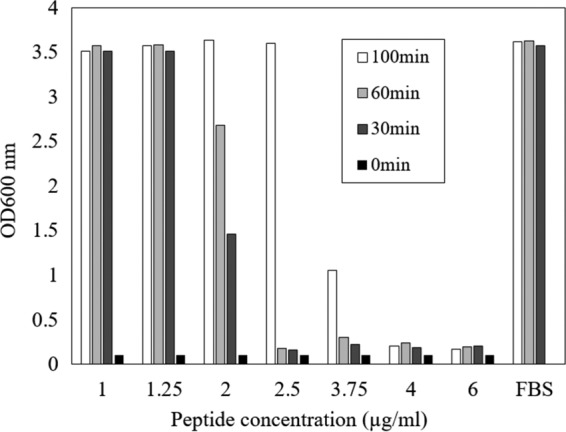
Stability of ΔPb-CATH4 in serum. ΔPb-CATH4 was incubated in 50% (vol/vol) fetal bovine serum (FBS) for 30, 60, and 100 min, and then antimicrobial activity against E. coli was assessed. After incubation for 30 and 60 min, activity was maintained at concentrations of 2.5 μg/ml and above. FBS with peptide and FBS without peptide from the 0-min time point were used as the negative and positive controls, respectively.
Low hemolytic activity of python cathelicidin.
The hemolytic activity of (Δ)Pb-CATH1, -3, and -4 against chicken erythrocytes was tested as a measure of cytotoxicity (Table 4). As shown in Table 4, the tested (Δ)Pb-CATHs showed low hemolytic activity compared to AMP melittin from honey bee venom as the positive control (17). The hemolytic rate of ΔPb-CATH4, which showed the strongest antimicrobial activity, was ∼10% at 64 μg/ml.
TABLE 4.
Hemolytic activity of ΔPb-CATH1, -3, and -4 against chicken erythrocytes
| Concn (μg/ml) | Hemolytic rate ± SD (%)a |
|||
|---|---|---|---|---|
| Pb-CATH1 | Pb-CATH3 | Pb-CATH4 | Melittin | |
| 4 | 0 | 0 | 0 | 13 ± 1.1 |
| 8 | 1.4 ± 0.4 | 0 | 1.2 ± 0.3 | 71.3 ± 5.2 |
| 16 | 5.1 ± 0.5 | 1.7 ± 0.5 | 3.2 ± 0.4 | 110.3 ± 1.5 |
| 32 | 7.3 ± 0.6 | 3.1 ± 0.5 | 6.5 ± 0.4 | 114.7 ± 1.7 |
| 64 | 12.2 ± 0.7 | 4.0 ± 0.2 | 10.7 ± 0.2 | 113.4 ± 0.2 |
Hemolytic activity was measured in triplicate.
Low cellular toxicity of python cathelicidin.
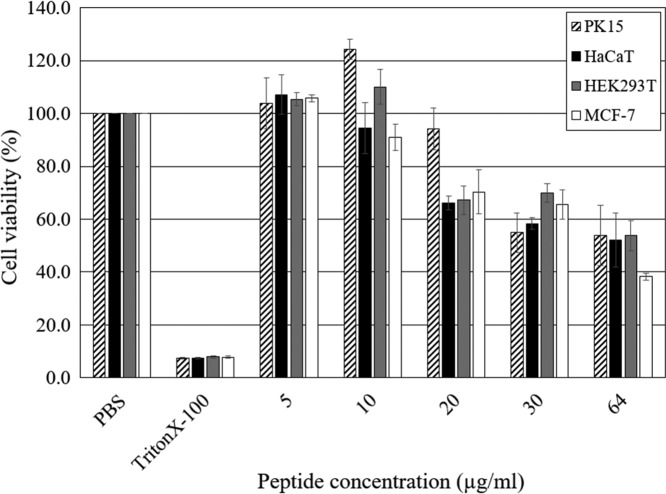
ΔPb-CATH4 showed the highest antimicrobial activity (geometric mean MIC = 2.0) against Gram-negative bacteria. In general, the cytotoxic activity of AMPs is positively correlated with antimicrobial activity (18). However, low cytotoxicity is a critical factor for consideration as a candidate for pharmaceutical use. The cytotoxicity of ΔPb-CATH4 was measured as the change in the viability of four different cell lines, porcine kidney fibroblasts, human kidney fibroblasts, human keratinocytes, and human breast cancer cells after 24 h of incubation with different concentrations (5 to 64 μg/ml) of peptide (Fig. 4). The results showed almost undetectable cytotoxicity for all cells at peptide concentrations of <10 μg/ml, which is about 4 times the MIC of ΔPb-CATH4. However, the cell viability was reduced to ∼50% at concentrations of >30 μg/ml, indicating cytotoxicity at high concentrations. One tested cancer cell line, MCF-7, did not show any difference in cytotoxicity in the presence of ΔPb-CATH4 compared to the other tested cell lines, indicating that ΔPb-CATH4 does not have any preferential effects on cancer cells.
FIG 4.
Effect of ΔPb-CATH4 on the viability of mammalian cell lines. Four different cell lines, PK15 (pig kidney fibroblasts), HaCaT (human keratinocytes), HEK293T (human kidney fibroblasts), and MCF-7 (human breast cancer cells), were treated with various concentrations of ΔPb-CATH4 (5, 10, 20, 30, and 64 μg/ml) for 24 h, and viability was assessed. PBS and Triton X-100 were used as the negative and positive controls, respectively. The data shown are the means and standard deviations from triplicate experiments.
Cell wall rupture and pore formation in ΔPb-CATH4-treated E. coli cells.
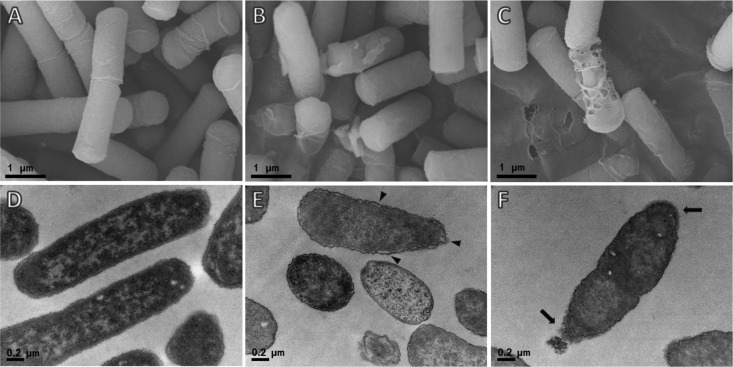
To address the mechanism underlying the antibacterial action of Pb-CATHs, we observed bacterial cells after treatment with ΔPb-CATH4 using electron microscopy. Field emission scanning electron microscopy (FE-SEM) and transmission electron microscopy (TEM) analysis of E. coli ATCC 25922 after ΔPb-CATH4 treatment showed morphological changes in the cell wall, including rupture and pore formation (Fig. 5B, C, E, and F), whereas untreated control cells showed intact cell membranes with smooth surfaces. Intracellular morphological observations of E. coli using TEM showed that ΔPb-CATH4-treated cells undergo the detachment of the outer membrane from the cytoplasmic zone, and the cytoplasm oozed out due to cell membrane breakage (Fig. 5E and F). However, control untreated cells showed dense cytoplasmic contents and clearly attached membranes.
FIG 5.
Electron micrographs showing the effects of ΔPb-CATH4 treatment on E. coli cells. Images were generated using a field emission scanning electron microscope (A, B, C) or using a transmission electron microscope (D, E, F). E. coli cells were incubated with ΔPb-CATH4 (at a final concentration of 2× MIC) for 2 h, and their cell morphology was observed by electron microscopy. (B and C) Ruptured cell walls and pores in the bacterial cell walls. (E and F) Outer membrane detachment (arrowheads) from the cytoplasmic zone and leakage of cytoplasm (arrows) induced by ΔPb-CATH4. (A and D) Untreated negative controls.
DISCUSSION
In this study, we report the genetic and biological characterization of cathelicidins in Python bivittatus. In our genome level analysis, we observed conserved signatures of AMPs in python. Expression of beta defensins and cathelicidins has been reported in reptiles (19, 20). In addition, other AMP-related proteins, including hepcidin (21), leap-2 (22), crotamine (23), leucocin (24), and omwaprin (25, 26), have been reported in diverse reptilian species. However, current evidence related to AMP expression in reptiles indicates that AMP families within the reptile class are diverse and their expression varies among reptilian species (27), which is probably due to the extreme biological diversity within the class.
Our results show that the AMP repertoire of python is quite limited compared to that of other reptilian species, as genes encoding cathelicidins were the only classical AMPs that we detected in the python genome. Interestingly, we were unable to identify any beta defensin family genes in the python genome even after analyzing the genome sequence (with 20× genome coverage) using all currently reported AMPs as queries in homology searches. However, other AMP-like protein families such as crotamines (23), flavin monoamine oxidases (28), phospholipase A2 (29), and snake waprins (26) were identified. The close relationship between beta defensins and crotamines has previously been described (30). The lack of classical defensins in the python could be due to the extreme sequence divergence of defensin-like genes.
Pb-CATH1 contains the previously reported amphipathic nonapeptide, KRFKKFFKK, which is nearly identical in all snake venom cathelin-related antimicrobial peptides (svCRAMPs), including crotalicidin, lachesicidin, batroxicidin, lutzicidin, BF-CRAMP, OH-CRAMP, NA-CRAMP, Pt-CRAMP1, and Pt-CRAMP2 (19, 31, 32) (see Fig. S2 in the supplemental material), suggesting that Pb-CATH1 is a member of a cathelicidin family common to snakes. However, the predicted mature peptide sequences of Pb-CATH3 and -4 were quite different from previously reported cathelicidin sequences. Interestingly, we identified a Pb-CATH3-like gene from the predicted cathelicidin-like sequence (XP_015669791.1), tentatively named CATH-like Pm 3 peptide, in the recently available genome of Protobothrops mucrosquamatus (Fig. S2), suggesting that Pb-CATH3 is a cathelicidin that is orthologous to peptides in other snakes.
However, we were not able to identify any Pb-CATH4-like sequences in searches against all available reptilian genome sequences. Previous reports have demonstrated the presence of an additional acidic-connecting peptide in other cathelicidins in snakes (31). Consistent with previous reports, Pb-CATH1 possesses an acidic connecting peptide. Interestingly, like mammalian cathelicidins (33, 34), Pb-CATH3 and -4 do not have this acidic domain.
ΔPb-CATH4 exhibited the most potent bactericidal activity among the Pb-CATH peptides tested in this study (Table 3). The arginine content of this peptide was 37.5% (Fig. 1), and it contains two tryptophan residues (8.3%), which have been shown to be important for strong antimicrobial activity (35). The importance of Trp and Arg has been extensively studied in indolicidin and tritrpticin. It was found that Trp preferentially interacts with the interfacial region of lipid bilayers in conjunction with Arg, which confers the cationic charges and hydrogen-bonding properties necessary for interaction with anionic lipopolysaccharide (LPS) (36, 37). These two amino acids in Pb-CATH4 participate in cation-π interactions with Trp to facilitate the entry of Arg into the hydrophobic lipid bilayer, thereby enhancing peptide-membrane interactions (38). Hence, Pb-CATH4 belongs to the Arg- and Trp-rich family of cathelicidins.
Interestingly, Pb-CATH showed activity with very high potency against S. Typhimurium, which causes typhoid and gastroenteritis in humans and animals. Pb-CATH4 also showed activity against B. cereus, which is a food pathogen causing foodborne illness. In addition, Pb-CATH4 showed a high level of activity against antibiotic-resistant clinical isolates of pathogens like E. coli and K. pneumoniae. Although our results are lacking in the number of isolates as well as in vivo and pharmacodynamics studies, Pb-CATH4 showed very promising data and could be exploited as a candidate for the development of alternatives to antibiotics.
A porcine cathelicidin, PG1, showed strong antibacterial activity against both Gram-positive and Gram-negative bacteria (39). However, while all three Pb-CATH peptides characterized in this study showed potent activity toward Gram-negative strains, only ΔPb-CATH4 showed mild antimicrobial activity against some Gram-positive bacteria (Table 2). The structure of the outer cell wall differs between Gram-positive and Gram-negative bacteria; the cell wall of Gram-negative microorganisms is largely composed of lipopolysaccharides (40), whereas the outer cell wall of Gram-positive bacteria is composed of peptidoglycan layers (41), suggesting that the Pb-CATH peptides might bind specifically to the LPS, although further studies are required to confirm this hypothesis. It has been reported that highly cationic peptides generally have a strong binding affinity for polyanionic LPS, which coincidentally inhibits the production of tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) by macrophages stimulated with LPS (42, 43), suggesting interactions between cationic peptides and LPS. The Pb-CATH peptides in our study had charges of +9 to +15 (Table 2); in comparison, other cathelicidins such as PG1, PMAP36, and LL37 have charges of +6 to +13 (44–46).
Numerous antimicrobial peptides have been shown to act by poration of the lipid membrane (47). SEM of E. coli after treatment with ΔPb-CATH4 (Fig. 5) showed cell wall disruption as well as clear holes in the cell membrane, indicating that the mechanism underlying the bactericidal activity of the peptide is toroidal pore formation. This is consistent with the results of previous studies and may contribute to the broad-spectrum activity of ΔPb-CATH4.
To our surprise, the Pb-CATH peptides showed lower cytotoxicity in a hemolytic assay than most reported cathelicidins (30). Most α-helical AMPs tend to be cytotoxic because of their high hydrophobicity (48). The ratio of hydrophobic to charged residues (H/C) for AMPs varies from 1:1 to 2:1 (41, 49), and a higher ratio is associated with lower hemolytic activity (41, 48). Pb-CATH3 exhibited just 4% hemolysis at 64 μg/ml (Table 4), and it has a H/C ratio of 1.1:1; thus, it belongs to the group with lower hydrophobic content. In general, greater hydrophobicity imparts broad-spectrum membranolytic activity (50). In BMAP-28, reducing hydrophobicity by replacing hydrophobic residues with hydrophilic analogues reduced cytotoxicity and decreased activity against Gram-positive bacteria but improved activity against Gram-negative bacteria compared to the parent peptide (51). Similarly, all the tested Pb-CATH peptides showed low cytotoxicity (Table 4), and their H/C ratios were between 1.1:1 and 1.5:1. This lower hydrophobicity may play a role in their membranolytic selectivity toward Gram-negative bacteria.
Conclusion.
We reported a genomewide analysis of the AMPs in the nonvenomous snake Python bivittatus and an evaluation of their biological activities, which led to the discovery of new reptilian cathelicidins with different structural characteristics than those of previously reported peptides. Their strong antimicrobial activity against Gram-negative bacteria, mild hemolytic activity and cytotoxicity, and considerable serum stability indicate that the Pb-CATHs characterized in this study could be promising candidates for further exploitation toward pharmaceutical applications.
MATERIALS AND METHODS
In silico identification of AMPs in the python genome.
The sequences of 2,488 nonredundant AMPs were downloaded from UniProtKB/Swiss-Prot using the query “antimicrobial peptide AND reviewed:yes.” Subsequently, 90 sequences (Table S1) belonging to Reptilia species were manually selected for further analyses. Then, the reptilian AMPs were compared against the genome and nonredundant protein sequence data sets for Python bivittatus (taxid 176946) using NCBI tblastn and blastp (http://blast.ncbi.nlm.nih.gov), respectively. Highly matched sequences (P < 0.001) were selected. Finally, the mature peptide regions in the predicted cathelicidin sequences of Python bivittatus were determined and named Pb-CATH peptides 1 to 5, where “Pb” represents Python bivittatus and “CATH” represents cathelicidin.
Phylogenetic analysis.
Multiple-sequence alignment of the conserved CLDs of 21 selected cathelicidins was carried out using MUSCLE (52). Annotations of the CLDs for the cathelicidins were obtained from the UniProt database (http://www.uniprot.org/). A phylogenetic tree was constructed using the maximum likelihood method in MEGA (version 7.0; www.megasoftware.net).
In silico analyses of putative AMP sequences.
The predicted Pb-CATH sequences in the python genome were used to identify the antimicrobial domains according to bactericidal propensity values (PV) using AMPA, an automated web server for the prediction of protein antimicrobial regions (http://tcoffee.crg.cat/apps/ampa/do). Analyses of hydrophobicity, net charge, and sequence similarities to existing AMPs were carried out using the APD3 antimicrobial peptide database (5). Subsequently, the secondary structures of Pb-CATH sequences were analyzed using the PSIPRED protein sequence analysis workbench (http://bioinf.cs.ucl.ac.uk/psipred/). Signal peptide predictions for Pb-CATH sequences were performed using SignalP4.1 (53).
Synthesis of predicted AMPs.
Truncated mature regions of the candidate AMPs ΔPb-CATH1 (N-RVKRFKKFFRKIKKGFRKIFKKTKIFIG-C), ΔPb-CATH4 (N-TRSRWRRFIRGAGRFARRYGWRIA-C), and Pb-CATH3 (N-HRVKRNGFRKFMRRLKKFFAGG-C) were generated via solid-phase peptide synthesis and purified via high-performance liquid chromatography (HPLC) using a commercial service (GeneScript, Piscataway Township, NJ, USA). Based on the secondary structure prediction, a few amino acids at the N and C termini were excluded from the predicted mature peptide sequences of Pb-CATH1 and Pb-CATH4.
Antimicrobial activity test.
Antimicrobial activity against ATCC reference strains of Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 29213), Pseudomonas aeruginosa (ATCC 27853), Salmonella Typhimurium (ATCC 14028), and Bacillus cereus (ATCC 10876; all from ATCC, Manassas, VA, USA), and E. coli and Klebsiella pneumoniae antibiotic resistance strains clinically isolated from Chonbuk National University Hospital, Jeonju, South Korea, was measured using the broth microdilution method. Ampicillin (Sigma-Aldrich, St. Louis, MO, USA) and gentamicin sulfate (Sigma-Aldrich) were used as the control. The MIC was determined using a colorimetric method with the Microbial Viability Assay kit-WST (Dojindo, Japan) according to the manufacturer's protocol. Briefly, a single colony of each bacterium was inoculated into 5 ml of Luria-Bertani (LB) medium and cultured overnight at 37°C. The overnight culture was diluted to 1 × 105 CFU/well in Mueller-Hinton (MH) broth and inoculated at 180 μl/well using fresh MH broth in a 96-well plate. Various concentrations (1 to 12 μg/ml) of the synthesized AMPs were generated by dilution with 10 μl of MH broth, added to each well, and incubated at 37°C for 6 h. Then, 10 μl of coloring reagent was added to each well, and the plate was incubated for 2 h. The plates were read at 450 nm using a microplate reader (xMark Microplate Absorbance Spectrophotometer; Bio-Rad, San Francisco, CA, USA). The MIC for Streptococcus iniae Korean Collection for Type Cultures (KCTC) 3657 was estimated by disk diffusion assay following CLSI guidelines because of its slow growth in liquid culture. Briefly, a single-colony-derived overnight culture of S. iniae (KCTC 3657) grown in brain heart infusion broth (BD Diagnostic Systems, Franklin Lakes, NJ, USA) was suspended in MH broth to a 1.0 McFarland standard (1.0 × 108 CFU/ml). The suspension was spread on cation-adjusted MH agar plates containing 5% horse blood (Hanil Komed, Seongnam, South Korea). Various concentrations (1 to 12 μg/ml) of the AMPs were applied to 6-mm paper disks (GE health care, USA). Similarly, ampicillin and gentamicin sulfate were prepared with various concentrations (0.25 to 4 μg/ml). Subsequently, the dried disks were placed on an agar plate and incubated at 37°C overnight. The diameter of the zone of inhibition was measured using Vernier calipers, and interpretation was carried out using the CLSI breakpoint guidelines.
In vitro serum stability assay.
Peptides were dissolved in 50% (vol/vol) fetal bovine serum (FBS; HyClone, Logan, UT) and incubated at 37°C. Aliquots were taken in duplicate after 30, 60, 100, and 120 min of incubation. Antimicrobial activity against E. coli (ATCC 25922) was assessed using the incubated samples as described above.
Hemolytic activity assay.
Freshly drawn chicken blood was collected in an EDTA tube, and plasma was removed by centrifugation. Erythrocytes were rinsed three times with phosphate-buffered saline (PBS; 35 mM phosphate, 150 mM NaCl, pH 7.4) and resuspended in PBS at a concentration of 2%. Then, 100 μl of the erythrocyte suspension was added to each well of a 96-well microtiter plate containing an equal volume of the synthesized python AMPs in PBS at a final concentration of 64 μg/ml. Triton X-100 (1%; Sigma-Aldrich) was used as a positive control for 100% hemolysis, and PBS was used as a negative control. Additionally, melittin (Sigma-Aldrich) from honey bee venom was used as a positive control. Plates were incubated at 37°C for 1 h. The release of hemoglobin into the supernatant was monitored as the change in absorbance at 550 nm using a microplate reader (Victor X4; Perkin-Elmer, MA, USA). Results from three independent experiments were pooled. Hemolysis was calculated as follows: hemolytic percentage (%) = 100 × (ODpeptide − ODnegative)/(ODpositive − ODnegative), where OD is the optical density.
In vitro cytotoxicity assay.
Various cell lines, including HaCaT (human keratinocytes), HEK293T (human embryonic kidney cells), MCF-7 (human mammary gland, breast cancer cells), and PK15 (pig kidney cells), were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone) containing 10% FBS (HyClone) and 1% penicillin-streptomycin (HyClone) in 5% CO2 at 37°C until 80% confluence. Cells were detached by adding Accutase (Innovative Cell Technologies, San Diego, CA, USA), and 1 × 104 to 4 × 104 cells were seeded into each well of a 96-well plate (Sigma-Aldrich). Cells were treated with serially diluted ΔPb-CATH4 (final concentrations, 5, 10, 20, 30, 40, and 64 μg/ml), and the plates were incubated for 24 h in 5% CO2 at 37°C. Triton X-100 (Sigma-Aldrich) was used as a positive control for 100% cell lysis, and untreated wells were used as a negative control. After incubation, the medium was removed from the well and 10 μl of coloring solution (Cell proliferation reagent wst-1; Sigma-Aldrich) and 100 μl of DMEM were added to the wells according to the manufacturer's protocol. Finally, the absorbances at 450 nm and 650 nm of each well were recorded with a microplate reader (xMark spectrophotometer; Bio-Rad). Cell viability was calculated as follows: cell viability (%) = (At − A0)/(Ac − A0) × 100, where At is the absorbance (at 450 nm) of the target sample, A0 is the background absorbance (at 650 nm), and Ac is the absorbance of the negative control. All experiments were carried out in triplicate.
Electron microscopy.
Cells at an OD600 of 0.2 were incubated with 2× MIC (2 μg/ml) of ΔPb-CATH4 for 2 h at 37°C. Then, the cells were collected by centrifugation at 3,500 rpm, and the pellets were washed twice with 1× PBS. Cells were fixed with 2% paraformaldehyde and 2.5% glutaraldehyde (Sigma-Aldrich) in 1× PBS overnight at 4°C. The cells were then washed three times with 1× PBS and postfixed with 1% osmium tetroxide (Sigma-Aldrich) in 1× PBS for 1.5 h at 4°C. For field emission scanning electron microscopy (FE-SEM), samples were washed with water and dehydrated with graded acetone (50%, 70%, 90%, and 100%) for 10 min each and dried with hexamethyldisilazane (Daejung Chemicals and Metals Co. Ltd., Siheung, South Korea) for 15 min. For observation, the prepared samples were sputter-coated with platinum using a Cressington sputter coater 108 auto (Cressington, Warford, UK) prior to imaging with a JEOL JSM-7500F FE-SEM (Welwyn Garden City, UK). For transmission electron microscopy (TEM), cells were washed twice with water and stained with 0.5% uranyl acetate (SPI Supplies, West Chester, PA, USA) for 30 min at 4°C. Samples were dehydrated and rinsed in propylene oxide (Sigma-Aldrich) three times for 10 min each. Cells were embedded in Spur resin (SPI supplies) according to the manufacturer's instructions. Then, 70-nm ultrathin sections were obtained using a microtome and stained with 2% uranyl acetate for 7 min. Stained sections were observed using a Hitachi H7650 TEM (Tokyo, Japan).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (116134-3) of the Ministry of Agriculture, Food, and Rural Affairs, Republic of Korea, and by the Science Research Center (2015R1A5A1009701) of the National Research Foundation of Korea, Republic of Korea. This work was also supported by the 2015 KU Brain Pool of Konkuk University.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00530-17.
REFERENCES
- 1.Park Y, Hahm KS. 2005. Antimicrobial peptides (AMPs): peptide structure and mode of action. J Biochem Mol Biol 38:507–516. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob Agents Chemother 43:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss GMH. 1962. On the venomous skin secretion of the orange speckled frog Bombina variegata. Toxicon 1:33–39. doi: 10.1016/0041-0101(62)90006-5. [DOI] [Google Scholar]
- 4.Zhao X, Wu H, Lu H, Li G, Huang Q. 2013. LAMP: a database linking antimicrobial peptides. PLoS One 8:e66557. doi: 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G, Li X, Wang Z. 2016. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin LM, Edwards MA, Li J, Yip CM, Deber CM. 2012. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J Biol Chem 287:7738–7745. doi: 10.1074/jbc.M111.303602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westerhoff HV, Juretic D, Hendler RW, Zasloff M. 1989. Magainins and the disruption of membrane-linked free-energy transduction. Proc Natl Acad Sci U S A 86:6597–6601. doi: 10.1073/pnas.86.17.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramamoorthy A, Thennarasu S, Lee DK, Tan A, Maloy L. 2006. Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys J 91:206–216. doi: 10.1529/biophysj.105.073890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopal R, Park JS, Seo CH, Park Y. 2012. Applications of circular dichroism for structural analysis of gelatin and antimicrobial peptides. Int J Mol Sci 13:3229–3244. doi: 10.3390/ijms13033229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuzaki K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta 1462:1–10. doi: 10.1016/S0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 11.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Y, Hughes AL, Ando J, Matsuda Y, Cheng JF, Skinner-Noble D, Zhang G. 2004. A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics 5:56. doi: 10.1186/1471-2164-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammami R, Fliss I. 2010. Current trends in antimicrobial agent research: chemo- and bioinformatics approaches. Drug Discov Today 15:540–546. doi: 10.1016/j.drudis.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Cavasotto CN, Phatak SS. 2009. Homology modeling in drug discovery: current trends and applications. Drug Discov Today 14:676–683. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Castoe TA, de Koning JA, Hall KT, Yokoyama KD, Gu W, Smith EN, Feschotte C, Uetz P, Ray DA, Dobry J, Bogden R, Mackessy SP, Bronikowski AM, Warren WC, Secor SM, Pollock DD. 2011. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol 12:406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. 2014. M100-S24. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI, Wayne, PA. [Google Scholar]
- 17.Tosteson MT, Holmes SJ, Razin M, Tosteson DC. 1985. Melittin lysis of red cells. J Membr Biol 87:35–44. [DOI] [PubMed] [Google Scholar]
- 18.Dempsey CE. 1990. The actions of melittin on membranes. Biochim Biophys Acta 1031:143–161. doi: 10.1016/0304-4157(90)90006-X. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Gan TX, Liu XD, Jin Y, Lee WH, Shen JH, Zhang Y. 2008. Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides 29:1685–1691. doi: 10.1016/j.peptides.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Alibardi L. 2013. Ultrastructural immunolocalization of beta-defensin-27 in granulocytes of the dermis and wound epidermis of lizard suggests they contribute to the anti-microbial skin barrier. Anat Cell Biol 46:246–253. doi: 10.5115/acb.2013.46.4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CH, Valore EV, Waring AJ, Ganz T. 2001. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 22.Sang Y, Ramanathan B, Minton JE, Ross CR, Blecha F. 2006. Porcine liver-expressed antimicrobial peptides, hepcidin and LEAP-2: cloning and induction by bacterial infection. Dev Comp Immunol 30:357–366. doi: 10.1016/j.dci.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Kerkis I, Silva Fde S, Pereira A, Kerkis A, Radis-Baptista G. 2010. Biological versatility of crotamine—a cationic peptide from the venom of a South American rattlesnake. Expert Opin Invest Drugs 19:1515–1525. doi: 10.1517/13543784.2010.534457. [DOI] [PubMed] [Google Scholar]
- 24.Yaraksa N, Anunthawan T, Theansungnoen T, Daduang S, Araki T, Dhiravisit A, Thammasirirak S. 2014. Design and synthesis of cationic antibacterial peptide based on Leucrocin I sequence, antibacterial peptide from crocodile (Crocodylus siamensis) white blood cell extracts. J Antibiot (Tokyo) 67:205–212. doi: 10.1038/ja.2013.114. [DOI] [PubMed] [Google Scholar]
- 25.Banigan JR, Mandal K, Sawaya MR, Thammavongsa V, Hendrickx AP, Schneewind O, Yeates TO, Kent SB. 2010. Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography. Protein Sci 19:1840–1849. doi: 10.1002/pro.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair DG, Fry BG, Alewood P, Kumar PP, Kini RM. 2007. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem J 402:93–104. doi: 10.1042/BJ20060318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hoek ML. 2014. Antimicrobial peptides in reptiles. Pharmaceuticals (Basel) 7:723–753. doi: 10.3390/ph7060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo C, Liu S, Yao Y, Zhang Q, Sun MZ. 2012. Past decade study of snake venom L-amino acid oxidase. Toxicon 60:302–311. doi: 10.1016/j.toxicon.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez JM, Lomonte B. 2013. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 62:27–39. doi: 10.1016/j.toxicon.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Falcao CB, de La Torre BG, Perez-Peinado C, Barron AE, Andreu D, Radis-Baptista G. 2014. Vipericidins: a novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids 46:2561–2571. doi: 10.1007/s00726-014-1801-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Chan JY, Rego JV, Chong CM, Ai N, Falcao CB, Radis-Baptista G, Lee SM. 2015. Rhodamine B-conjugated encrypted vipericidin nonapeptide is a potent toxin to zebrafish and associated with in vitro cytotoxicity. Biochim Biophys Acta 1850:1253–1260. doi: 10.1016/j.bbagen.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Hong J, Liu X, Yang H, Liu R, Wu J, Wang A, Lin D, Lai R. 2008. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS One 3:e3217. doi: 10.1371/journal.pone.0003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tossi A, Scocchi M, Zanetti M, Storici P, Gennaro R. 1995. PMAP-37, a novel antibacterial peptide from pig myeloid cells. cDNA cloning, chemical synthesis and activity. Eur J Biochem 228:941–946. [DOI] [PubMed] [Google Scholar]
- 34.Gallo RL, Kim KJ, Bernfield M, Kozak CA, Zanetti M, Merluzzi L, Gennaro R. 1997. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J Biol Chem 272:13088–13093. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- 35.Chan DI, Prenner EJ, Vogel HJ. 2006. Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta 1758:1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Killian JA, Salemink I, de Planque MR, Lindblom G, Koeppe RE II, Greathouse DV. 1996. Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane alpha-helical peptides: importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry 35:1037–1045. doi: 10.1021/bi9519258. [DOI] [PubMed] [Google Scholar]
- 37.Jing W, Demcoe AR, Vogel HJ. 2003. Conformation of a bactericidal domain of puroindoline a: structure and mechanism of action of a 13-residue antimicrobial peptide. J Bacteriol 185:4938–4947. doi: 10.1128/JB.185.16.4938-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yau WM, Wimley WC, Gawrisch K, White SH. 1998. The preference of tryptophan for membrane interfaces. Biochemistry 37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 39.Steinberg DA, Hurst MA, Fujii CA, Kung AH, Ho JF, Cheng FC, Loury DJ, Fiddes JC. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother 41:1738–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lugtenberg B, Van Alphen L. 1983. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim Biophys Acta 737:51–115. doi: 10.1016/0304-4157(83)90014-X. [DOI] [PubMed] [Google Scholar]
- 41.Tossi A, Sandri L, Giangaspero A. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4–30. [DOI] [PubMed] [Google Scholar]
- 42.Scott MG, Yan H, Hancock RE. 1999. Biological properties of structurally related alpha-helical cationic antimicrobial peptides. Infect Immun 67:2005–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gough M, Hancock RE, Kelly NM. 1996. Antiendotoxin activity of cationic peptide antimicrobial agents. Infect Immun 64:4922–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scocchi M, Zelezetsky I, Benincasa M, Gennaro R, Mazzoli A, Tossi A. 2005. Structural aspects and biological properties of the cathelicidin PMAP-36. FEBS J 272:4398–4406. doi: 10.1111/j.1742-4658.2005.04852.x. [DOI] [PubMed] [Google Scholar]
- 45.Sayyed-Ahmad A, Kaznessis YN. 2009. Determining the orientation of protegrin-1 in DLPC bilayers using an implicit solvent-membrane model. PLoS One 4:e4799. doi: 10.1371/journal.pone.0004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J 341(Part 3):501–513. [PMC free article] [PubMed] [Google Scholar]
- 47.Leontiadou H, Mark AE, Marrink SJ. 2006. Antimicrobial peptides in action. J Am Chem Soc 128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 48.Blondelle SE, Houghten RA. 1992. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry 31:12688–12694. doi: 10.1021/bi00165a020. [DOI] [PubMed] [Google Scholar]
- 49.Pane K, Durante L, Crescenzi O, Cafaro V, Pizzo E, Varcamonti M, Zanfardino A, Izzo V, Di Donato A, Notomista E. 2017. Antimicrobial potency of cationic antimicrobial peptides can be predicted from their amino acid composition: application to the detection of “cryptic” antimicrobial peptides. J Theor Biol 419:254–265. doi: 10.1016/j.jtbi.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. 2007. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother 51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skerlavaj B, Gennaro R, Bagella L, Merluzzi L, Risso A, Zanetti M. 1996. Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J Biol Chem 271:28375–28381. doi: 10.1074/jbc.271.45.28375. [DOI] [PubMed] [Google Scholar]
- 52.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.