ABSTRACT
Contamination of environmental waters by extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli (ESBLEC) is of great concern. Wastewater treatment plants (WWTPs) and hospitals release large amounts of ESBLEC into the environment. In the present study, we isolated ESBLEC strains from wastewater collected from a WWTP and a hospital in Japan and performed whole-genome sequencing to characterize these strains. Genomic analysis of 54 strains (32 from the WWTP and 22 from hospital wastewater) revealed the occurrence of clinically important clonal groups with extraintestinal pathogenic E. coli status in the WWTP and hospital wastewater. Fine-scale phylogenetic analysis was performed to further characterize 15 sequence type 131 (ST131) complex strains (11 from the WWTP and 4 from hospital wastewater). These ST131 complex strains were comprised of the following different subgroups: clade A (n = 2), C1-M27 (n = 8), and C1 (non-C1-M27) (n = 1) for strains from the WWTP and clade A (n = 2), C1-M27 (n = 1), and C1 (non-C1-M27) (n = 1) for strains from hospital wastewater. The results indicate that ESBLEC strains belonging to clinically important lineages, including the C1-M27 clade, may disseminate into the environment through wastewater, highlighting the need to monitor for antibiotic resistance in wastewater.
KEYWORDS: ESBL, Escherichia coli, wastewater, whole-genome sequencing
INTRODUCTION
The occurrence of antibiotic-resistant bacteria in the environment increases global health risks. Of great concern is extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli (ESBLEC) because some E. coli strains are pathogenic (1) and treatment options are limited for ESBLEC infections (2). Human intestinal carriage of ESBLEC is well documented in community and clinical settings (3, 4). Therefore, wastewater treatment plants (WWTPs) and hospitals are considered to be major sources of ESBLEC released into the environment (5). In fact, the presence of ESBLEC in hospital wastewater and the inflow and outflow of WWTPs has been well documented in previous studies (6–8). However, data are limited with respect to detailed genetic characteristics of ESBLEC in wastewater.
Clinical ESBLEC isolates have often been characterized at the sequence type (ST) level by using multilocus sequence typing (MLST), and some pandemic clonal lineages of ESBLEC have been identified in this way. Among them, a clonal lineage of ST131 is of particular concern because this clone is the predominant lineage among drug-resistant extraintestinal pathogenic E. coli (ExPEC) strains worldwide (9, 10). Recent studies based on whole-genome sequencing showed that ST131 can be divided into three clades, namely, A/H41, B/H22, and C/H30 (11, 12). H41, H22, and H30 indicate the fimH (type 1 fimbrial adhesin gene) allele type, and most strains belonging to each clade are known to carry the corresponding fimH allele. Clade C contains C0, C1, and C2, which can be defined based on the positions in the whole-genome phylogeny (13). Previous studies reported that the blaCTX-M-15-harboring C2/H30Rx is highly responsible for the pandemic of ExPEC strains that carry ESBLs (9, 11, 12). Recently, we described a novel ST131 C1 subclade with blaCTX-M-27, named C1-M27, by analysis of clinical ST131 strains, and we determined that this subclade is prevalent among Japanese ESBL-producing ST131 isolates and is also contributing to the global spread of ST131 (14). Importantly, some previous studies have detected ESBLEC belonging to ST131 in wastewater (15, 16). Dolejska et al. (15) detected CTX-M-15-producing E. coli strains belonging to B2-O25b-ST131 in treated wastewater, and Colomer-Lluch et al. (16) reported the presence of CTX-M-15-producing O25b:H4-B2-ST131 strains in raw urban sewage. However, those studies did not perform a whole-genome single nucleotide polymorphism (SNP)-based phylogenetic analysis. In fact, limited information exists regarding the fine-scale phylogeny of environmental ST131 strains. In particular, the reports describing E. coli belonging to C1-M27 are almost entirely restricted to clinical isolates (14). Detection and characterization of environmental E. coli strains belonging to this clade are needed to better understand the molecular epidemiology and reservoirs of the C1-M27 clade.
In the present study, we performed whole-genome sequencing and analysis of ESBLEC isolated from wastewater collected in Japan to examine the genetic characteristics of the detected strains and determine the presence of clinically important ESBLEC lineages, including the C1-M27 clade, in wastewater. A whole-genome approach was adopted because it enables us to obtain comprehensive information on genetic characteristics, such as virulence gene profiles, antibiotic resistance determinants, and fine-scale phylogeny.
RESULTS AND DISCUSSION
Detection and isolation of ESBLEC.
During the study period, we collected 10 samples from the WWTP and 10 samples from hospital wastewater. All samples tested positive for E. coli and ESBLEC (see Table S1 in the supplemental material for concentrations of total E. coli and ESBLEC CFU in each sample). The average proportion of ESBLEC CFU among total E. coli CFU was 4.2% (minimum, 2.3%; maximum, 9.3%) for the WWTP samples, which is relatively high compared to previous studies (0.4% to 2.3%) (7). Conversely, the average proportion was 3.5% (minimum, 0.2%; maximum, 11.3%) for the hospital wastewater, and this value is relatively low compared to previous studies (3.8% to 13.6%) (7). Wastewater treatment processes can increase the proportion of resistant bacteria because the presence of antibiotics used in human medicine in wastewater poses selective pressures and the high cell density sustained by a nutrient-rich environment can promote the transfer of antibiotic resistance genes (5, 17). These may be reasons for the relatively high proportion of ESBLEC CFU in the WWTP samples.
In total, 32 strains from the WWTP and 29 strains from the hospital wastewater were isolated. Four strains were identified as Citrobacter freundii (genomic analysis also confirmed this), and three strains were identified as redundant strains (i.e., strains isolated from the same sample, belonging to the same ST, and carrying the same antimicrobial resistance genes). Therefore, we removed these seven strains, leaving 32 strains from the WWTP and 22 strains from the hospital wastewater for further analysis (see Data Set S1 in the supplemental material for information on these 54 strains).
Phylogenetic analysis.
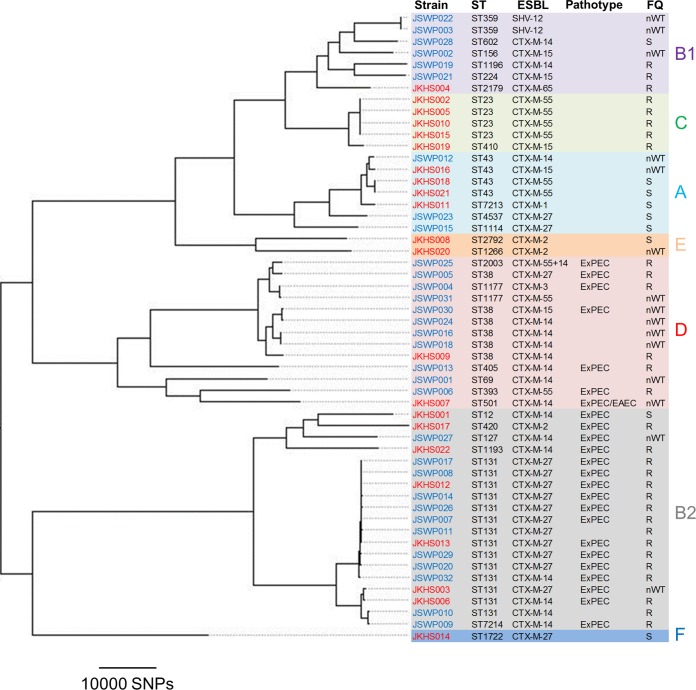
A whole-genome SNP-based tree was constructed using kSNP3 (Fig. 1). ESBLEC strains are scattered throughout the phylogenetic tree, representing seven phylogenetic groups (A, B1, B2, C, D, E, and F). MLST identified 28 STs, including two novel STs (ST7213 and ST7214). ST131 (n = 10) was the most prevalent ST among the WWTP isolates. One WWTP isolate belonged to ST7214, which is a single-locus variant (SLV) of ST131. ST131 (n = 4) and ST23 (n = 4) were the most prevalent STs among the strains obtained from the hospital wastewater. Clonal overlaps for ST38, ST43, and ST131 were observed between the WWTP and hospital wastewater isolates, suggesting that ESBLEC belonging to these STs may be prevalent in both community and clinical settings. It should be noted that we did not collect ESBLEC randomly from each plate but selected colonies based on their morphologies to represent the genetic diversity of ESBLEC strains in a sample. Therefore, the actual clonal compositions of ESBLEC isolates in the wastewater samples may differ from those that we observed in the present study.
FIG 1.
Phylogeny of 54 ESBLEC strains. A parsimony tree was constructed based on SNP loci occurring in at least 50% of the strains. The tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Colors of the strain names reflect the isolation sources, i.e., blue for the WWTP and red for hospital wastewater. Phylogenetic groups (A, B1, B2, C, D, E, and F) are indicated with different colors. The FQ column indicates ciprofloxacin susceptibilities for each isolate (S, susceptible; nWT, non-wild type according to the ECOFF criteria; R, resistant according to the clinical breakpoint).
Pathotyping of the 54 strains revealed that 24 (44.4%) strains had ExPEC status. These ExPEC strains belonged to phylogenetic groups B2 and D, which is congruent with observations that these phylogenetic groups are associated with human extraintestinal infections (18). Furthermore, these ExPEC strains included those belonging to clinically important clonal groups, such as ST12, ST38, ST127, ST131, ST393, and ST405 (10), posing a public health concern. One of the 24 ExPEC strains, JKHS007, also had enteroaggregative E. coli (EAEC) status due to possession of aggR. A previous study reported human-derived E. coli belonging to ST38 with characteristics of both uropathogenic E. coli and EAEC (19). JKHS007 belonged to ST501, which is associated with EAEC from human sources, according to EnteroBase. Further analysis of sequence data revealed that JKHS007 carried ExPEC-associated genes, such as sitABCD (iron/manganese transport) as well as iutA and kpsM II (two of the five key markers defining ExPEC). The cooccurrence of ExPEC and EAEC-associated genes in JKHS007 indicates potential emergence of an ExPEC/EAEC hybrid pathotype in wastewater.
Phenotypic and genotypic resistance.
Phenotypic resistance was determined by microdilution. Nonsusceptibility/non-wild-type rates ranged from 0% (amoxicillin-clavulanic acid, piperacillin-tazobactam, imipenem, meropenem, amikacin, colistin, and fosfomycin) to 100% (ampicillin, cefazolin, cefpodoxime, cefotaxime, cefepime, and aztreonam) (see Fig. S1 in the supplemental material). The nonsusceptibile/non-wild-type rate for each antibiotic was similar between the WWTP isolates and the hospital wastewater isolates. However, we did not calculate the statistical significance because our ESBLEC collection was possibly biased due to the nonrandom selection of colonies as noted above.
Even though care should be taken in interpreting the results, the proportion of strains resistant to quinolones was quite high among our 54 ESBLEC strains (81.5% for nalidixic acid and 55.6% for ciprofloxacin according to the clinical breakpoints). Remarkably, 19 (79.2%) strains with ExPEC status were resistant to ciprofloxacin (Fig. 1), which is partly due to the predominance of strains belonging to ST131 clade C (this point is further discussed in the following section). All of the strains resistant to nalidixic acid carried at least one mutation in the quinolone resistance-determining region (QRDR), and all but two strains resistant to ciprofloxacin carried two mutations in the QRDR of gyrA and at least one mutation in the QRDR of parC, which is congruent with our previous study (20). The remaining two ciprofloxacin-resistant isolates carried one or two mutations in QRDRs in combination with quinolone resistance genes, such as aac(6′)-Ib-cr, qnrS1, qnrS2, oqxA, and oqxB.
All of the ESBLEC strains carried ESBL genes, and no plasmid-mediated AmpC genes or chromosomal ampC promoter/attenuator mutations that can result in ampC overexpression were detected. Among our ESBLEC strains, blaCTX-M-14 (n = 18) was the most prevalent followed by blaCTX-M-27 (n = 15) and blaCTX-M-55 (n = 9). One isolate carried both blaCTX-M-14 and blaCTX-M-55. Importantly, 45 (83.3%) strains carried genes conferring resistance to non-β-lactam antibiotics. Cross-resistance of ESBLEC to other classes of antibiotics is of particular concern. A high level of cross-resistance in environmental ESBLEC was also observed in previous studies (21, 22). This cross-resistance may be partly due to the coexistence of ESBL genes with other resistance genes on the same plasmids (23). Detection of ESBL genes, other resistance genes, and plasmid replicons in the same contig would support this hypothesis, but this was hampered by the short read length in the present study.
Further characterization of ST131 complex strains.
ESBLEC strains belonging to ST131 and its SLV were further analyzed to determine the ST131 clade/subclade of each strain. To gain insights into the fine-scale phylogeny of ST131 complex (STC131) strains, we constructed a core parsimony tree using 15 STC131 strains detected in the present study and 61 STC131 strains analyzed in our previous study (14). Strain SE15 was also included as a reference strain for clade A. kSNP3 identified 8,897 core SNPs in 77 strains. The constructed tree is shown in Fig. 2. The tree comprises three clades (A, B, and C) and four subgroups (C0, C1, C1-M27, and C2) within clade C, findings consistent with previous studies (13, 14). Among the STC131 strains detected in the present study, four strains belonged to clade A and 11 strains belonged to clade C. Congruent with previous studies, strains belonging to clade C were characterized by the fimH30 allele and LNIV (S83L and D87N in GyrA and S80I and E84V in ParC) genotypes in QRDRs. Importantly, 9 of 11 clade C strains were placed in the C1-M27 clade in the phylogenetic tree. Because clade A strains lack some of the genomic regions shared among clade B and clade C strains, we also constructed a core SNP-based tree by excluding clade A strains to increase the number of informative SNP sites (see Fig. S2 in the supplemental material). The strains assigned to each ST131 clade/subclade in Fig. S2 were the same as those in Fig. 2, confirming the placement of the nine strains in the C1-M27 clade. All of these strains carried the C1-M27 clade-specific prophage-like region (M27PP1) (14) and C1-M27 clade-unique SNPs that we identified in a recent study (24). Based on these results, these nine strains (eight from the WWTP and one from hospital wastewater) were defined as belonging to the C1-M27 clade. Strains belonging to subclade C2, which is largely responsible for the global pandemic of ESBL-carrying ExPEC, were not detected in the present study. Interestingly, our recent study showed that the C1-M27 clade accounted for 38.7% of Japanese ESBL-producing ST131 isolates and was the most prevalent ST131 clade among these isolates (24). It should be noted that this clade is also present among clinical ST131 isolates from Thailand, Australia, Canada, and the United States (14), and a recent study also showed the prevalence of the C1-M27 clade in the fecal carriage of children in France (25), indicating contribution of this clade to the global spread of ST131. Although our ESBLEC collection may not represent the actual clonal compositions of ESBLEC in wastewater, the prevalence of this clade among the collection is a concern. As is the case with most clinical C1-M27 isolates, all of the C1-M27 isolates detected in the present study carried blaCTX-M-27, and eight of these isolates were typed as virotype C. We further investigated the presence of ExPEC-associated virulence genes in clinical and environmental C1-M27 strains (see Fig. S3 in the supplemental material). Results indicate that the environmental C1-M27 strains carry similar (or even the same) sets of virulence genes compared to clinical C1-M27 strains, although the environmental strains tended to lack certain virulence genes, such as papB. Further studies are needed to assess the virulence potential of environmental C1-M27 strains.
FIG 2.
Core SNP-based phylogenetic tree of STC131 strains. Colors of the strain names reflect the isolation sources, i.e., blue for the WWTP, red for hospital wastewater, and black for those analyzed in our previous study and SE15. Most strains in black were clinical isolates. IEH71520 was obtained from vacuum cleaner dust collected from the home of a case patient (50). SE15 was obtained from the feces of a healthy adult (51). S100EC was obtained from a rectal swab in a prospective study of returned travelers (12, 52). The QRDR column indicates amino acids of GyrA codons 83 and 87 and ParC codons 80 and 84. The ST131 clades/subclades are shown in different colors. JSWP007 carried a fimH allele that is different from fimH30 by one nucleotide. NT, nontypeable.
In conclusion, genomic analysis of ESBLEC strains from the WWTP and hospital wastewater revealed the presence of clinically important clones among ESBLEC strains. The apparent high prevalence of fluoroquinolone resistance among these ESBLEC strains is of great concern. Core SNP-based phylogenetic analysis revealed the presence of the C1-M27 clade among our STC131 strains. This study highlights the need to monitor for antibiotic-resistant bacteria in wastewater.
MATERIALS AND METHODS
Sample collection and isolation of ESBLEC.
The municipal WWTP and hospital from which the wastewater samples were collected are in the Kansai region of Japan. In October 2015, 10 samples per location were collected on different occasions from WWTP and hospital wastewater. Samples from the WWTP were collected from effluent from the final settling tanks after biological (activated sludge) treatment. Samples from the hospital consisted of untreated wastewater. A detailed description of the sampling procedure is provided in the supplemental material. Samples were processed using the membrane filter method with XM-G agar (Nissui, Tokyo, Japan) for enumeration of total E. coli and with chromID ESBL agar (bioMérieux, Lyon, France) for enumeration and isolation of ESBLEC. For each sample, up to four colonies showing an E. coli profile on the chromID ESBL plate were selected (care was taken to select colonies showing different morphologies if possible). In total, 32 and 29 isolates were obtained from the WWTP and hospital wastewater, respectively. The isolates were stored at −85°C in 35% glycerol.
Species identification and antibiotic susceptibility testing.
Species identification was performed with the matrix-assisted laser desorption ionization (MALDI) Biotyper Compass 4.1 (Bruker Daltonics GmbH, Bremen, Germany). Antibiotic susceptibility testing was performed by the microdilution method using the dry plate Eiken assay (Eiken, Tokyo, Japan) according to CLSI guidelines (26). The susceptibility testing included the following 25 antibiotics: ampicillin, amoxicillin-clavulanic acid, ampicillin-sulbactam, piperacillin-tazobactam, cefazolin, cefpodoxime, cefotaxime, ceftazidime, cefepime, cefoxitin, imipenem, meropenem, aztreonam, nalidixic acid, ciprofloxacin, gentamicin, tobramycin, amikacin, kanamycin, trimethoprim-sulfamethoxazole, tetracycline, minocycline, chloramphenicol, colistin, and fosfomycin. Results were interpreted according to the current epidemiological cutoff (ECOFF) values (http://mic.eucast.org/Eucast2/) as well as 2015 CLSI criteria (26). Intermediate susceptibility to each antibiotic was considered to be resistance. ESBL production was confirmed following the CLSI guidelines (26). We used cefotaxime, ceftazidime, and cefpodoxime disks with and without clavulanate (Eiken) for the confirmatory test (27).
Genome sequencing and assembly.
DNA was extracted from each isolate using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany), and sequencing libraries were prepared using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA). Each library was sequenced on an Illumina MiSeq instrument for 600 cycles (300-bp paired-end reads) to achieve an average sequencing depth of 80. Raw reads from each sample were trimmed using ERNE-FILTER (28) and assembled using SPAdes v3.10.0 (29). Assemblies were improved using Pilon (30).
Genomic analysis.
Assembled contigs were subjected to the following analysis. Acquired resistance genes were detected using the ResFinder antimicrobial resistance gene database (31) and the ARG-ANNOT database (32). Chromosomal ampC promoter/attenuator mutations that can result in ampC overexpression were analyzed as previously described (33). Mutations in quinolone resistance-determining regions (QRDRs) of gyrA and parC were analyzed according to Aoike et al. (34). Plasmid replicons were detected using PlasmidFinder (35). MLST in silico was performed using the MLST web tool (36) and EnteroBase (http://enterobase.warwick.ac.uk). Phylogenetic groups were determined as described previously (20). The fimH allele types, which enable further discrimination of strains belonging to the same ST, were determined according to the classification system established previously (37). Parsimony trees based on SNPs in whole-genome data were constructed using kSNP3 (38, 39). Virulence genes were detected using an in-house database compiled from the Virulence Factors Database (VFDB) (40) and by literature review (41–47). E. coli pathotypes were defined based on the presence of specific virulence genes (48). ST131 virotypes were determined as described previously (49).
Accession number(s).
Sequence data obtained in the present study have been deposited in the DDBJ Sequence Read Archive database under DDBJ accession number DRA005619.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Kyoto University Research Funds for Young Scientists.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00564-17.
REFERENCES
- 1.Erb A, Sturmer T, Marre R, Brenner H. 2007. Prevalence of antibiotic resistance in Escherichia coli: overview of geographical, temporal, and methodological variations. Eur J Clin Microbiol Infect Dis 26:83–90. doi: 10.1007/s10096-006-0248-2. [DOI] [PubMed] [Google Scholar]
- 2.Pitout JD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Bano J, Lopez-Cerero L, Navarro MD, Diaz de Alba P, Pascual A. 2008. Faecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli: prevalence, risk factors and molecular epidemiology. J Antimicrob Chemother 62:1142–1149. doi: 10.1093/jac/dkn293. [DOI] [PubMed] [Google Scholar]
- 4.Stromdahl H, Tham J, Melander E, Walder M, Edquist PJ, Odenholt I. 2011. Prevalence of faecal ESBL carriage in the community and in a hospital setting in a county of Southern Sweden. Eur J Clin Microbiol Infect Dis 30:1159–1162. doi: 10.1007/s10096-011-1202-5. [DOI] [PubMed] [Google Scholar]
- 5.Brechet C, Plantin J, Sauget M, Thouverez M, Talon D, Cholley P, Guyeux C, Hocquet D, Bertrand X. 2014. Wastewater treatment plants release large amounts of extended-spectrum beta-lactamase-producing Escherichia coli into the environment. Clin Infect Dis 58:1658–1665. doi: 10.1093/cid/ciu190. [DOI] [PubMed] [Google Scholar]
- 6.Korzeniewska E, Korzeniewska A, Harnisz M. 2013. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol Environ Saf 91:96–102. doi: 10.1016/j.ecoenv.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Hocquet D, Muller A, Bertrand X. 2016. What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93:395–402. doi: 10.1016/j.jhin.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Ojer-Usoz E, Gonzalez D, Garcia-Jalon I, Vitas AI. 2014. High dissemination of extended-spectrum beta-lactamase-producing Enterobacteriaceae in effluents from wastewater treatment plants. Water Res 56:37–47. doi: 10.1016/j.watres.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 11.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura Y, Pitout JD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradford PA, Motyl MR, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolejska M, Frolkova P, Florek M, Jamborova I, Purgertova M, Kutilova I, Cizek A, Guenther S, Literak I. 2011. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J Antimicrob Chemother 66:2784–2790. doi: 10.1093/jac/dkr363. [DOI] [PubMed] [Google Scholar]
- 16.Colomer-Lluch M, Mora A, Lopez C, Mamani R, Dahbi G, Marzoa J, Herrera A, Viso S, Blanco JE, Blanco M, Alonso MP, Jofre J, Muniesa M, Blanco J. 2013. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J Antimicrob Chemother 68:758–765. doi: 10.1093/jac/dks477. [DOI] [PubMed] [Google Scholar]
- 17.Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. 2013. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 18.Kohler CD, Dobrindt U. 2011. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301:642–647. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Chattaway MA, Jenkins C, Ciesielczuk H, Day M, DoNascimento V, Day M, Rodriguez I, van Essen-Zandbergen A, Schink AK, Wu G, Threlfall J, Woodward MJ, Coldham N, Kadlec K, Schwarz S, Dierikx C, Guerra B, Helmuth R, Mevius D, Woodford N, Wain J. 2014. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg Infect Dis 20:1935–1937. doi: 10.3201/eid2011.131845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. 2017. Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl Environ Microbiol 83:e02703-16. doi: 10.1128/AEM.02703-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zurfluh K, Hachler H, Nuesch-Inderbinen M, Stephan R. 2013. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol 79:3021–3026. doi: 10.1128/AEM.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaak H, de Kruijf P, Hamidjaja RA, van Hoek AH, de Roda Husman AM, Schets FM. 2014. Prevalence and characteristics of ESBL-producing E. coli in Dutch recreational waters influenced by wastewater treatment plants. Vet Microbiol 171:448–459. doi: 10.1016/j.vetmic.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Gniadkowski M. 2001. Evolution and epidemiology of extended-spectrum beta-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect 7:597–608. doi: 10.1046/j.1198-743x.2001.00330.x. [DOI] [PubMed] [Google Scholar]
- 24.Matsumura Y, Pitout JDD, Peirano G, DeVinney R, Noguchi T, Yamamoto M, Gomi R, Matsuda T, Nakano S, Nagao M, Tanaka M, Ichiyama S. Rapid identification of different Escherichia coli ST131 clades. Antimicrob Agents Chemother, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birgy A, Bidet P, Levy C, Sobral E, Cohen R, Bonacorsi S. 2017. CTX-M-27-producing Escherichia coli of sequence type 131 and clade C1-M27, France. Emerg Infect Dis 23:885. doi: 10.3201/eid2305.161865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Fujita S, Yosizaki K, Ogushi T, Uechi K, Takemori Y, Senda Y. 2011. Rapid identification of gram-negative bacteria with and without CTX-M extended-spectrum beta-lactamase from positive blood culture bottles by PCR followed by microchip gel electrophoresis. J Clin Microbiol 49:1483–1488. doi: 10.1128/JCM.01976-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. 2013. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 8:e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peter-Getzlaff S, Polsfuss S, Poledica M, Hombach M, Giger J, Bottger EC, Zbinden R, Bloemberg GV. 2011. Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J Clin Microbiol 49:2924–2932. doi: 10.1128/JCM.00091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoike N, Saga T, Sakata R, Yoshizumi A, Kimura S, Iwata M, Yoshizawa S, Sugasawa Y, Ishii Y, Yamaguchi K, Tateda K. 2013. Molecular characterization of extraintestinal Escherichia coli isolates in Japan: relationship between sequence types and mutation patterns of quinolone resistance-determining regions analyzed by pyrosequencing. J Clin Microbiol 51:1692–1698. doi: 10.1128/JCM.03049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 42.Johnson JR, Stell AL, Scheutz F, O'Bryan TT, Russo TA, Carlino UB, Fasching C, Kavle J, Van Dijk L, Gaastra W. 2000. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun 68:1587–1599. doi: 10.1128/IAI.68.3.1587-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco M, Schumacher S, Tasara T, Zweifel C, Blanco JE, Dahbi G, Blanco J, Stephan R. 2005. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joffre E, von Mentzer A, Abd El Ghany M, Oezguen N, Savidge T, Dougan G, Svennerholm AM, Sjoling A. 2015. Allele variants of enterotoxigenic Escherichia coli heat-labile toxin are globally transmitted and associated with colonization factors. J Bacteriol 197:392–403. doi: 10.1128/JB.02050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joffre E, von Mentzer A, Svennerholm AM, Sjoling A. 2016. Identification of new heat-stable (STa) enterotoxin allele variants produced by human enterotoxigenic Escherichia coli (ETEC). Int J Med Microbiol 306:586–594. doi: 10.1016/j.ijmm.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Vidal M, Kruger E, Duran C, Lagos R, Levine M, Prado V, Toro C, Vidal R. 2005. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol 43:5362–5365. doi: 10.1128/JCM.43.10.5362-5365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomi R, Matsuda T, Fujimori Y, Harada H, Matsui Y, Yoneda M. 2015. Characterization of pathogenic Escherichia coli in river water by simultaneous detection and sequencing of 14 virulence genes. Environ Sci Technol 49:6800–6807. doi: 10.1021/acs.est.5b00953. [DOI] [PubMed] [Google Scholar]
- 49.Mora A, Dahbi G, Lopez C, Mamani R, Marzoa J, Dion S, Picard B, Blanco M, Alonso MP, Denamur E, Blanco J. 2014. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One 9:e87025. doi: 10.1371/journal.pone.0087025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kutumbaka KK, Han S, Mategko J, Nadala C, Buser GL, Cassidy MP, Beldavs ZG, Weissman SJ, Morey KE, Vega R, Samadpour M. 2014. Draft genome sequence of blaNDM-1-positive Escherichia coli O25b-ST131 clone isolated from an environmental sample. Genome Announc 2:e00462-14. doi: 10.1128/genomeA.00462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toh H, Oshima K, Toyoda A, Ogura Y, Ooka T, Sasamoto H, Park SH, Iyoda S, Kurokawa K, Morita H, Itoh K, Taylor TD, Hayashi T, Hattori M. 2010. Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J Bacteriol 192:1165–1166. doi: 10.1128/JB.01543-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers BA, Kennedy KJ, Sidjabat HE, Jones M, Collignon P, Paterson DL. 2012. Prolonged carriage of resistant E. coli by returned travellers: clonality, risk factors and bacterial characteristics. Eur J Clin Microbiol Infect Dis 31:2413–2420. doi: 10.1007/s10096-012-1584-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.