ABSTRACT
The emergence of fluoroquinolone (FQ)-resistant mutants of Legionella pneumophila in infected humans was previously reported using a next-generation DNA sequencing (NGS) approach. This finding could explain part of the therapeutic failures observed in legionellosis patients treated with these antibiotics. The aim of this study was to develop digital PCR (dPCR) assays allowing rapid and accurate detection and quantification of these resistant mutants in respiratory samples, especially when the proportion of mutants in a wild-type background is low. We designed three dPCRgyrA assays to detect and differentiate the wild-type and one of the three gyrA mutations previously described as associated with FQ resistance in L. pneumophila: at positions 248C→T (T83I), 259G→A (D87N), and 259G→C (D87H). To assess the performance of these assays, mixtures of FQ-resistant and -susceptible strains of L. pneumophila were analyzed, and the results were compared with those obtained with Sanger DNA sequencing and real-time quantitative PCR (qPCR) technologies. The dPCRgyrA assays were able to detect mutated gyrA sequences in the presence of wild-type sequences at up to 1:1,000 resistant/susceptible allele ratios. By comparison, Sanger DNA sequencing and qPCR were less sensitive, allowing the detection of gyrA mutants at up to 1:1 and 1:10 ratios, respectively. When testing 38 respiratory samples from 23 legionellosis patients (69.6% treated with an FQ), dPCRgyrA detected small amounts of gyrA mutants in four (10.5%) samples from three (13.0%) patients. These results demonstrate that dPCR is a highly sensitive alternative to quantify FQ resistance in L. pneumophila, and it could be used in clinical practice to detect patients that could be at higher risk of therapeutic failure.
KEYWORDS: digital PCR, antibiotic resistance, fluoroquinolones, gyrA, Legionella pneumophila
INTRODUCTION
Legionella pneumophila, a Gram-negative, facultative, intracellular bacterium, is the causative agent of legionellosis, a severe pneumonia associated with mortality rates ranging from 5% to 25% (1). The diagnosis of this disease mainly relies on a urinary antigen test and culture and PCR testing of respiratory samples. The first-line drugs for the treatment of legionellosis are the macrolides and the fluoroquinolones (FQ) (2). In vitro antibiotic susceptibility testing of Legionella species strains is currently not recommended on a routine basis, especially because no standardized method is available from the CLSI (Clinical and Laboratory Standards Institute) or the EUCAST (European Committee on Antimicrobial Susceptibility Testing). However, therapeutic failures and relapses are still reported (3–5), FQ-resistant mutants of L. pneumophila have been readily selected in vitro (6–9), and FQ-resistant mutants have also recently been found in legionellosis patients (10, 11). Indeed, Shadoud et al. recently characterized the in vivo selection of such FQ-resistant mutants of L. pneumophila in two legionellosis patients treated with these antibiotics (11).
Bacterial resistance to quinolones of clinical relevance is most often related to mutations in the genes encoding type II topoisomerases, also called DNA gyrase (encoded by gyrA and gyrB genes) and type IV topoisomerase (encoded by parC and parE genes) (12). Resistance-conferring mutations occur within specific regions of these genes, called the quinolone resistance-determining regions (QRDRs). The in vitro selection of L. pneumophila strains with high-level resistance to quinolones has been mainly related to mutations affecting codons 83 and 87 of the gyrA QRDR (7–9), although mutations in gyrB and parC were also described (9). Only mutations at codon position 83 of the gyrA gene [gyrA(83)] have been reported so far in vivo (10, 11). Using a next-generation DNA sequencing (NGS) approach, Shadoud et al. previously demonstrated that an L. pneumophila mutant population harboring the 248C→T (T83I) mutation was rapidly selected in vivo in two legionellosis patients treated with an FQ, representing from 1.05% of the total L. pneumophila lung population at the time of diagnosis up to 94% after a few days of FQ treatment (11). It can be hypothesized that a higher proportion of mutated gyrA sequences at the time of legionellosis diagnosis may represent a higher risk of FQ treatment failure. Also, the selection of the gyrA-mutated population by the administration of an FQ may occur faster when the proportion of mutants over wild-type strains is high at the time of treatment onset.
The prevalence of the in vivo selection of L. pneumophila FQ-resistant mutants in legionellosis patients treated with these antibiotics remains to be established. It cannot be determined by traditional phenotypic methods for several reasons. (i) Legionella cultures are negative in most legionellosis patients because of the fastidious nature of this bacterium. (ii) L. pneumophila isolation is even more difficult to obtain in patients under antibiotic treatment, even when patients experience treatment failure or relapses. (iii) A mixture of antibiotic-susceptible and a few resistant strains of L. pneumophila would certainly result in selecting the most susceptible population for antibiotic susceptibility testing. (iv) It is necessary to evaluate the respective proportion of such susceptible and resistant populations to evaluate the potential risk of treatment failure.
Digital PCR (dPCR) is an innovative PCR technology developed in the 2000s based on the partition of the sample to be analyzed into thousands to millions of individual PCRs. A major advantage of dPCR over real-time quantitative PCR (qPCR) is an increased sensitivity for the detection of a few mutants mixed with wild-type DNA sequences (13). dPCR has wide clinical applications in the oncology field (14) and ongoing applications in noninvasive prenatal diagnosis (15, 16) and organ transplant rejection monitoring (17). For infectious diseases, dPCR technology has been mainly used for the diagnosis of viral diseases (13), the quantification of bacteria in clinical samples (18–21), the detection and quantification of antibiotic resistance genes (22, 23), and the detection and quantification of bacterial toxins (24).
The aim of the present study was to develop dPCRgyrA assays for the detection and quantification of FQ-resistant mutants of L. pneumophila in respiratory samples collected from legionellosis patients.
RESULTS
Sanger sequencing of the gyrA QRDR of the mutant-LPP mixtures.
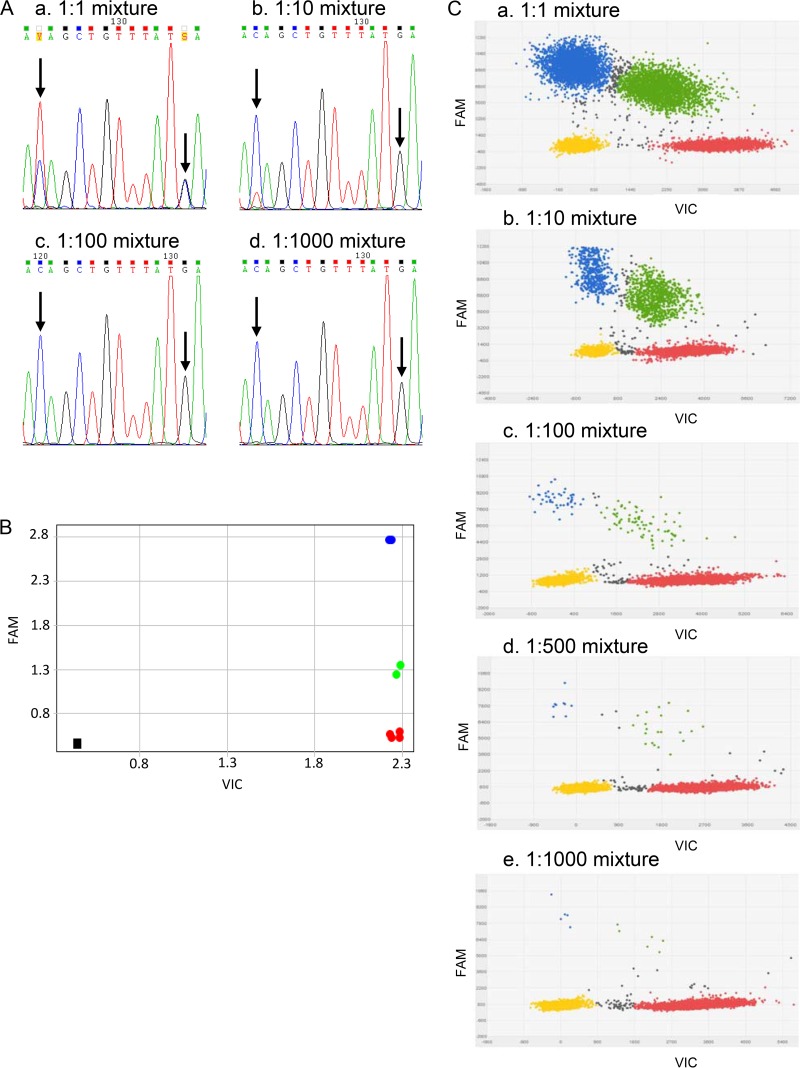
To assess the performance of Sanger sequencing, mixtures of FQ-resistant and -susceptible strains of L. pneumophila were analyzed. The mutated gyrA QRDR sequences were all detected at a 1:1 mutant/LPP (L. pneumophila serogroup 1 wild-type strain) ratio. By contrast, the mutated gyrA sequences were only occasionally detected at a 1:10 ratio and were not detected at 1:100 and 1:1,000 ratios. Figure 1A shows the sequencing chromatograms of the gyrA QRDR sequences for the D87H LPP mutant (LPPI5)-LPP mixtures. Similar results were obtained for the T83I LPP mutant (LPPI1)-LPP and the D87N LPP mutant (LPPI4)-LPP mixtures (data not shown). The Sanger sequencing sensitivity was evaluated at a 1:1 ratio (50% of mutated gyrA sequences).
FIG 1.
Comparison of Sanger sequencing, qPCRgyrA, and dPCRgyrA for mutant sequence detection in the LPPI5-LPP mixtures. (A) Sanger sequencing for the LPPI5-LPP mixtures: a, 1:1 mixture; b, 1:10 mixture; c, 1:100 mixture; d, 1:1,000 mixture. In the LPPI5-LPP 1:1 mixture, C/T and G/C heterozygous peaks were detected. In the LPPI5-LPP 1:10 mixture, a weak T peak was associated with the C peak, but the second mutation was not detected. For the 1:100 and 1:1,000 mixtures, sequencing chromatograms corresponded to the wild-type sequence. (B) qPCRgyrA allelic discrimination plot for the LPPI5-LPP mixtures. qPCRgyrA detected mutant sequences for 1:1 (in blue) and 1:10 mixtures (in green). The 1:100 (in red) and 1:1,000 (in red) mixtures were qualified as wild-type by qPCRgyrA. Black square is NTC. (C) dPCRgyrA scatter plots for the LPPI5:LPP mixtures: a, 1:1 mixture; b, 1:10 mixture; c, 1:100 mixture; d, 1:500 mixture; e, 1:1,000 mixture. Yellow dots represent wells with no amplification signal. Red dots represent wells with the VIC signal (corresponding to the wild-type gyrA QRDR sequences). Blue dots represent wells with the FAM signal (corresponding to mutated gyrA QRDR sequences). Green dots represent wells with the FAM plus VIC signal. Gray dots represent undetermined reaction wells. dPCRgyrA detected mutant sequences in up to 1:1,000 mixture.
Validation of the qPCRgyrA assays.
The results of the experiments conducted for the validation of the three qPCRgyrA assays are shown in the supplemental material (Fig. S1, S2, and S3). The three assays were highly specific for their targets, although weak fluorescence signals were detected with the wild-type and mutant probes in the absence of the corresponding targets for the qPCRgyrA-D87N assay and with the wild-type probe for the qPCRgyrA-T83I assay. An adjustment of the thresholds allowed us to circumvent these nonspecific reactions. No signal was obtained with the no-template controls (NTCs).
Study of the mutant-LPP mixtures using the qPCRgyrA assays.
To assess the performance of qPCRgyrA, mixtures of FQ-resistant and -susceptible strains of L. pneumophila were analyzed. The presence of mutated gyrA QRDR sequences was detected at 1:1 and 1:10 mutant/LPP ratios. By contrast, the mixtures at the 1:100 and 1:1,000 ratios were reported as wild-type DNA sequences by qPCRgyrA testing. The results for the LPPI5-LPP mixtures are shown in Fig. 1B. Similar results were obtained for the LPPI1-LPP and LPPI4-LPP mixtures (data not shown). The qPCRgyrA sensitivity was evaluated at a 1:10 ratio (10% of mutated gyrA sequences).
Validation of the dPCRgyrA assays.
The results of the experiments conducted for the validation of the dPCRgyrA assays are shown in Table S3. As expected, dPCRgyrA quantified on average 1,000 target copies/μl. The three assays were highly specific for their targets, although the weak cross-hybridization reactions detected for qPCRgyrA assays were also visible in dPCRgyrA experiments as slight background noise. For the dPCRgyrA-T83I assay, the background noise was observed only for the mutant probe at 0.43 target copies/μl. For the dPCRgyrA-D87N assay, the background noise was observed with mutant and wild-type probes up to 0.69 target copies/μl. For the dPCRgyrA-D87H assay, the background noise was also observed with mutant and wild-type probes up to 1.27 target copies/μl. This small background noise was circumvented by the fixation of thresholds above these nonspecific signals. No signal was observed with the NTCs.
Study of the mutant-LPP mixtures using the dPCRgyrA assays.
To assess the performance of dPCRgyrA, mixtures of FQ-resistant and -susceptible strains of L. pneumophila were analyzed. The percentages of mutated gyrA QRDR sequences in the mutant-LPP mixtures were calculated using the formula: mutant percentage = [mutant copies/(mutant copies + wild copies)] × 100. dPCRgyrA was able to detect the presence of mutated gyrA QRDR sequences at up to 1:1,000 mutant/LPP ratios. Taking into account the previously defined thresholds, the dPCRgyrA sensitivity was evaluated at a 1:1,000 ratio (0.1% of mutated gyrA sequences) for the dPCRgyrA-T83I and dPCRgyrA-D87N assays, and at a 1:500 ratio (0.2% of mutated gyrA sequences) for the dPCRgyrA-D87H assay. The results are shown in Table 1 for all the mixtures and in Fig. 1C for the LPPI5-LPP mixtures. Similar results were obtained for the LPPI1-LPP and LPPI4-LPP mixtures (data not shown).
TABLE 1.
dPCRgyrA results for the mutant-LPP mixturesa
| Mutant strain (mutation) | Mutant/LPP ratio | No. of copies/μl |
Mean % of mutants (SD) | |||||
|---|---|---|---|---|---|---|---|---|
| Wild type (VIC) |
Mutant (FAM) |
|||||||
| Chip 1 | Chip 2 | Mean (SD) | Chip 1 | Chip 2 | Mean (SD) | |||
| LPPI1 (T83I) | 1:1 | 660.31 | 482.67 | 571.49 (125.61) | 655.22 | 651.94 | 653.58 (2.32) | 53.63 (5.41) |
| 1:10 | 1,601.80 | 1,550.60 | 1,576.20 (36.20) | 119.84 | 129.20 | 124.52 (6.62) | 7.33 (0.52) | |
| 1:100 | 1,393.90 | 1,385.60 | 1,389.75 (5.87) | 9.88 | 6.87 | 8.38 (2.13) | 0.60 (0.15) | |
| 1:500 | 1,534.70 | 1,369.30 | 1,452.00 (116.96) | 4.83 | 2.31 | 3.57 (1.78) | 0.24 (0.10) | |
| 1:1,000 | 1,249.90 | 1,325.00 | 1,287.45 (53.10) | 0.83 | 1.27 | 1.05 (0.31) | 0.08 (0.02) | |
| LPPI4 (D87N) | 1:1 | 434.52 | 432.43 | 433.48 (1.48) | 365.80 | 353.98 | 359.89 (8.36) | 45.36 (0.49) |
| 1:10 | 1,075.90 | 1,091.20 | 1,083.55 (10.82) | 81.92 | 80.50 | 81.21 (1.00) | 6.97 (0.14) | |
| 1:100 | 1,353.00 | 1,303.70 | 1,328.35 (34.86) | 9.88 | 8.43 | 9.16 (1.03) | 0.68 (0.06) | |
| 1:500 | 1,412.70 | 1,450.50 | 1,431.60 (26.73) | 1.83 | 2.66 | 2.25 (0.59) | 0.16 (0.04) | |
| 1:1,000 | 1,462.10 | 1,418.80 | 1,440.45 (30.62) | 0.94 | 0.87 | 0.91 (0.05) | 0.06 (0.002) | |
| LPPI5 (D87H) | 1:1 | 633.68 | 650.10 | 641.89 (11.61) | 510.10 | 533.82 | 521.96 (16.77) | 44.84 (0.35) |
| 1:10 | 1,306.80 | 1,299.30 | 1,303.05 (5.30) | 98.79 | 95.77 | 97.28 (2.14) | 6.95 (0.12) | |
| 1:100 | 1,099.90 | 1,284.50 | 1,192.20 (130.53) | 7.89 | 8.45 | 8.17 (0.40) | 0.68 (0.04) | |
| 1:500 | 1,189.40 | 1,112.90 | 1,151.15 (54.09) | 2.30 | 1.82 | 2.06 (0.34) | 0.18 (0.02) | |
| 1:1,000 | 1,311.20 | 1,282.70 | 1,296.95 (20.15) | 1.33 | 0.87 | 1.10 (0.33) | 0.08 (0.02) | |
The results of each chip are presented, followed by the means and the standard deviations (SDs) from the duplicates.
Validation of the dPCRgyrA-T83I assay for application to human respiratory samples.
dPCRgyrA-T83I assay specificity was evaluated by testing DNA extracts from other bacterial species and Legionella-negative respiratory samples. The dPCRgyrA-T83I assay showed no cross-reactivity against any of the non-Legionella strains or against the non-pneumophila Legionella species, except a weak signal with the L. parisiensis strain. No cross-reactivity was observed with the culture-positive but Legionella-free respiratory samples.
The linearity and interpretative criteria of a dPCRgyrA-T83I result were determined by testing a range of an L. pneumophila DNA concentrations (from 1.9 to 19,000 copies/μl as determined with the NanoDrop 2000c spectrophotometer) using the dPCRgyrA-T83I assay and Legionella sp. qPCR16S and L. pneumophila qPCRmip tests. The results from experiments conducted to determine the linearity and interpretative criteria of the dPCRgyrA-T83I assay are presented in Table S4. First, for the L. pneumophila DNA concentration at 1.9 DNA copies/μl as determined using the NanoDrop spectrophotometer, the dPCRgyrA-T83I assay quantified 0.49 copies of Legionella DNA/μl. This concentration corresponded to the background noise of the dPCRgyrA-T83I assay (which was evaluated as 0.43 target copies/μl). For this concentration, the Legionella sp. qPCR16S and L. pneumophila qPCRmip quantification cycle (Cq) values were nearly 36 cycles. Consequently, to obtain a dPCRgyrA-T83I signal above the background noise, we decided that only clinical samples with a Legionella sp. qPCR16S or L. pneumophila qPCRmip Cq of less than or equal to 35 cycles would be considered appropriate for dPCRgyrA-T83I testing. Moreover, we considered that, because the proportion of mutant sequences in a mixed mutant–wild-type population of L. pneumophila can be very low, a minimum total DNA concentration would be necessary to obtain a reliable evaluation of the percentage of mutant sequences. Therefore, we decided to set the minimum total DNA concentration required at 4 total DNA copies/μl (as determined by dPCRgyrA-T83I) to obtain an accurate analysis of the percentage of L. pneumophila gyrA mutants.
Correlation between dPCRgyrA and NGS results for respiratory samples.
The dPCRgyrA-T83I assay was applied to DNA samples obtained from six legionellosis patients previously studied with NGS (11). In this previous work, NGS identified the in vivo selection of a T83I mutation in two patients (patients #2 and #4 [patient designations are from reference 11]), with proportions of mutated sequences ranging from 1.05% to 94%. dPCRgyrA confirmed the T83I mutation in all the samples collected from these two patients, in proportions ranging from 0.38% to 99.67%. Unfortunately, there was not a sufficient amount of material remaining from the sample collected from patient #2 at day 4 (the day after legionellosis diagnosis) to be tested by dPCR. For the four remaining patients, NGS found lower percentages of T83I mutations, ranging from 0.023% to 0.19%, which were considered nonsignificant (11). dPCRgyrA did not find any T83I mutations in these four patients. Table 2 compares the results obtained with the NGS and dPCRgyrA methods.
TABLE 2.
Concordance between dPCRgyrA and NGS assays for clinical samples
| Patient no. | Sampling daya | FQ treatment | % of T83I mutants from: |
|
|---|---|---|---|---|
| NGSb | dPCRgyrA-T83I | |||
| #2 | 0 | No | 2.9 | 1.68 |
| 4 | Yes | 94 | NPc | |
| #4 | 0 | Yes | 1.05 | 0.38 |
| 2 | Yes | NP | 81.65 | |
| 3 | Yes | 75 | NP | |
| 5 | Yes | 85 | 99.67 | |
| Negative for NGS (n = 4) | 0 | No | 0.023 to 0.19 | 0 |
Sampling day is given according to the day of legionellosis diagnosis.
As determined by Shadoud et al. (11).
NP, not performed.
Analysis of respiratory samples from other legionellosis patients.
Using the dPCRgyrA-T83I assay, 38 respiratory samples collected from 23 severe legionellosis patients were retrospectively analyzed. These samples corresponded to the previously defined selection criteria for a dPCRgyrA analysis, namely, a positive Legionella sp. qPCR16S or L. pneumophila qPCRmip test with a Cq of less than or equal to 35 cycles.
dPCRgyrA gave a significant amplification signal (DNA concentration higher than 4 copies/μl, as previously defined) for 34 (89.5%) samples from 20 (87.0%) patients. The results of the dPCR for these samples are presented in Table 3. For three samples, mutant sequences in proportions less than those of the dPCRgyrA-T83I assay sensitivity (0.1%) and/or background noise (0.43 FAM copies/μl) were found; these results were considered uninterpretable. For four samples collected in three patients, low percentages (0.13 to 1.13%) of T83I mutations were found. These three patients were treated with levofloxacin at the dose of 500 mg twice a day from the day of legionellosis diagnosis.
TABLE 3.
dPCRgyrA-T83I assay results for clinical samples with a significant amplification signal
| Patient no. (n = 20) | No. of respiratory samples (n = 34) |
Sampling daya | % of T83I mutants | |
|---|---|---|---|---|
| FQ | No FQ | |||
| 4–20 | 19 | 7 | 0 or uninterpretableb | |
| 1 | 1 | 0 | 0 | |
| 1 | 10 | 0 | ||
| 1 | 15 | 0 | ||
| 1 | 22 | 0.13 | ||
| 2 | 1 | 1 | 0 | |
| 1 | 2 | 0.43 | ||
| 1 | 5 | 1.13 | ||
| 3 | 1 | 5 | 0.83 | |
Sampling day is given according to the day of legionellosis diagnosis.
Three samples with mutant sequences in proportions inferior to dPCRgyrA-T83I assay sensitivity (0.1%) and/or background noise (0.43 FAM copies/μl).
DISCUSSION
It has commonly been assumed that L. pneumophila could not develop FQ resistance. However, Bruin et al. recently reported the isolation from a legionellosis patient of an L. pneumophila strain displaying a ciprofloxacin MIC of 2 mg/liter, higher than the epidemiological cutoff (1 mg/liter) they previously defined (10). Although no FQ resistance breakpoint has been currently established for Legionella, this strain was considered FQ-resistant because it was associated with a poor response to the antibiotic therapy and a prolonged hospitalization of the patient.
More recently, using an NGS approach, Shadoud et al. showed the in vivo selection of an FQ-resistant subpopulation of this pathogen in two legionellosis patients (11). Indeed, NGS allowed the detection over time of increasing percentages (from 1.05% to 94%) of gyrA(83) mutations in respiratory samples from these legionellosis patients (11). In these two patients, the most probable situation was the presence of a low level of resistant mutants that could have been missed by phenotypic tests but that dramatically expanded after the administration of an FQ, leading to therapeutic failure (25). Defining ciprofloxacin resistance from molecular data is challenging. In this study, we considered FQ-resistant L. pneumophila strains harboring gyrA(83) mutations for three main reasons. First, these mutants were previously shown to display an 8-fold increase in ciprofloxacin MIC in comparison with their wild-type parental strain (9). Second, the strain isolated by Bruin et al. (10) also harbored a gyrA(83) mutation. Third, such a mutation is well known to be associated with high-level FQ resistance in many other Gram-negative bacterial species (12, 26, 27) and occurs at a high rate (12).
Consequently, it is of clinical interest to assess the presence of FQ-resistant mutants in an L. pneumophila population infecting a given legionellosis patient to adapt the antibiotic therapy. However, standard culture-based antibiotic susceptibility testing methods are not appropriate for L. pneumophila because of the fastidious nature of this pathogen. Therefore, the development of molecular techniques allowing the detection of FQ-resistant subpopulations of L. pneumophila in respiratory samples is warranted. Shadoud et al. previously demonstrated the usefulness of NGS for the detection, identification, and quantification of specific gyrA mutants within an L. pneumophila population (11). However, NGS technology remains expensive, and data interpretation is complex and time-consuming. This technique is not yet used on a routine basis in bacteriology laboratories. By contrast, dPCR is less expensive, easier to perform, and can also detect and quantify rare mutant DNA sequences in a mixed mutant–wild-type population.
In this work, we developed dPCRgyrA assays to detect and quantify FQ-resistant subpopulations of L. pneumophila in respiratory samples. As a proof of concept, three dPCRgyrA assays targeting the gyrA QRDR mutations previously characterized as responsible for FQ resistance in L. pneumophila were developed (8, 9, 11). The dPCRgyrA-T83I assay targeted the 248C→T mutation (T83I substitution), a mutation systematically observed in evolved FQ-resistant clones selected in vitro by Almahmoud et al. (9). It was also described in vitro by Jonas et al. (8) and in vivo in two legionellosis patients by Shadoud et al. (11). The 248C→T mutation was found alone or combined with other mutations in gyrA at codon position 87 (9). Thus, we designed two dPCRgyrA assays, the dPCRgyrA-D87N assay and the dPCRgyrA-D87H assay, targeting the mutations 259G→A and 259G→C, respectively.
A validation of the three dPCRgyrA assays showed high sensitivities. dPCRgyrA was able to detect gyrA mutant sequences mixed with wild-type sequences up to mutant/susceptible allele ratios of 1:1,000 (0.1%). By comparison, the Sanger sequencing and qPCR tests were less sensitive, because they detected mutant sequences only at 1:1 (50%) and 1:10 (10%) ratios, respectively. Consequently, the Sanger sequencing and qPCR tests may not be able to detect antibiotic-resistant subpopulations of L. pneumophila in the early stage of infection in legionellosis patients. The dPCRgyrA sensitivity levels observed in this study are similar to those previously reported by Pholwat et al. for the detection and quantification of isoniazid [katG(315) mutation], rifampin [rpoB(531) mutation], FQ [gyrA(94,95) mutations], and aminoglycoside [rrs(1401) mutation] resistance in Mycobacterium tuberculosis (sensitivity from 1% to 0.1%) (23). In the literature, the maximum sensitivity reported for dPCR is 0.001% (one mutant sequence for 100,000 wild-type sequences) (28–30). Such a high sensitivity may be obtained with a higher number of individual reactions (e.g., more than 1,000,000 individual reactions) and/or a different dPCR technique (droplet dPCR versus dPCR on a chip). dPCRgyrA could be considered a highly accurate method, because the percentages of mutant sequences quantified in the mixtures were very similar to the theoretical proportions. dPCRgyrA also produced highly reproducible results, since the quantitations of mutant sequences between the two chip replicates were very similar.
Currently, the resistance to FQ in L. pneumophila strains infecting humans has only been associated with gyrA(83) mutations (10, 11). Therefore, we applied the dPCRgyrA-T83I assay to respiratory samples collected from legionellosis patients. Additional validation steps for the dPCRgyrA-T83I assay were performed beforehand. Specificity was assessed by testing bacterial species belonging to genera other than Legionella and Legionella species other than L. pneumophila. No cross-amplification was observed, except a slight cross-reactivity with L. parisiensis. However, only three cases of human infection with this species have been reported so far in the literature (31–33). We also determined that only respiratory samples with Legionella sp. qPCR16S or L. pneumophila qPCRmip Cq values of 35 cycles or less were considered eligible for a dPCRgyrA analysis because of a sufficient DNA amount. In addition, we considered 4 total DNA copies/μl as a threshold for reliable interpretation of the percentage of gyrA mutant sequences.
We then applied the dPCRgyrA-T83I assay to clinical samples from legionellosis patients previously studied with NGS (11). dPCRgyrA confirmed the in vivo selection of an FQ-resistant mutant in patient #4, with 0.38% T83I mutants at day 0 (D0) (versus 1.05% for NGS) and 99.67% at D5 (versus 85% for NGS).
We then retrospectively analyzed 38 clinical samples from 23 legionellosis patients using the dPCRgyrA-T83I assay. No sample harbored a majority of gyrA T83I mutants. This result was not unexpected, because FQ resistance selection in L. pneumophila seems a rare phenomenon (11) and most respiratory samples were collected early in the course of legionellosis for the majority of these patients, precluding visualization of in vivo selection of FQ-resistant mutants under FQ therapy. However, small amounts of gyrA(83) mutants (0.13 to 1.68%) were detected in four (10.5%) samples from three (13.0%) patients. Although higher than the cutoffs determined, these low percentages of mutant gyrA sequences must be interpreted with caution, because they may correspond to false-positive results of the dPCRgyrA assays, as well as for NGS (11). Alternatively, they may correspond to the presence of a low level of FQ-resistant mutants in the Legionella population infecting these patients. We only studied clinical samples collected on a routine basis for diagnostic purposes. The possibility of collecting several respiratory samples in legionellosis patients under FQ therapy would make it possible to better monitor the kinetics of the percentage of mutants. Indeed, an increase in the percentage of gyrA mutants would indicate an in vivo emergence of FQ-resistant mutants of L. pneumophila, and could be an alert to the possibility of the occurrence of a therapeutic failure. A demonstration was conducted with patient #4, for which dPCRgyrA detected 0.38% T83I mutants at D0 and 81.65% at D2, suggesting that the low level of mutant strain at D0 likely reflected true FQ-resistant mutants rather than false-positive results. This legionellosis patient might have been infected with a mixed L. pneumophila population containing a low level of T83I mutants, or these mutations might have occurred rapidly after infection.
The incidence of in vivo selection of FQ resistance in L. pneumophila is probably underestimated given that only a single FQ-resistant strain of L. pneumophila has been isolated so far in the clinical setting (10). dPCR exhibits a sensitivity level as good as that of NGS, but it is more adapted for routine diagnostic use. It is an inexpensive, easy-to-handle, rapid technology, and data interpretation is simple. Consequently, a good strategy for the characterization of antibiotic resistance in fastidious microorganisms would be to use NGS for the identification of the gene mutations involved, and then to develop dPCR tests to detect these resistances more easily. Although not found by Shadoud et al. (11) using NGS, it would be particularly interesting to look for the gyrA 247A→G mutation (T83A) previously described by Bruin et al. for the FQ-resistant strain isolated in a respiratory sample (10). Similarly, other mutations in DNA gyrase [e.g., gyrA(83), gyrA(87), and gyrB] and in topoisomerase IV-encoding genes should be searched for using dPCR, although they were only described in vitro in L. pneumophila (7–9). It would then be possible to develop multiplex dPCR assays to detect the most common mutations responsible for FQ resistance. Indeed, multiplex dPCR assays have already been developed, for example, in cancerology for KRAS mutations (34).
In conclusion, we developed dPCRgyrA assays to detect and quantify FQ-resistant subpopulations of L. pneumophila harboring mutations in the gyrA QRDR. We demonstrated that dPCR is a really powerful tool displaying high sensitivity, reproducibility, and precision and that this technique is suitable for daily practice in bacteriology laboratories. The major clinical interest of this technique was highlighted by confirming the in vivo emergence of FQ-resistant mutants of L. pneumophila in respiratory samples from a legionellosis patient, as previously characterized using NGS. The dPCR approach would be useful to set up a multicenter study of a larger cohort of patients to further define the prevalence of FQ-resistant mutants in legionellosis patients and evaluate their potential clinical impact. In case gyrA mutations in L. pneumophila are associated with a worsened prognosis, dPCR would be a useful diagnostic tool to predict the effectiveness of the FQ therapy in legionellosis patients.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The L. pneumophila strains used in this study are listed in Table 4. The L. pneumophila sg 1 strain Paris CIP107629T (referred to as LPP) was used as an FQ-susceptible control. The FQ-resistant mutants LPPI1, LPPI4, and LPPI5 were derived from LPP by homologous recombination introducing the three main gyrA mutations previously selected and characterized in vitro, including the mutations 248C→T (T83I) for LPPI1, 248C→T and 259G→A (D87N) for LPPI4, and 248C→T and 259G→C (D87H) for LPPI5 (9).
TABLE 4.
L. pneumophila strains used in this study
| Strain | GyrA mutation description |
FQ susceptibility (moxifloxacin MIC [mg/liter]b) | Reference or source | |
|---|---|---|---|---|
| Amino acid | Nucleotide | |||
| LPPa | Wild type | Wild type | Susceptible (0.0625) | NRC for Legionella (Lyon, France) |
| LPPI1 | GyrA83 (T83I) | 248C→T | Resistant (0.5) | 9 |
| LPPI4 | GyrA83 (T83I) plus GyrA87 (D87N) | 248C→T plus 259G→A | Resistant (0.5) | 9 |
| LPPI5 | GyrA83 (T83I) plus GyrA87 (D87H) | 248C→T plus 259G→C | Resistant (0.5) | 9 |
LPP, L. pneumophila serogroup 1 strain Paris CIP107629T.
According to Almahmoud et al. (9).
For specificity purposes, 20 reference or clinical strains belonging to bacterial species other than Legionella sp. (see Table S1 in the supplemental material) and 19 strains of Legionella species other than L. pneumophila, provided by the French National Reference Center (NRC) for Legionella (see Table S2), were used.
Legionella strains were grown on BMPA medium (Oxoid, Cambridge, UK), corresponding to buffered charcoal yeast extract medium plus antimicrobial agents, at 37°C in a 5% CO2-enriched atmosphere for 3 days. FQ-resistant Legionella strains were grown in a biosafety level 3 laboratory. The other bacterial species were grown on Columbia agar supplemented with 5% sheep blood or on chocolate agar supplemented with 5% PolyVitex (bioMérieux, Marcy l'Etoile, France) at 37°C in a 5% CO2-enriched atmosphere for 24 h.
Patients and clinical samples.
First, eight respiratory samples from six patients, previously studied with NGS (11), were analyzed by dPCR. These samples came from a previous single-center cohort of legionellosis patients admitted to an intensive care unit at Grenoble University Hospital (France) between 2006 and 2011. Samples from two patients that were previously detected as positive for T83I FQ-resistant mutants of L. pneumophila using NGS served as positive controls (11). NGS-negative samples from four patients served as negative controls (11). These controls were used for the validation of the dPCRgyrA assay developed targeting the T83I mutation.
Then, 38 respiratory samples from 23 patients with unknown NGS status were retrospectively evaluated for the presence of the T83I mutation. These patients were admitted to an intensive care unit at Grenoble University Hospital or Lyon University Hospital (France) between 2011 and 2016. They included 20 male patients and three female patients, with a mean age of 56 years (range, 23 to 83 years). During hospitalization, lower respiratory tract (LRT) (i.e., bronchoalveolar lavage fluid, bronchial aspirations, tracheal aspirations, or sputum) and urine samples were collected for diagnostic purposes. LRT samples were tested using routine bacterial cultures, Legionella cultures, and Legionella qPCR tests (qPCR16S for all Legionella species and qPCRmip for L. pneumophila, as previously described [35]) and then frozen at −80°C until used for this study. The 23 patients were selected because of positive Legionella qPCR tests (qPCR16S and qPCRmip) (see Results), while 21 patients also had positive urinary antigen tests for L. pneumophila sg 1, and 18 patients had positive cultures allowing isolation of L. pneumophila sg 1. All these patients were considered to be infected with an L. pneumophila strain because of the positive qPCRmip tests on LRT samples. Sixteen (69.6%) of the 23 patients were treated with an FQ.
In addition, to validate the clinical use of the dPCRgyrA targeting the T83I mutation, 10 respiratory samples from patients suffering from pneumonia other than legionellosis were used. These samples were negative by Legionella qPCR tests (qPCR16S and qPCRmip), while commensal and pathogenic bacterial species were detected by routine bacterial cultures.
The clinical sample collections of Grenoble and Lyon University Hospitals were declared to the French Ministry of Education and Research (numbers DC-2008-677 and DC-2008-176, respectively), and an ethics committee (CPP Sud-Est V) gave the authorization to use them for research purposes. Patient information was provided through a hospital medical booklet, and only nonopposition on the part of the patients was needed.
DNA extraction.
For bacterial strains, colonies were harvested and suspended in Tris-EDTA buffer solution (Sigma-Aldrich, St. Louis, MO, USA). For FQ-resistant Legionella strains, bacteria were inactivated by heating at 95°C for 15 min before DNA extraction. DNA was extracted using the QIAamp DNA minikit (Qiagen, Courtaboeuf, France), according to the manufacturer's recommendations.
DNA concentrations of LPP, LPPI1, LPPI4, and LPPI5 were assessed using Qubit fluorometric quantitation (Thermo Fisher Scientific, Carlsbad, CA, USA) according to the manufacturer's recommendations and adjusted to 1 ng/μl. Then mixtures of DNA from LPPI1, LPPI4, or LPPI5 with DNA from the wild-type LPP strain were prepared at different mutant/LPP ratios, namely, 1:1, 1:10, 1:100, 1:500, and 1:1,000.
DNA concentrations of suspensions of strains belonging to non-Legionella species and Legionella species other than L. pneumophila were assessed using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific), according to the manufacturer's recommendations.
For the respiratory samples, DNA extraction was performed using either the EZ1 DNA tissues kit (Qiagen) or the MagNA Pure compact nucleic acid isolation kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturers' instructions.
Sanger sequencing of the L. pneumophila gyrA QRDR.
The mutant-LPP mixtures were tested using a PCR-sequencing approach to assess the sensitivity of this technique for detecting mutated gyrA QRDR sequences in the presence of wild-type sequences.
A 383-bp fragment of the gyrA QRDR was amplified using the primers LgyrALubS7 (5′-GCGATGAGTGTCATTGTAG-3′) and LgyrALubASR2bis (5′-GTTTCATCATAGTTAGGGCTAAAATCAAC-3′) as previously described (11). Sequencing was performed using the same primers and the dye terminator method with the DTCS quick start kit (Beckman Coulter, Villepinte, France) on a CEQ 8000 sequencer (Beckman Coulter) as previously described (9). Amplicons were sequenced from both ends. The sequences obtained were visualized and analyzed with Chromas Lite (version 2.1.1; Technelysium) and aligned with Seaview4 (36).
Primers and probes for qPCRgyrA and dPCRgyrA.
We followed the MIQE guidelines for nomenclature and qPCR protocol development (37). For qPCRgyrA and dPCRgyrA experiments, primers and probes targeting the wild-type and mutated sequences of the gyrA QRDR were designed using the custom TaqMan assay design tools (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA). Three assays were developed to detect the wild-type gyrA sequence and the gyrA QRDR mutations previously selected in vitro in L. pneumophila (9), including 248C→T (T83I), 259G→A (D87N), and 259G→C (D87H).
Primers and probes were provided by Life Technologies. Each assay mix contained PCR primers and two hydrolysis MGB probes, including a VIC-labeled probe complementary to the wild-type gyrA QRDR sequence and a FAM-labeled probe complementary to one of the three mutated gyrA QRDR sequences (Table 5).
TABLE 5.
Primers and probes for qPCRgyrA and dPCRgyrA experiments
| Assay target | Primer or probea | Sequence (5′→3′)b |
|---|---|---|
| T83I | T83I-F | ATGTCATCGGTAAATACCATCCTCAC |
| T83I-R | GGTTGGGCCATACGAACAATG | |
| T83I-wt | VIC-AAACAGCTGTATCCCC | |
| T83I-mt | FAM-AAACAGCTATATCCCC | |
| D87N | D87N-F | GTCATCGGTAAATACCATCCTCACG |
| D87N-R | CGATTAAAAGGTAGCGCATGGAAAA | |
| D87N-wt | VIC-CAATGGTGTCATAAACA | |
| D87N-mt | FAM-ACAATGGTGTTATAAACA | |
| D87H | D87H-F | GTCATCGGTAAATACCATCCTCACG |
| D87H-R | CGATTAAAAGGTAGCGCATGGAAAA | |
| D87H-wt | VIC-CAATGGTGTCATAAACA | |
| D87H-mt | FAM-CAATGGTGTGATAAACA |
wt, wild-type; mt, mutant.
Targeted nucleotides are underlined.
qPCRgyrA validation and study of the mutant-LPP mixtures.
Validation of the three qPCRgyrA assays for the specific detection of the wild-type and mutated (T83I, D87N, or D87H) gyrA QRDR sequences was performed by testing the LPP, LPPI1, LPPI4, and LPPI5 strains with the three assays. Each PCR run included NTCs to validate the absence of false-positive results caused by exogenous DNA contamination, autohydrolysis of the probes, or nonspecific hybridization.
Then, the mutant-LPP mixtures (at 1:1, 1:10, 1:100, and 1:1,000 ratios) were tested using the qPCRgyrA assays, in duplicate, to assess the sensitivity of this technique to detect mutated gyrA QRDR sequences in the presence of wild-type sequences.
Each of the PCR mixtures (25 μl) contained 12.5 μl of TaqMan genotyping master mix (Life Technologies, Thermo Fisher Scientific), 0.625 μl of 40× TaqMan assay mix (36 μM each primer, 8 μM wild-type probe, and 8 μM mutant probe), and 11.875 μl of diluted DNA (i.e., 5 to 10 ng of DNA per reaction). The qPCRgyrA assays were performed using a StepOnePlus system (Life Technologies, Thermo Fisher Scientific) with an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 92°C for 15 s and annealing and extension at 60°C for 1 min, and a final step of incubation at 60°C for 30 s. The results were analyzed at the endpoint with the StepOne software v2.3 (Life Technologies, Thermo Fisher Scientific).
dPCRgyrA validation and study of the mutant-LPP mixtures.
As above, validation of the three dPCRgyrA assays for the specific detection of the wild-type gyrA QRDR sequence, T83I, D87N, and D87H mutations was performed by testing the LPP, LPPI1, LPPI4, and LPPI5 strains with the three assays. NTCs were also added to each run.
Then the mutant-LPP mixtures (at 1:1, 1:10, 1:100, 1:500, and 1:1,000 ratios) were analyzed in duplicates by the dPCRgyrA assays to assess the sensitivity of this technique to detect mutated gyrA QRDR sequences in the presence of wild-type sequences.
dPCRgyrA-T83I assay validation for application to human respiratory samples.
Additional specificity testing, evaluation of linearity, and determination of interpretative criteria for a dPCRgyrA result were performed for the dPCRgyrA-T83I assay.
Given that respiratory samples may contain DNA from a number of commensal or pathogenic microorganisms other than L. pneumophila, as well as human DNA, additional specificity testing for the dPCRgyrA-T83I assay was performed. Therefore, 20 DNA extracts from non-Legionella bacterial species commonly isolated from respiratory samples (Table S1) and 19 DNA extracts from Legionella species other than L. pneumophila (Table S2) were tested. In addition, 10 DNA extracts from respiratory samples collected in nonlegionellosis patients containing commensal or pathogenic bacteria, but no Legionella sp. as determined by negative Legionella sp. qPCR16S and L. pneumophila qPCRmip tests, were tested.
The Legionella DNA concentration in clinical samples is difficult to determine accurately, and thus it can hardly be standardized for dPCR testing. Therefore, it was necessary to evaluate the linearity of the dPCRgyrA-T83I assay and the minimum DNA concentration required in a respiratory sample to obtain interpretable results. For this purpose, a range of L. pneumophila genome concentrations (from 1.9 to 19,000 genome copies/μl, as determined using the NanoDrop 2000c spectrophotometer) were tested using the dPCRgyrA-T83I assay as well as the Legionella sp. qPCR16S and L. pneumophila qPCRmip assays.
dPCRgyrA-T83I assay on respiratory samples.
The dPCRgyrA-T83I assay was applied to DNA extracts from respiratory samples collected from legionellosis patients.
The manufacturers' instructions specify that for any dPCR reaction, the maximum precision is obtained at 1.6 copies of target DNA per reaction. Therefore, DNA extracts obtained from bacterial strains tested (except L. pneumophila DNA of the concentration range) were diluted to reach the recommended concentrations, i.e., from 0.6 to 1.6 target DNA copies per reaction well, corresponding to 200 to 2,000 copies/μl in the final dPCR reaction mix. By contrast, the DNA extracts obtained from the clinical samples were tested by dPCR without diluting the samples.
Each of the dPCRgyrA mixtures (15 μl) contained 7.5 μl of 2× QuantStudio 3D digital PCR master mix (Life Technologies, Thermo Fisher Scientific), 0.375 μl of 40× TaqMan assay mix (containing 36 μM each primer, 8 μM wild-type probe, and 8 μM mutant probe), and 7.125 μl of diluted DNA. A QuantStudio 3D digital PCR 20K chip (Life Technologies, Thermo Fisher Scientific) was loaded with 14.5 μl of the PCR mixtures with a QuantStudio 3D digital PCR chip loader. dPCRgyrA was performed on a GeneAmp PCR system 9700 (Life Technologies, Thermo Fisher Scientific) with an initial denaturation step at 98°C for 2 min, followed by 40 cycles of denaturation at 98°C for 30 s, annealing at 55°C for 10 s, and extension at 60°C for 30 s, and a final extension step at 60°C for 2 min.
Chips were read with the QuantStudio 3D digital PCR instrument (Life Technologies, Thermo Fisher Scientific), and the results were analyzed with the QuantStudio 3D AnalysisSuite software (Life Technologies, Thermo Fisher Scientific).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the French Centre National de la Recherche Scientifique, the Université Grenoble Alpes, and the Direction de la Recherche Clinique et de l'Innovation (DRCI) Centre Hospitalier Universitaire Grenoble Alpes. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conception of the work was performed by S.B. and A.H. Experiments were performed by A.H. Analysis of the data was performed by S.B., A.H., and M.B. Clinical information about the patients was provided by S.J., L.B., and C.S. The manuscript was written by A.H., S.B., and M.M. All authors reviewed the manuscript before submission.
All the authors have no conflict of interest to declare.
We thank Charles Coutton who loaned the StepOnePlus system and Xavier Fonrose for his assistance in the qPCR data interpretation. We also thank Dominique Dewolf, from Thermo Fisher Scientific, for her technical assistance during this study. We also thank Linda Northrup for editing the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00628-17.
REFERENCES
- 1.Mykietiuk A, Carratalà J, Fernández-Sabé N, Dorca J, Verdaguer R, Manresa F, Gudiol F. 2005. Clinical outcomes for hospitalized patients with Legionella pneumonia in the antigenuria era: the influence of levofloxacin therapy. Clin Infect Dis 40:794–799. doi: 10.1086/428059. [DOI] [PubMed] [Google Scholar]
- 2.Griffin AT, Peyrani P, Wiemken T, Arnold F. 2010. Macrolides versus quinolones in Legionella pneumonia: results from the Community-Acquired Pneumonia Organization international study. Int J Tuberc Lung Dis 14:495–499. [PubMed] [Google Scholar]
- 3.Kurz RW, Graninger W, Egger TP, Pichler H, Tragl KH. 1988. Failure of treatment of Legionella pneumonia with ciprofloxacin. J Antimicrob Chemother 22:389–391. doi: 10.1093/jac/22.3.389. [DOI] [PubMed] [Google Scholar]
- 4.Schindel C, Siepmann U, Han S, Ullmann AJ, Mayer E, Fischer T, Maeurer M. 2000. Persistent Legionella infection in a patient after bone marrow transplantation. J Clin Microbiol 38:4294–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Reilly KMA, Urban MA, Barriero T, Betts RF, Trawick DR. 2005. Persistent culture-positive Legionella infection in an immunocompromised host. Clin Infect Dis 40:e87–e89. doi: 10.1086/429826. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen K, Bangsborg JM, Høiby N. 2000. Susceptibility of Legionella species to five antibiotics and development of resistance by exposure to erythromycin, ciprofloxacin, and rifampicin. Diagn Microbiol Infect Dis 36:43–48. doi: 10.1016/S0732-8893(99)00095-4. [DOI] [PubMed] [Google Scholar]
- 7.Linde H, Morgenroth K, Barabas S, Lehn N. 2002. The amino acid substitution Asp87Tyr in GyrA of Legionella pneumophila Sg 1 is associated with elevated MICs to nalidixic acid but not to ciprofloxacin or moxifloxacin in vitro. Int J Med Microbiol 292:137–138. [Google Scholar]
- 8.Jonas D, Engels I, Hartung D, Beyersmann J, Frank U, Daschner FD. 2003. Development and mechanism of fluoroquinolone resistance in Legionella pneumophila. J Antimicrob Chemother 51:275–280. doi: 10.1093/jac/dkg054. [DOI] [PubMed] [Google Scholar]
- 9.Almahmoud I, Kay E, Schneider D, Maurin M. 2009. Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J Antimicrob Chemother 64:284–293. doi: 10.1093/jac/dkp173. [DOI] [PubMed] [Google Scholar]
- 10.Bruin JP, Koshkolda T, IJzerman EPF, Lück C, Diederen BMW, Den Boer JW, Mouton JW. 2014. Isolation of ciprofloxacin-resistant Legionella pneumophila in a patient with severe pneumonia. J Antimicrob Chemother 69:2869–2871. doi: 10.1093/jac/dku196. [DOI] [PubMed] [Google Scholar]
- 11.Shadoud L, Almahmoud I, Jarraud S, Etienne J, Larrat S, Schwebel C, Timsit J-F, Schneider D, Maurin M. 2015. Hidden selection of bacterial resistance to fluoroquinolones in vivo: the case of Legionella pneumophila and humans. EBioMedicine 2:1179–1185. doi: 10.1016/j.ebiom.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41:S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 13.Hall Sedlak R, Jerome KR. 2014. The potential advantages of digital PCR for clinical virology diagnostics. Expert Rev Mol Diagn 14:501–507. doi: 10.1586/14737159.2014.910456. [DOI] [PubMed] [Google Scholar]
- 14.Day E, Dear PH, McCaughan F. 2013. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods 59:101–107. doi: 10.1016/j.ymeth.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Fan HC, Quake SR. 2007. Detection of aneuploidy with digital polymerase chain reaction. Anal Chem 79:7576–7579. doi: 10.1021/ac0709394. [DOI] [PubMed] [Google Scholar]
- 16.Barrett AN, McDonnell TCR, Chan KCA, Chitty LS. 2012. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin Chem 58:1026–1032. doi: 10.1373/clinchem.2011.178939. [DOI] [PubMed] [Google Scholar]
- 17.Beck J, Bierau S, Balzer S, Andag R, Kanzow P, Schmitz J, Gaedcke J, Moerer O, Slotta JE, Walson P, Kollmar O, Oellerich M, Schütz E. 2013. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem 59:1732–1741. doi: 10.1373/clinchem.2013.210328. [DOI] [PubMed] [Google Scholar]
- 18.Roberts CH, Last A, Molina-Gonzalez S, Cassama E, Butcher R, Nabicassa M, McCarthy E, Burr SE, Mabey DC, Bailey RL, Holland MJ. 2013. Development and evaluation of a next-generation digital PCR diagnostic assay for ocular Chlamydia trachomatis infections. J Clin Microbiol 51:2195–2203. doi: 10.1128/JCM.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devonshire AS, Honeyborne I, Gutteridge A, Whale AS, Nixon G, Wilson P, Jones G, McHugh TD, Foy CA, Huggett JF. 2015. Highly reproducible absolute quantification of Mycobacterium tuberculosis complex by digital PCR. Anal Chem 87:3706–3713. doi: 10.1021/ac5041617. [DOI] [PubMed] [Google Scholar]
- 20.Talarico S, Safaeian M, Gonzalez P, Hildesheim A, Herrero R, Porras C, Cortes B, Larson A, Fang FC, Salama NR. 2016. Quantitative detection and genotyping of Helicobacter pylori from stool using droplet digital PCR reveals variation in bacterial loads that correlates with cagA virulence gene carriage. Helicobacter 21:325–333. doi: 10.1111/hel.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian L, Song H, Cai W. 2016. Determination of Bifidobacterium and Lactobacillus in breast milk of healthy women by digital PCR. Benef Microbes 7:559–569. doi: 10.3920/BM2015.0195. [DOI] [PubMed] [Google Scholar]
- 22.Kelley K, Cosman A, Belgrader P, Chapman B, Sullivan DC. 2013. Detection of methicillin-resistant Staphylococcus aureus by a duplex droplet digital PCR assay. J Clin Microbiol 51:2033–2039. doi: 10.1128/JCM.00196-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pholwat S, Stroup S, Foongladda S, Houpt E. 2013. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One 8:e57238. doi: 10.1371/journal.pone.0057238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhaegen B, De Reu K, De Zutter L, Verstraete K, Heyndrickx M, Van Coillie E. 2016. Comparison of droplet digital PCR and qPCR for the quantification of shiga toxin-producing Escherichia coli in bovine feces. Toxins (Basel) 8:pii=E157. doi: 10.3390/toxins8050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komp Lindgren P, Karlsson A, Hughes D. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother 47:3222–3232. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong A, Kassen R. 2011. Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa. Microbiology 157:937–944. doi: 10.1099/mic.0.046870-0. [DOI] [PubMed] [Google Scholar]
- 28.Heyries KA, Tropini C, Vaninsberghe M, Doolin C, Petriv OI, Singhal A, Leung K, Hughesman CB, Hansen CL. 2011. Megapixel digital PCR. Nat Methods 8:649–651. doi: 10.1038/nmeth.1640. [DOI] [PubMed] [Google Scholar]
- 29.Pekin D, Skhiri Y, Baret J-C, Le Corre D, Mazutis L, Salem CB, Millot F, El Harrak A, Hutchison JB, Larson JW, Link DR, Laurent-Puig P, Griffiths AD, Taly V. 2011. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 30.Ma J, Li N, Guarnera M, Jiang F. 2013. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomark Insights 8:127–136. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo Presti F, Riffard S, Vandenesch F, Reyrolle M, Ronco E, Ichai P, Etienne J. 1997. The first clinical isolate of Legionella parisiensis, from a liver transplant patient with pneumonia. J Clin Microbiol 35:1706–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igel L, Helbig JH, Lück PC. 2004. Isolation and characterization of a nonfluorescent strain of Legionella parisiensis. J Clin Microbiol 42:2877–2878. doi: 10.1128/JCM.42.6.2877-2878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han XY, Ihegword A, Evans SE, Zhang J, Li L, Cao H, Tarrand JJ, El-Kweifi O. 2015. Microbiological and clinical studies of legionellosis in 33 patients with cancer. J Clin Microbiol 53:2180–2187. doi: 10.1128/JCM.00380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janku F, Huang HJ, Fujii T, Shelton DN, Madwani K, Fu S, Tsimberidou AM, Piha-Paul SA, Wheler JJ, Zinner RG, Naing A, Hong DS, Karp DD, Cabrilo G, Kopetz ES, Subbiah V, Luthra R, Kee BK, Eng C, Morris VK, Karlin-Neumann GA, Meric-Bernstam F. 2016. Multiplex KRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction. Ann Oncol 28:642–650. doi: 10.1093/annonc/mdw670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurin M, Hammer L, Gestin B, Timsit JF, Rogeaux O, Delavena F, Tous J, Epaulard O, Brion JP, Croizé J. 2010. Quantitative real-time PCR tests for diagnostic and prognostic purposes in cases of legionellosis. Clin Microbiol Infect 16:379–384. doi: 10.1111/j.1469-0691.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- 36.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 37.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.