ABSTRACT
The skin microbiome is a complex ecosystem with important implications for cutaneous health and disease. Topical antibiotics and antiseptics are often employed to preserve the balance of this population and inhibit colonization by more pathogenic bacteria. However, despite their widespread use, the impact of these interventions on broader microbial communities remains poorly understood. Here, we report the longitudinal effects of topical antibiotics and antiseptics on skin bacterial communities and their role in Staphylococcus aureus colonization resistance. In response to antibiotics, cutaneous populations exhibited an immediate shift in bacterial residents, an effect that persisted for multiple days posttreatment. By contrast, antiseptics elicited only minor changes to skin bacterial populations, with few changes to the underlying microbiota. While variable in scope, both antibiotics and antiseptics were found to decrease colonization by commensal Staphylococcus spp. by sequencing- and culture-based methods, an effect which was highly dependent on baseline levels of Staphylococcus. Because Staphylococcus residents have been shown to compete with the skin pathogen S. aureus, we also tested whether treatment could influence S. aureus levels at the skin surface. We found that treated mice were more susceptible to exogenous association with S. aureus and that precolonization with the same Staphylococcus residents that were previously disrupted by treatment reduced S. aureus levels by over 100-fold. In all, the results of this study indicate that antimicrobial drugs can alter skin bacterial residents and that these alterations can have critical implications for cutaneous host defense.
KEYWORDS: Staphylococcus aureus, antimicrobial agents, skin, microbiome
INTRODUCTION
Antimicrobial drugs are commonly employed to inhibit the growth of pathogenic microorganisms. However, these interventions are rarely narrow in spectrum, instead acting on a range of bacterial species in our commensal microbiota (1). A number of studies have elucidated this effect in gut microbial populations, describing a dramatic reorganization of resident communities (2). This includes decreased bacterial diversity and outgrowth by previously minor contributors (3–5). Importantly, these alterations can persist for months to years posttreatment (6–8) and also affect a number of host functions, including metabolism, immunity, and transcriptional regulation (9, 10).
Despite these findings, few studies have assessed the impact of antimicrobial drugs at alternative body sites, such as the skin. Rather, the majority of research at this site has been devoted to a subset of easily cultured microorganisms studied in isolation (11). This includes MIC tests of pathogenic skin bacteria, as well as exogenous colonization studies in which nonresident test microorganisms are applied to the skin prior to treatment (12). While these results are often applied more broadly, their main purpose is to inform the effect of antimicrobial drugs on transient infectious bacteria rather than on more stable members of the community (13). As such, few studies have truly assessed the impact of antimicrobial drugs on inhabitant cutaneous populations. This dearth of research is especially notable given the frequency with which humans disrupt skin bacterial communities in both clinical and nonclinical settings. Indeed, the intent of most antiseptics is to sterilize the skin by employing agents with nonspecific mechanisms of action (14), with little regard for their effect on the resident microbiota.
While culture-independent surveys have recently illuminated the complexity of the skin microbiota (15–17), its necessity for normal function and disease remains unclear. One postulated function includes a role in colonization resistance, whereby members of the commensal microbiota could protect the host from infection by opportunistic and pathogenic skin microorganisms (18). This particular process has been well documented in the gut. Here, numerous studies have highlighted the ability of bacterial residents to impair colonization by pathogenic bacteria through immune activation, nutrient exclusion, and the production of toxic metabolites (19). Antibiotics have also been shown to shift the resident microbiota and render hosts more susceptible to certain pathogenic bacteria (20). This includes studies of the sporulating bacterium Clostridium difficile, which can recur repeatedly in response to antibiotic treatment but can also be controlled in most patients following the administration of fecal material from healthy unaffected donors (21–23). Importantly, this particular effect is not isolated to C. difficile, as a number of bacterial pathogens, including vancomycin-resistant Enterococcus and Salmonella enterica, have been shown to exploit newly available niches in response to treatment as well (24–26).
Similar to tests in the gut, recent studies have begun to assess the potential for skin microorganisms to play a role in colonization resistance. This includes defense against Staphylococcus aureus by unique strains of S. epidermidis (27), S. lugdunensis (28), and most recently, S. hominis (29). Here, it was found that certain individuals are colonized by host-specific Staphylococcus strains with the ability to alter S. aureus colonization patterns. While these studies also suggest that a removal of resident bacteria with antimicrobial agents could promote S. aureus colonization, no study to date has assessed this hypothesis in detail. Indeed, the long-term impact of topical antimicrobial drugs on skin bacterial communities and their ability to alter colonization patterns by S. aureus competitors remains largely unknown.
Here, we report this missing link by assessing the effect of antibiotics and antiseptics on the resident skin microbiota through a comparative time-series analysis. We report a differential impact of treatment on skin bacterial inhabitants, with the greatest disturbances elicited by a broad-spectrum triple antibiotic cocktail of bacitracin, neomycin, and polymyxin B. By contrast, we report a relatively muted effect of antiseptics, with only modest alterations to overall bacterial community structure. Despite these differences, we identified a conserved decrease in the levels of Staphylococcus residents regardless of treatment, a result that was strongly influenced by baseline levels of Staphylococcus.
Because commensal Staphylococcus spp. have been shown to impair colonization by the skin pathogen Staphylococcus aureus, we further evaluated this antimicrobial effect in the context of S. aureus colonization resistance. We show that treatment can promote exogenous association with S. aureus and that the same Staphylococcus residents disrupted by treatment are also capable of S. aureus competition, decreasing S. aureus levels by over 100-fold in precolonization experiments. In all, our results demonstrate that antimicrobial drugs can elicit long-term shifts in skin bacterial communities and that treatment with these agents has key implications for host susceptibility to pathogens such as S. aureus.
RESULTS
Topical antibiotic treatment alters skin bacterial residents.
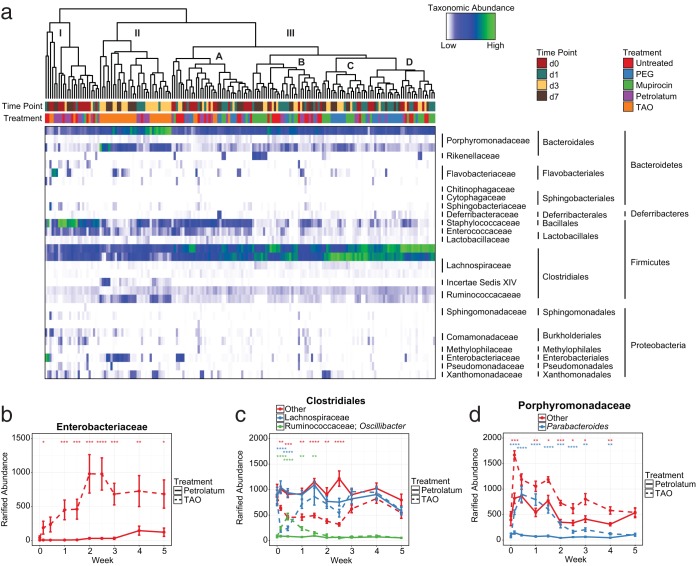
To assess the impact of topical antibiotics on the skin microbiota, we began by treating the dorsal skin of SKH-1 hairless mice twice daily for 1 week with the narrow-spectrum antibiotic mupirocin, a broad spectrum triple antibiotic ointment ([TAO] bacitracin, neomycin, polymyxin B), or their respective vehicles, polyethylene glycol (PEG) and petrolatum (see Fig. S1a in the supplemental material). These particular antibiotics were chosen for their range of activities as well as their extensive use as both therapeutic and prophylactic agents in both clinical and nonclinical settings (30). In all, antibiotics led to durable changes in skin bacterial residents, with populations forming three distinct clusters (I to III) and four subclusters (IIIA to IIID) (Fig. 1a). Interestingly, clusters I and IIIA were composed largely of baseline and early time point samples high in Staphylococcus, while treatment with antibiotics led to sustained decreases in Staphylococcus (Fig. S1b) and alternative clustering patterns. By contrast, cluster II was composed almost entirely of TAO-treated mice, a group that exhibited significant increases in Enterobacteriaceae, Porphyromadaceae, and Ruminococcaceae, as well as significant decreases in Lachnospiraceae and certain taxa classified more generally within the Clostridiales family (Fig. 1b to d). This distinction led to a marked absence of TAO-treated mice from clusters IIIB to IIID and, similar to that seen with Staphylococcus, was sustained for multiple weeks posttreatment.
FIG 1.
Topical antibiotics induce long-term shifts to skin microbial residents. (a) Heat map of rarified abundances for the 30 most common phylotypes on murine skin in response to treatment with polyethylene glycol (PEG), mupirocin, petrolatum, or triple antibiotic ointment (TAO). Dendrograms represent hierarchical clustering of Euclidean distances using complete agglomeration. Horizontal bars above the map designate treatment and time point features for individual mice. (b to d) Breakdown and longitudinal analysis of rarified abundances for Enterobacteriaceae (b), Clostridiales (c), and Porphyromonadaceae (d). Data are presented as individual mice (a) or means ± standard errors of the means (SEMs) (b to d). Statistical significance was determined at each time point by the Wilcoxon rank sum test (Mann Whitney U test). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Unlike TAO-treated mice, those administered mupirocin displayed community shifts largely in line with those treated with the vehicle PEG. Indeed, while these mice exhibited significant increases in Alistipes and decreases in Oscillibacter and Staphylococcus (Fig. S1b and c), these minor changes were not enough to elicit separate clustering patterns among the two treatment groups. These particular changes also displayed similar kinetics to those of bacterial taxa in TAO-treated mice, including immediate increases in rarified abundance and sustained posttreatment effects, underscoring the difficulties faced by skin communities when attempting to reacclimate upon treatment cessation.
An analysis of bacterial burden revealed a contrasting effect of antibiotics on absolute abundance as well. While mupirocin led to the characteristic decreases often associated with antibiotic treatment, TAO treatment resulted in increases in bacterial load at numerous time points as measured by 16S rRNA gene quantitative PCR (qPCR) (Fig. S1d). These findings further highlight the impact of antibiotic treatment on skin communities and suggest that the changes elicited by TAO may also be due to increases in the overall numbers of certain bacteria and not just their relative proportions.
Topical antibiotics shift bacterial community structure.
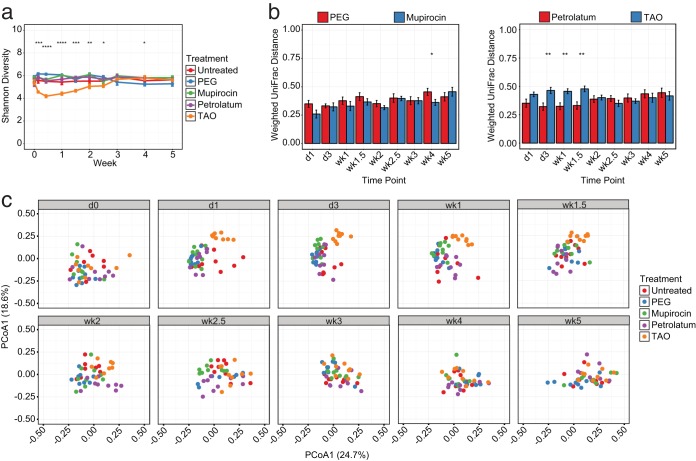
To better quantify these results at the community level, we next evaluated the diversity of bacterial populations over time. Similar to taxonomic analyses, we observed a relative stability in untreated mice and those treated with PEG, mupirocin, and petrolatum when testing alpha diversity metrics such as Shannon diversity, which takes into account the richness and evenness of taxa (Fig. 2a). By contrast, those treated with TAO exhibited an immediate and significant decrease in diversity starting after a single day (d1) of treatment, an effect that was maintained for greater than 1 week posttreatment. This was also recapitulated when evaluating community similarity by the weighted UniFrac metric, which assesses population differences based on abundance and phylogeny. When comparing each mouse to their baseline (d0) samples, we observed significantly greater differences in the TAO-treated group than in the vehicle-treated mice, a trend not shared by those administered mupirocin (Fig. 2b). Additional visualization of these samples by principal coordinate analysis further confirmed these results, as distinct clustering patterns were observed when comparing TAO-treated mice to other treatment groups (Fig. 2c).
FIG 2.
Triple antibiotic ointment alters skin bacterial diversity. (a) Shannon diversity measurements of murine bacterial communities following treatment with antibiotics and vehicles over time. (b) Weighted UniFrac distances comparing longitudinal time points to baseline communities of bacterial residents in treated and untreated mice. (c) Principal coordinate analyses of weighted UniFrac distances for murine bacterial communities over time. Data are presented as means ± SEMs (a, b) or individual mice (c). Statistical significance was determined at each time point by the Kruskal-Wallis rank sum test (a) or Wilcoxon rank sum test (Mann Whitney U test) (b). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Previously, others have shown similarities in the functional composition of a population despite differences in community membership and structure (31). To evaluate whether antibiotic treatment could lead to changes in the functional potential of skin inhabitants, we also utilized the PICRUSt software package (32) to infer metagenomic content of our populations. Specifically, PICRUSt analysis focuses on chromosomally encoded conserved differences among species as a method to approximate functional disparities. We found that treatment with antibiotics and vehicles led to a number of significant differences in genes predicted to be associated with metabolism, signaling, transport, and biosynthesis, among others (see Fig. S2). As such, the potential exists that by shifting the residents of the cutaneous microbiota, treatment may shift the functional capabilities of these populations as well.
Antiseptic treatment elicits only modest changes to skin bacterial community structure.
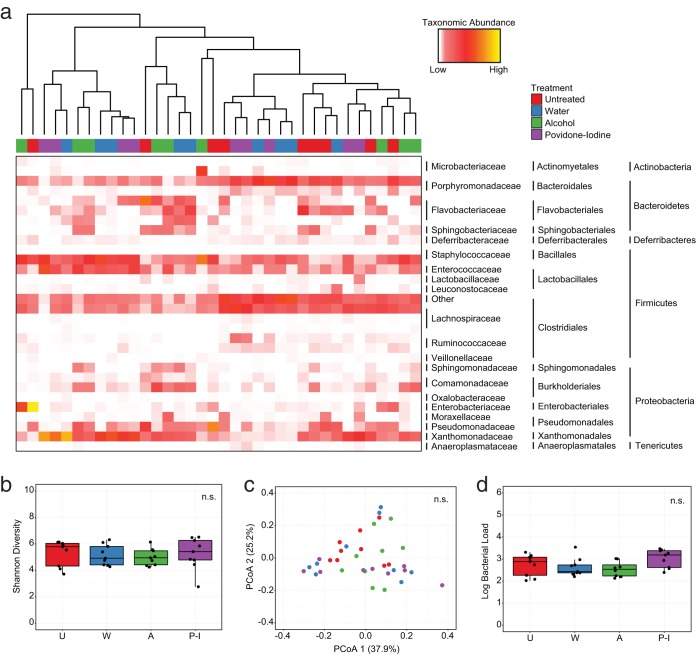
Following our tests with antibiotic regimens, we next endeavored to evaluate the impact of antiseptics, a more promiscuous class of antimicrobials, on the skin microbiome. We reasoned that these topical interventions should provide an even greater impetus for community disruption due to their indiscriminate mechanisms and proven efficacy in clinical settings (14). To evaluate this hypothesis, we treated mice with the common clinical antiseptics alcohol (80% ethanol) and povidone-iodine (10%) and compared this to mice treated with water or untreated controls (see Fig. S3a). Surprisingly, we observed no clustering of mice in response to antisepsis when taking into account major taxonomic groups at even the earliest (d1) posttreatment time point (Fig. 3a). Furthermore, when comparing the relative abundances of individual taxa following treatment, we detected no significant differences among treated mice and untreated controls (see Table S1). To evaluate whether subtle differences could contribute to a disruption at the population level, we also tested the diversity of communities in response to treatment. Similar to our taxonomic analyses, we found that antiseptic treatment resulted in no significant differences to Shannon Diversity (Fig. 3b), nor could we detect significant clustering by treatment using beta diversity metrics such as weighted UniFrac at d1 posttreatment (Fig. 3c). To assess whether we had missed decreases in absolute abundance by focusing our analyses on the relative proportions of taxa, we also tested the impact of treatment on the bacterial load of communities. Once again, we observed no significant differences between treated and untreated mice (Fig. 3d), further underscoring the stability of cutaneous bacterial communities in response to antiseptic treatment.
FIG 3.
Antiseptic treatment does not significantly alter skin bacterial community structure. (a) Heat map of rarified abundances for the 30 most common phylotypes on murine skin following treatment with water, alcohol, or povidone-iodine at d1 posttreatment. Dendrograms represent hierarchical clustering of Euclidean distances using complete agglomeration. Horizontal bar above the map designates treatments for individual mice. (b) Shannon diversity of murine bacterial communities in response to treatment. (c) Weighted UniFrac principal-coordinate analysis representing differences in murine bacterial populations following treatment. (d) Bacterial load comparison of treated and untreated mice calculated by 16S rRNA gene content at the skin surface. U, untreated; W, water; A, alcohol; P-I, povidone-iodine. Treatments were compared by Kruskal-Wallis rank sum test (b, d) or the adonis statistical test for community similarity (c).
As this result was particularly surprising, we also compared bacterial phylotypes at baseline to their d1 counterparts. This enabled us to evaluate whether treatment could shift populations in a conserved manner, thus explaining the modest effects seen between regimens at d1 posttreatment. However, when comparing the abundances of major taxonomic groups, we once again observed relatively few changes from d0 to d1 in response to treatment. Only Staphylococcus results differed significantly, and only in response to alcohol treatment (Table S2). Interestingly, this effect was strongly dependent upon starting communities, as mice with higher baseline levels of Staphylococcus were more strongly disrupted than those with lower baseline levels, regardless of treatment (Fig. S3b.). In all, this indicates that antiseptics elicit a more muted response in skin bacterial populations, but that their effects may be dependent upon starting communities.
Culture-based studies recapitulate sequence analyses of skin microbiota dynamics.
Our finding that most antiseptics elicited only minor changes to the resident skin microbiota was particularly surprising given the wealth of data describing their benefit in clinical settings. To address this discrepancy, we next sought to validate our findings using culturable skin inhabitants. Specifically, Staphylococcus spp. were chosen as a proxy because of their established response to topical antimicrobials in the clinic and their importance to human health. These bacteria were also the only inhabitants to vary in response to both antibiotics and antiseptics in our sequencing experiments and thus represented the best opportunity to verify our results in a culture setting.
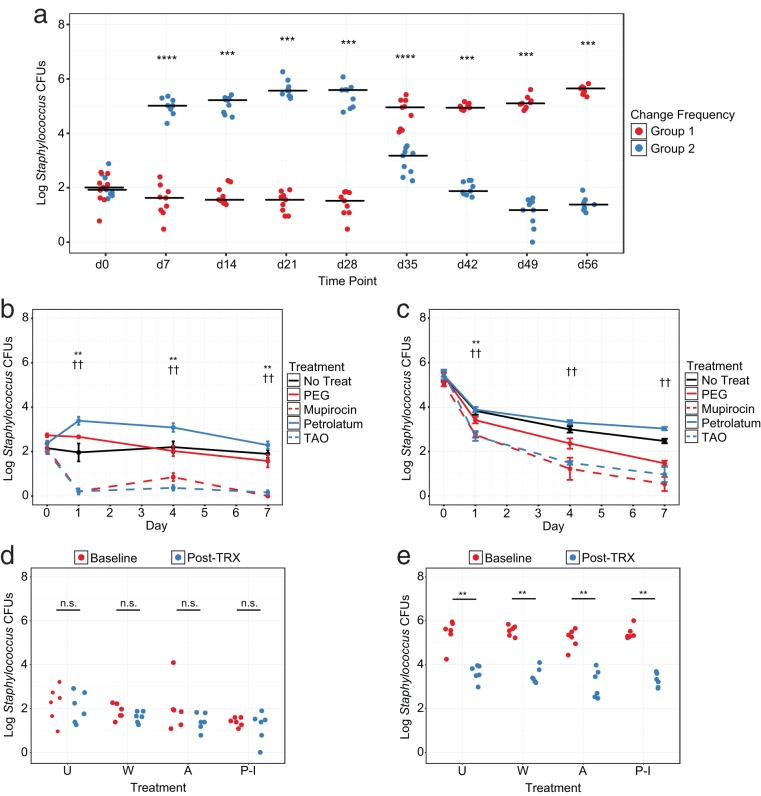
Because our antiseptic experiments showed antibacterial effects were dependent upon baseline communities, we began by designing a system to control Staphylococcus levels in murine populations. Specifically, we observed that mice housed in cages changed once per week displayed a significant elevation in Staphylococcus levels (high Staphylococcus [HS]) compared with those changed more frequently (low Staphylococcus [LS]) (Fig. 4a). When controlled over time, this effect could be maintained for multiple weeks and had the potential for reversibility, as mice swapped from frequent to infrequent cage changes rapidly converted to the alternate phenotype. Cage change frequency and monitoring thus presented the opportunity to maintain Staphylococcus at distinct levels prior to treatment.
FIG 4.
Antimicrobial treatment alters resident Staphylococcus colonization in a baseline-dependent manner. (a) Murine resident Staphylococcus CFUs in response to cage change frequency over time. Group 1 mice were changed every other day and group 2 mice were changed once per week at the start. Groups were switched to the alternate regimen at d28. Data are presented as individual mice with median bars. (b and c) Murine resident Staphylococcus CFUs in response to antibiotic treatment starting at low (b) or high (c) baseline levels. Statistical comparisons were made between polyethylene glycol (PEG) and mupirocin (*) or petrolatum and triple antibiotic ointment (TAO) (†). Data are presented as means ± SEMs. (d and e) Murine resident Staphylococcus CFUs in response to antiseptic treatment starting at low (d) or high (e) baseline levels. Data are presented as individual mice at baseline and at d1 posttreatment. U, untreated; W, water; A, alcohol; P-I, povidone-iodine. Statistical significance was determined by Wilcoxon rank sum test (Mann Whitney U test). *, P < 0.05; **, P < 0.01; ††, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To evaluate the impact of antimicrobial drugs on culturable Staphylococcus, we began by housing mice in cages with frequent or infrequent changes and then treating with PEG, mupirocin, petrolatum, or TAO. Similar to sequencing experiments, antibiotic treatment led to a significant decrease in Staphylococcus starting at d1 posttreatment regardless of starting community, although this effect was more pronounced in LS mice (Fig. 4b and c). Interestingly, while we also observed a gradual decrease of Staphylococcus in response to PEG treatment, petrolatum-treated LS mice displayed increased Staphylococcus colonization at early time points and elevated levels of Staphylococcus compared with those in untreated controls in HS mice. Because our sequencing results revealed similar decreases in Staphylococcus in response to treatment with antibiotics but not petrolatum, this represents a reproducible mechanism in multiple testing protocols.
To assess this effect in the context of antiseptics, a separate cohort of HS and LS mice was next treated with water, alcohol, or povidone-iodine and compared with untreated controls. Unlike those treated with antibiotics, no significant differences in Staphylococcus were observed in LS mice following treatment with water, alcohol, or povidone-iodine compared to baseline colonization at d1 posttreatment (Fig. 4d). Moreover, while Staphylococcus colonization in HS mice was significantly decreased following treatment, untreated mice with a single cage change exhibited an almost identical reduction in colonization, confirming that a change in environment can also have significant impacts on bacterial communities (Fig. 4e). In all, these experiments indicate that antibiotics and antiseptics have distinct effects on skin bacterial residents and that the magnitude of this response can vary depending upon starting communities.
Antimicrobial drugs reduce colonization by Staphylococcus aureus competitors.
After confirming our sequencing results with culture experiments, we next endeavored to explore the ramifications of cutaneous bacterial community disruption. As previous studies have suggested a role for the skin microbiota, and specifically resident Staphylococcus spp., in S. aureus colonization resistance (27–29), we chose this particular commensal-pathogen pair for further analysis. We were particularly attracted by the ability of antimicrobial drugs to shift communities for multiple days posttreatment, suggesting a window in which S. aureus could access the skin unencumbered by competing residents or antimicrobial drugs. As alcohol was found to have relatively minor effects on skin bacterial residents, with the exception of Staphylococcus spp., we first tested whether treatment with this antiseptic could promote S. aureus association. Specifically, mice were treated with alcohol, similar to in previous experiments, and then exogenously associated with S. aureus 1 day posttreatment. As hypothesized, we observed a slight but significant increase in S. aureus levels in treated mice compared with that in untreated controls, indicating a reduction in colonization resistance in response to treatment (Fig. 5a).
FIG 5.
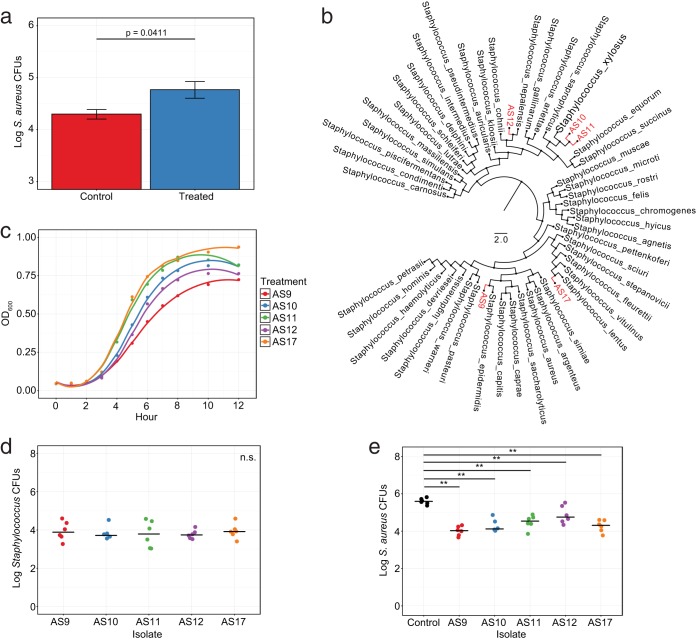
Resident Staphylococcus can reduce colonization by Staphylococcus aureus. (a) Staphylococcus aureus CFUs following exogenous administration in mice pretreated with alcohol or in untreated controls. (b) Phylogenetic tree of 16S rRNA gene diversity using approximate-maximum-likelihood to compare murine Staphylococcus residents (red) to known Staphylococcus isolates from the RDP database (black). (c) Growth curve analysis of resident Staphylococcus isolates at an optical density of 600 nm (OD600). (d) Enumeration of Staphylococcus isolate CFUs following exogenous administration to mouse dorsa. (e) S. aureus CFU levels following precolonization of mouse dorsa with resident Staphylococcus isolates. Data are presented as means ± SEMs (a) or with median bars (d, e). Statistical significance was determined by Wilcoxon rank sum test (Mann Whitney U test). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Because this effect could also be the result of additional factors, including previously unidentified microbial inhabitants, we next profiled individual Staphylococcus isolates that were reduced by antimicrobial treatment in our previous experiments. We reasoned that if these bacteria were the true source of colonization resistance, then adding them back to the skin should reduce S. aureus association in kind. Following phenotypic analysis and full-length 16S rRNA gene sequencing, we isolated five unique resident Staphylococcus genotypes, namely, AS9, AS10, AS11, AS12, and AS17. Comparing these to reference sequences within the Ribosomal Database Project (RDP) (33), we identified four distinct species and two strain level variants: S. epidermidis (AS9), S. xylosus (AS10 and AS11), S. nepalensis (AS12), and S. lentus (AS17) (Fig. 5b). Interestingly, while each of these bacteria fell within the Staphylococcus genus, they also had considerable genomic variability within the 16S rRNA gene region, suggesting a relative permissivity at the skin surface for these particular taxa (see Fig. S4).
To assess the colonization potential of each isolate, we next compared their growth dynamics under various conditions. When comparing growth in enriched medium, we observed distinct differences among isolates, with AS17 S. lentus and AS11 S. xylosus displaying the most robust expansion kinetics (Fig. 5c). By contrast, AS9 S. epidermidis appeared to replicate the slowest and exhibited the most gradual exponential curve. AS10 S. xylosus and AS12 S. nepalensis both displayed intermediate growth patterns. To further evaluate colonization potential, we assessed the ability of these isolates to colonize murine dorsa in vivo. Specifically, mice were housed in frequently changed cages to reduce endogenous Staphylococcus and then were epicutaneously inoculated every other day for 1 week to promote association with individual Staphylococcus isolates. Despite variable growth dynamics in vitro, all isolates colonized mice to an equal titer in vivo, suggesting conserved undefined factors to promote colonization at the skin surface (Fig. 5d).
As each of these isolates displayed notable colonization when added to murine hosts, we further tested all five to see whether they could also represent potential S. aureus competitors. To evaluate the ability of each isolate to restrict S. aureus colonization, we precolonized mice with each Staphylococcus resident, similar to the method described above, and then challenged them with S. aureus 1 day later. While isolates exhibited various levels of competition, all resulted in significant decreases to S. aureus association compared with that in uncolonized mice (Fig. 5e). Indeed, most mice exhibited greater than 10-fold reductions in S. aureus, and many, including those precolonized with S. epidermidis, were capable of decreasing S. aureus by levels greater than 100-fold. In all, this shows that skin bacterial residents can compete with S. aureus at the skin surface and that their removal can impact S. aureus colonization potential.
DISCUSSION
Given the expansive use of topical antibiotics and antiseptics, it is somewhat surprising that longitudinal studies to evaluate their effects on a community-wide scale are not more common. Here, we report that antimicrobial drugs can elicit significant changes to skin bacterial community membership and structure, albeit to various degrees. We also demonstrate that these alterations can have important consequences for colonization resistance and the skin pathogen Staphylococcus aureus.
Previous work has focused extensively on antibiotics and the gut microbiota. These studies have highlighted the ability of antimicrobials to disrupt bacterial communities and the consequences of these drugs on host physiology (34). One such example includes the elimination of colonization resistance, leading to increased susceptibility to bacterial infections (35). By altering the structure of bacterial populations in the gut, antibiotics can shift the balance in favor of more infectious microorganisms (19). Clostridium difficile is perhaps the best-studied representation of this effect (36). However, additional pathogens such as vancomycin-resistant Enterococcus and Salmonella enterica can also exploit newly available niches and cause disease (37, 38). As a result, the true question has transcended beyond whether or not antimicrobial drugs can promote pathogenicity to how best to mediate these unintended consequences.
The first step in such ventures is the elucidation of antimicrobial effects on a community-wide scale. While studies of the gut have been vital to this endeavor, we present the skin as an additional body site worthy of consideration. In our investigations, triple antibiotic ointment (TAO) was found to provoke the greatest response in microbial residence, with a significant decrease in bacterial diversity and domination by previously minor contributors. While these changes originated as a result of treatment-specific effects, they often endured, and in some cases were enhanced, following treatment cessation. This indicates that disrupted resident skin bacteria may also undergo multiple levels of succession prior to community stabilization, similar to that in the gut (39).
In accordance with their mechanisms of action, we also found the overall effect of mupirocin to be relatively minor compared with that of TAO. While TAO led to profound increases in bacteria from multiple families, including Enterobacteriaceae and Porphyromonadaceae, mupirocin produced relatively minor shifts in less-abundant taxa such as Alistipes and Oscillibacter. This finding is particularly notable as certain members of the Enterobacteriaceae and Porphyromonadaceae families have known intrinsic resistance mechanisms against TAO components, such as polymyxin B (40, 41). This could also explain the increase in overall bacterial load seen in mice following TAO administration, as certain bacteria may thrive when given access to a newly available cutaneous niche.
Perhaps most surprisingly, we also report a relatively muted impact of antiseptics on the skin microbiota, with alcohol and povidone-iodine both failing to shift baseline communities in a significant manner. While it is tempting to explain this finding as an inability of 16S rRNA gene sequencing to distinguish between live and dead bacteria, we find this conclusion unlikely in the context of results from our studies and from those before us. Indeed, our ability to detect differences in TAO-treated mice within 1 day of treatment provides strong evidence to the contrary. Others have also reported similar community responses to both decolonization protocols (42) and mild and antibacterial soaps (43), further validating this assertion.
Rapid repopulation of the skin could also explain our perceived lack of effect in response to antiseptic stress. However, as our study and those before us employed relatively early posttreatment samplings, we find it unlikely that residents could recolonize the skin in such a short period of time. Indeed, many of the bacteria observed in our experiments have been shown to exhibit particularly slow growth dynamics in previous examinations (44, 45). However, repopulation is likely shaped by both the magnitude of change and the environment. As such, future work will be necessary to establish a more complete understanding of this process as it relates to skin bacterial dynamics.
With this in mind, it is important to note that multiple studies have shown a reduction of certain culturable skin inhabitants in response to antisepsis. This includes residents from the commonly studied genus Staphylococcus, often chosen for its ease of use in culture-based experiments (46, 47). In line with these findings, we also observed a decrease in Staphylococcus residents in our sequencing and culture studies. However, we note that, because this bacterium was only one member of the larger community, this decline did not lead to shifts in overall population structure. As such, we hypothesize that the true utility of antiseptics may lie in their ability to disrupt a particular subset of microorganisms at the skin surface, while leaving the underlying community relatively unchanged.
Interestingly, Staphylococcus residents also exhibited distinct baseline-dependent dynamics in response to antiseptic treatment during our sequencing experiments. Specifically, we observed that mice with high levels of Staphylococcus responded more readily to treatment than mice with low levels of colonization. This suggested a nuanced impact of antiseptics on certain bacterial inhabitants, whereby treatment effects could vary depending upon starting communities. To verify this hypothesis, we developed a system in which Staphylococcus could be tested for antimicrobial susceptibility at both high and low colonization levels. As anticipated, we found the efficacy of antiseptics to be highly dependent upon baseline communities. Mice with low levels of Staphylococcus at baseline (LS) exhibited little to no decline in Staphylococcus, while those in mice with high levels (HS) were reduced by approximately 100-fold. Importantly, we observed a similar effect in control HS mice, suggesting that higher levels of Staphylococcus are less stable in general, and thus represent atypical colonization. By contrast, the inability of antiseptics to reduce Staphylococcus in LS mice indicates a relative stability in this community and a population capable of resisting the short-term stressors of antisepsis. We believe these studies have important implications for antimicrobial efficacy, particularly in the case of human skin, as humans are likely exposed to a greater number of transient microorganisms than are laboratory mice housed in more controlled environments (48).
When comparing antibiotic and antiseptic treatments, we observed that a standard course of antibiotics was more capable of community disruption than was acute antisepsis. While these are the most commonly employed regimens in the clinic, further research should also evaluate the effects of long-term antiseptic treatments on the skin microbiota as well as other delivery mechanisms. Indeed, the potential exists that consistent exposure to antiseptics through alternative means may have a more significant impact on skin inhabitants due to increased contact time or bioavailability. This is especially important when considering the rise of decolonization practices in the clinic, a procedure employing multiday prophylactic antibiotic and antiseptic treatments to remove resident Staphylococcus species (49, 50). While these methods efficiently remove endogenous S. aureus from the nares and extranasal body sites, they likely alter the underlying skin microbiota in kind. Without proper recolonization, these interventions could feasibly elicit long-term shifts in the skin microbiota, similar to that in our experiments, and promote infection by more dangerous hospital- and community-acquired pathogens (51–53).
To assess this very possibility, we investigated the potential of treatment to promote S. aureus colonization at the skin surface in our mouse model. In response to treatment, we observed a significant increase in S. aureus levels compared with that in untreated controls following exogenous association, suggesting an increase in cutaneous permissivity. As previous studies have illustrated the role of certain Staphylococcus spp. to compete with S. aureus for colonization (27–29), we proceeded by testing the ability of murine Staphylococcus isolates to compete with S. aureus. Specifically, we chose Staphylococcus residents that were disrupted by antibiotic and antiseptic treatment in our previous experiments for further analysis. This enabled us to determine whether these particular bacterial residents were responsible for the decrease in colonization resistance and to confirm the ability of antimicrobial drugs to alter communities with the potential for S. aureus competition. Importantly, we found that all isolates were capable of protecting against S. aureus association, with a number of mice exhibiting reductions in S. aureus levels by over 100-fold. These results support the notion that antimicrobial drugs can impact S. aureus colonization resistance and argue for enhanced stewardship in the context of posttreatment recovery.
In all, we describe the importance of antimicrobial drugs to skin bacterial community dynamics. By detecting unique changes in the microbiota in response to topical antibiotics and antiseptics, we present the skin as a body site capable of reproducible disruptions and fluctuations in colonization resistance. For this reason and others, we further advocate for the judicious use of antibiotics and antiseptics, as well as increased monitoring of bacterial populations, to combat the unintentional consequences which can follow cutaneous perturbations.
MATERIALS AND METHODS
Mice.
Six-week-old female SKH-1 immunocompetent hairless mice were purchased from Charles River and acclimated for at least 2 weeks prior to testing. Throughout experimentation, mice were housed on ALPHA-Dri bedding and given ad libitum access to autoclaved food and water. Mice treated with the same antimicrobial drug or exogenous Staphylococcus strains were housed together to avoid mixing, and at least two cages were used per condition to assess caging effects. All cages were changed three to four times per week during the course of a study unless otherwise noted. All mouse procedures were performed under protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Antimicrobial treatment and sample collection.
For experiments involving antibiotics, mice were treated every 12 h for 7 days on the dorsum with mupirocin (2% in polyethylene glycol), a triple antibiotic ointment (bacitracin, 400 U; neomycin, 3.5 mg; polymyxin B, 5,000 U in petrolatum), or their respective vehicles, polyethylene glycol (PEG 400 and PEG 3350) and petrolatum. Mice were swabbed longitudinally as described in Fig. S1a in the supplemental material, with collections occurring prior to morning applications during treatment to minimize experimental disruptions. For experiments involving antiseptics, mice were treated on the dorsum with UltraPure water (MoBio), alcohol (80% ethanol), or povidone-iodine (Betadine, 10%) every 8 h, three times in total. Mice were swabbed as described in Fig. S3a, with d1 collections occurring 4 h after the final treatment. At least three cages of three mice each were used for all conditions to evaluate caging effects. All treatments were applied with sterile UV-irradiated cotton swabs (Beauty 360; CVS), and samples were collected with sterile foam-tipped applicators (Puritan). A standard topical inoculum of approximately 150 μl per mouse was utilized for both antibiotic and antiseptic experiments. All swabs were stored at −20°C prior to extraction.
Bacterial DNA isolation and 16S rRNA gene sequencing and qPCR.
Bacterial DNA was extracted as described previously (54). Briefly, Ready-Lyse lysozyme solution (Epicentre), bead beating, and heat shock at 65°C were used to lyse cells. The Invitrogen PureLink kit was used for DNA extraction. During our testing, the V4 region of the 16S rRNA gene was found to better approximate murine skin communities than V1V3. PCR and sequencing of the V4 region were thus performed using 150-bp paired-end chemistry and the barcoded primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (55) on the Illumina MiSeq platform. Accuprime high-fidelity Taq polymerase was used for PCR cycling conditions of 94°C for 3 min, followed by 35 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s, and ending with 72°C for 10 min. For bacterial load comparisons, 16S rRNA genes were amplified by qPCR using Fast SYBR green master mix (Fisher Scientific) and the qPCR optimized primers 533F (5′-GTGCCAGCAGCCGCGGTAA-3′) and 902R (5′-GTCAATTCITTTGAGTTTYARYC-3′, where I represents deoxyinosine and Y and R represent degenerate bases [C/T and A/G, respectively]). Samples were compared to standard curves generated from known concentrations of serially diluted bacterial DNA to calculate burden.
Microbiome analysis.
Raw sequences were assembled, demultiplexed, and trimmed to yield 24,026,791 total high-quality V4 reads. Sequences were then further processed using QIIME 1.7.0 prior to downstream analyses (56). Briefly, sequences were de novo clustered into operational taxonomic units (OTUs) based on 97% similarity by UClust (57), and taxonomy was assigned to the most abundant representative sequence per cluster using the RDP classifier (58). Sequences were aligned by PyNAST (59), and chimeric sequences were removed using ChimeraSlayer (60) along with those identified as “Unclassified,” “Bacteria;Other,” or “Cyanobacteria.” Singletons were also removed in addition to any OTU found at greater than 1% abundance in at least 50% of kit and environmental control samples to eliminate potential contaminating sequences. All antiseptics, antibiotics, and vehicles were similarly sequenced and evaluated for possible contaminating sequences. All samples were rarified to 5,000 sequences/sample corresponding to an average Good's coverage of 0.95/sample, and samples below this cutoff were removed from downstream analyses. Alpha and beta diversity matrices were calculated in QIIME, and statistical analysis and visualization were performed in the R statistical computing environment (61). Heat maps were constructed by condensing all OTUs above 0.1% to the top 30 taxonomic identifications. The PICRUSt bioinformatics software package was used to infer functional content of bacterial communities (32).
Caging effects.
Mice were housed three per cage with three cages per group, and cages were randomly assigned to be changed every other day (frequently) or once per week (infrequently) for 4 weeks. Swabs were taken every 7 days prior to changes of the infrequent group and cultured for Staphylococcus residents on mannitol salt agar ([MSA] Acumedia) overnight at 37°C. At day 28 (d28), mice from each cohort were reassigned to the alternate groups and swabbed for an additional 4 weeks to evaluate normalization.
Antimicrobials and alternate Staphylococcus communities.
Mice were assigned to frequent or infrequent cage changes prior to treatment to generate low Staphylococcus and high Staphylococcus communities, respectively, and treated as described above. During experimentation, all cages were changed on a frequent schedule with untreated mice representing controls. Swabs were taken at baseline, d1, day 4 (d4), and day 7 (d7) for antibiotic-treated mice and at baseline and 4 h posttreatment for antiseptic-treated mice. Samples were cultured on MSA overnight at 37°C to enumerate Staphylococcus numbers.
Staphylococcus isolation, sequencing, and phylogenetic tree.
To obtain a more complete profile of our Staphylococcus isolates, phenotypically distinct Staphylococcus colonies were picked from MSA plates following culture from murine dorsa prior to and after antimicrobial treatment. DNA was extracted from colonies as described above, and DNA was PCR-amplified using full-length 16S rRNA gene primers (27F and 1492R). The primary PCR conditions used were 98°C for 3 min, 35 cycles of 95°C for 45 s, 56°C for 60 s, and 72°C for 90 s, and 72°C for 10 min. Full-length 16S rRNA gene sequencing was performed by Sanger sequencing, and resident Staphylococcus isolates were compared to known Staphylococcus 16S rRNA genes downloaded from the RDP database (33). Phylogenetic trees were generated by FastTree (62) and visualized in FigTree v1.4.3.
Growth curves.
Staphylococcus isolates were grown at 37°C in liquid Luria broth (Fisher Scientific) for 12 h with shaking at 300 rpm. Samples were taken every hour and optical density was determined at 600 nm (OD600) using the BioTek Synergy HT plate reader.
Exogenous Staphylococcus colonization and S. aureus competition.
Staphylococcus isolates were grown overnight in liquid Luria broth (Fisher Scientific) at 37°C and 300 rpm. On the following day, isolates were subcultured, incubated to achieve log growth, and resuspended in phosphate-buffered saline (PBS) to acquire 108 CFU/ml inoculums. Titers were validated by culture and OD600 measurements. Two cages of three mice each were frequently changed to reduce levels of endogenous Staphylococcus and were monoassociated at the dorsum with 200 μl of Staphylococcus isolate inoculum using a sterile swab. Applications of Staphylococcus suspensions were repeated every other day over the course of 1 week for a total of four applications. Mice were then swabbed 1 day following the fourth application, and swabs were cultured on MSA overnight at 37°C for CFU enumeration. S. aureus strain 502A with selective streptomycin resistance was chosen for S. aureus competition studies because of its proven efficiency in skin colonization and its potential for pathogenicity (63, 64). S. aureus was grown in a manner similar to that of Staphylococcus isolates and applied 1 day posttreatment or 1 day postmonoassociation with individual Staphylococcus isolates. Control mice were administered PBS only. Mice were then swabbed the following day for S. aureus, and swabs were cultured on LB agar with streptomycin for selective CFU enumeration.
Accession number(s).
16S rRNA sequence reads have been deposited in the NCBI Short Read Archive under BioProject no. PRJNA383404. Sequences of Staphylococcus isolates have been deposited in GenBank under accession numbers MF286534 to MF286538.
Supplementary Material
ACKNOWLEDGMENTS
We thank Penn Next Generation Sequencing Core for sequencing support, the Penn Medicine Academic Computing Services for computing support, Jeffrey Weiser for the kind gift of S. aureus strain 502A engineered with streptomycin resistance, and members of the Grice laboratory for their underlying contributions.
Funding for this work was provided by the National Institutes of Health, National Institute of Arthritis, Musculoskeletal, and Skin Diseases (R00AR060873 and R01AR066663 to E.A.G.). A.J.S. is supported by a Department of Defense National Defense Science and Engineering Graduate fellowship.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Defense.
The authors declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00774-17.
REFERENCES
- 1.Blaser MJ. 2016. Antibiotic use and its consequences for the normal microbiome. Science 352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langdon A, Crook N, Dantas G. 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. 2010. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. 2005. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol 43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jernberg C, Lofmark S, Edlund C, Jansson JK. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 8.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108(Suppl 1):S4554–S4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgun A, Dzutsev A, Dong X, Greer RL, Sexton DJ, Ravel J, Schuster M, Hsiao W, Matzinger P, Shulzhenko N. 2015. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut 64:1732–1743. doi: 10.1136/gutjnl-2014-308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampf G, Kramer A. 2004. Epidemiologic background of hand hygiene and evaluation of the most important agents for scrubs and rubs. Clin Microbiol Rev 17:863–893. doi: 10.1128/CMR.17.4.863-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rotter M, Sattar S, Dharan S, Allegranzi B, Mathai E, Pittet D. 2009. Methods to evaluate the microbicidal activities of hand-rub and hand-wash agents. J Hosp Infect 73:191–199. doi: 10.1016/j.jhin.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Echols K, Graves M, LeBlanc KG, Marzolf S, Yount A. 2015. Role of antiseptics in the prevention of surgical site infections. Dermatol Surg 41:667–676. doi: 10.1097/DSS.0000000000000375. [DOI] [PubMed] [Google Scholar]
- 14.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2014. Biogeography and individuality shape function in the human skin metagenome. Nature 514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannigan GD, Grice EA. 2013. Microbial ecology of the skin in the era of metagenomics and molecular microbiology. Cold Spring Harb Perspect Med 3:a015362. doi: 10.1101/cshperspect.a015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. 2014. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 21.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. 2013. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 4:125–135. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, Young VB. 2014. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 5:e00893-14. doi: 10.1128/mBio.00893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes 2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 28.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brotz-Oesterhelt H, Grond S, Peschel A, Krismer B. 2016. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 29.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim JN, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DY, Gallo RL. 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spann CT, Tutrone WD, Weinberg JM, Scheinfeld N, Ross B. 2003. Topical antibacterial agents for wound care: a primer. Dermatol Surg 29:620–626. doi: 10.1046/j.1524-4725.2003.29143.x. [DOI] [PubMed] [Google Scholar]
- 31.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willing BP, Russell SL, Finlay BB. 2011. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 35.Littman DR, Pamer EG. 2011. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seekatz AM, Young VB. 2014. Clostridium difficile and the microbiota. J Clin Invest 124:4182–4189. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, Taur Y, Jenq RR, van den Brink MR, Xavier JB, Pamer EG. 2013. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. 2009. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun 77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterfreund GL, Vandivier LE, Sinha R, Marozsan AJ, Olson WC, Zhu J, Bushman FD. 2012. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS One 7:e46966. doi: 10.1371/journal.pone.0046966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coats SR, To TT, Jain S, Braham PH, Darveau RP. 2009. Porphyromonas gingivalis resistance to polymyxin B is determined by the lipid A 4′-phosphatase, PGN_0524. Int J Oral Sci 1:126–135. doi: 10.4248/IJOS.09062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burnham CA, Hogan PG, Wallace MA, Deych E, Shannon W, Warren DK, Fritz SA. 2016. Topical decolonization does not eradicate the skin microbiota of community-dwelling or hospitalized adults. Antimicrob Agents Chemother 60:7303–7312. doi: 10.1128/AAC.00925-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Two AM, Nakatsuji T, Kotol PF, Arvanitidou E, Du-Thumm L, Hata TR, Gallo RL. 2016. The cutaneous microbiome and aspects of skin antimicrobial defense system resist acute treatment with topical skin cleansers. J Investig Dermatol 136:1950–1954. doi: 10.1016/j.jid.2016.06.612. [DOI] [PubMed] [Google Scholar]
- 44.Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, Bercik P, Surette MG. 2016. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med 8:72. doi: 10.1186/s13073-016-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziemer CJ. 2014. Newly cultured bacteria with broad diversity isolated from eight-week continuous culture enrichments of cow feces on complex polysaccharides. Appl Environ Microbiol 80:574–585. doi: 10.1128/AEM.03016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durani P, Leaper D. 2008. Povidone-iodine: use in hand disinfection, skin preparation and antiseptic irrigation. Int Wound J 5:376–387. doi: 10.1111/j.1742-481X.2007.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rochon-Edouard S, Pons JL, Veber B, Larkin M, Vassal S, Lemeland JF. 2004. Comparative in vitro and in vivo study of nine alcohol-based handrubs. Am J Infect Control 32:200–204. doi: 10.1016/j.ajic.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, Sekaly RP, Jenkins MK, Vezys V, Haining WN, Jameson SC, Masopust D. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R, CDC Prevention Epicenters Program, AHRQ DECIDE Network and Healthcare-Associated Infections Program. 2013. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hetem DJ, Bootsma MC, Bonten MJ. 2016. Prevention of surgical site infections: decontamination with mupirocin based on preoperative screening for Staphylococcus aureus carriers or universal decontamination? Clin Infect Dis 62:631–636. doi: 10.1093/cid/civ990. [DOI] [PubMed] [Google Scholar]
- 51.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 52.Syed AK, Ghosh S, Love NG, Boles BR. 2014. Triclosan promotes Staphylococcus aureus nasal colonization. mBio 5:e01015. doi: 10.1128/mBio.01015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, Grice EA. 2016. Skin microbiome surveys are strongly influenced by experimental design. J Investig Dermatol 136:947–956. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):S4516–S4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 58.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 62.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Light IJ, Sutherland JM, Schott JE. 1965. Control of a Staphylococcal outbreak in a nursery, use of bacterial interference. JAMA 193:699–704. doi: 10.1001/jama.1965.03090090005001. [DOI] [PubMed] [Google Scholar]
- 64.Houck PW, Nelson JD, Kay JL. 1972. Fatal septicemia due to Staphylococcus aureus 502A. Report of a case and review of the infectious complications of bacterial interference programs. Am J Dis Child 123:45–48. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.