ABSTRACT
Nucleot(s)ide analogues and peginterferon (PEG-IFN) treatment are the only approved therapies for chronic hepatitis B virus (HBV) infection. However, complete eradication of the virus, as indicated by persistent loss of hepatitis B surface antigen (HBsAg), is rare among treated patients. This is due to long-term persistence of the HBV genome in infected hepatocytes in the form of covalently closed circular DNA (cccDNA). In this study, we investigated whether administration of a large dose of a nucleoside analogue in combination with PEG-IFN can achieve long-term loss of HBsAg in human hepatocyte chimeric mice. Mice were treated with a high dose of entecavir and/or PEG-IFN for 6 weeks. High-dose combination therapy with both drugs resulted in persistently negative HBV DNA in serum. Although small amounts of HBV DNA and cccDNA (0.1 and 0.01 copy/cell, respectively) remained in the mouse livers, some of the mice remained persistently negative for serum HBV DNA at 13 weeks after cessation of the therapy. Serum HBsAg and hepatitis B core-related antigen (HBcrAg) continued to decrease and eventually became negative at 12 weeks after cessation of the therapy. Analysis of the HBV genome in treated mice showed accumulation of G-to-A hypermutation and CpG III island methylation. Persistent loss of serum HBV DNA and loss of HBV markers by high-dose entecavir and PEG-IFN combination treatment in chimeric mice suggests that control of HBV can be achieved even in the absence of a cellular immune response.
KEYWORDS: hepatitis B virus, cccDNA, human hepatocyte chimeric mouse, hypermutation, methylation, DNA methylation
INTRODUCTION
Hepatitis B virus (HBV) infection is the most common chronic viral infection in the world (1), affecting at least 250 million people. Individuals with chronic HBV infection are at greater risk of liver fibrosis, cirrhosis, hepatocellular carcinoma (HCC), and death due to complications resulting from liver disease (2). Current therapy for chronic hepatitis B is limited to treatment with nucleot(s)ide analogues (NAs) or peginterferon (PEG-IFN). These treatments reduce serum HBV DNA levels, improve liver histology (3–5), and reduce the risk of liver failure and HCC development (6, 7); however, cure is rare in these individuals due to the stable, long-term persistence of viral DNA in hepatocytes in the form of covalently closed circular DNA (cccDNA) (8, 9).
NAs, such as lamivudine, adefovir, entecavir, and tenofovir disoproxil fumarate, act mainly by inhibiting reverse transcription of the pregenomic RNA into HBV DNA and have no direct effect on cccDNA (10). Consequently, serum HBV DNA levels often rebound to pretreatment levels after cessation of NA treatment (11, 12). Unlike NAs, IFN has both antiviral and immunomodulatory activity and inhibits both HBV transcription and replication (13). PEG-IFN monotherapy has been reported to result in hepatitis B e antigen (HBeAg) seroconversion in 29 to 32% of patients and in hepatitis B surface antigen (HBsAg) loss in 3 to 7% of patients at 24 weeks after completion of treatment (11, 14). Combination therapy with both an NA and PEG-IFN-α-2a has been shown to result in greater viral decline and HBeAg loss than NA therapy alone (15–18) and resulted in reduced HBV cccDNA in chronic hepatitis B patients (19, 20). Therefore, we hypothesized that combination therapy with high doses of NA and PEG-IFN could eliminate HBV infection in an immunodeficient animal model.
Human hepatocyte chimeric mice, prepared from urokinase-type plasminogen activator (uPA) transgenic severe combined immunodeficiency (SCID) mice transplanted with human hepatocytes (21, 22), serve as a mouse model for HBV infection and are useful for the study of virology and evaluation of anti-HBV drugs (23). In this study, we investigated the effect of high-dose entecavir and PEG-IFN combination treatment on HBV replication using HBV-infected human hepatocyte chimeric mice. We also analyzed intrahepatic HBV methylation following interferon therapy.
RESULTS
Effect of entecavir and PEG-IFN on HBV-infected mice.
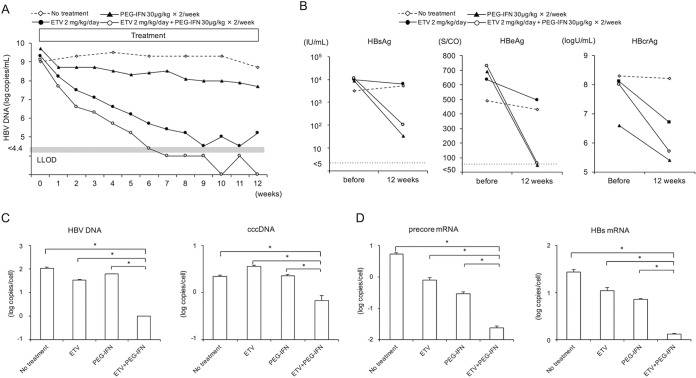
HBV-infected mice were treated for 12 weeks with 2 mg/kg of entecavir daily and/or with 30 μg/kg of PEG-IFN twice weekly. PEG-IFN monotherapy resulted in a modest 1.7-log reduction in the serum HBV DNA level, but the HBV DNA level remained high (Fig. 1A). In the mouse treated with entecavir alone, the serum HBV DNA level decreased by 4.0 logs but did not become undetectable. However, in the mouse treated with both PEG-IFN and entecavir, serum HBV DNA levels decreased rapidly and fell below the detectable level (4.4 log copies/ml) at 7 weeks. Serum HBsAg, HBeAg, and hepatitis B core-related antigen (HBcrAg) values in the mouse treated with entecavir alone for 12 weeks decreased slightly, but in the mice treated with PEG-IFN either alone or in combination with entecavir (Fig. 1B), these values decreased significantly. After 12 weeks of treatment, we extracted DNA and RNA from three samples of liver tissues from each mouse and measured intrahepatic HBV DNA and cccDNA levels and the expression levels of HBV transcripts. Intrahepatic HBV DNA and cccDNA levels in HBV-infected mice were 2.1 ± 1.9 and 0.25 ± 0.04 log copies/cell, respectively (Fig. 1C). The observed baseline intrahepatic HBV DNA and cccDNA levels in HBV-infected mice were similar to those in chronic hepatitis B patients (17, 24). Intrahepatic HBV DNA and cccDNA levels in mice treated with either entecavir or PEG-IFN alone did not differ significantly from those in untreated mice. In contrast, the levels of intrahepatic HBV DNA and cccDNA were significantly reduced in the mouse treated with entecavir and PEG-IFN (Fig. 1C). Consistent with a reduction in cccDNA levels, intrahepatic expression levels of the transcription products precore mRNA and HBs mRNA were also significantly reduced in the mouse that received both entecavir and PEG-IFN (Fig. 1D).
FIG 1.
Effect of antivirals on HBV-infected mice. Four mice were treated with or without (no treatment) 2 mg/kg of entecavir (ETV) alone daily or 30 μg/kg of PEG-IFN alone or entecavir plus PEG-IFN twice weekly for 12 weeks. (A) Time course of serum HBV DNA titer in each mouse. (B) Serum HBsAg, HBeAg, and HBcrAg levels in each mouse at baseline and after 12 weeks of treatment. (C) Intrahepatic HBV DNA and HBV cccDNA levels in each mouse. An HBV-infected mouse without treatment (no treatment) was also analyzed. Data are presented as the mean ± standard deviation (SD) for three samples of liver tissue from each mouse. *, P < 0.05. (D) Intrahepatic precore mRNA and HBs mRNA. HBcrAg, hepatitis B core-related antigen. LLOD, lower limit of detection.
Effect of high-dose combination treatment with entecavir and PEG-IFN on HBV-infected mice.
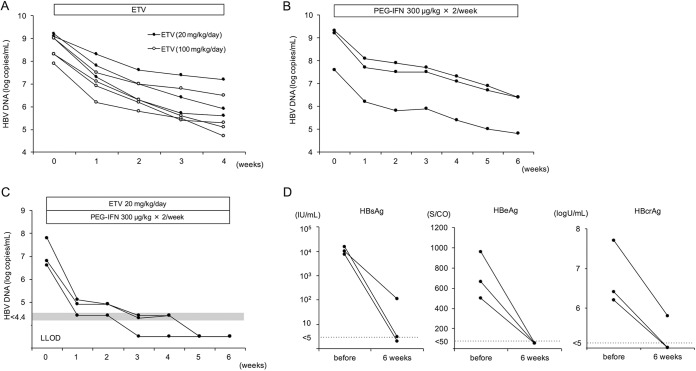
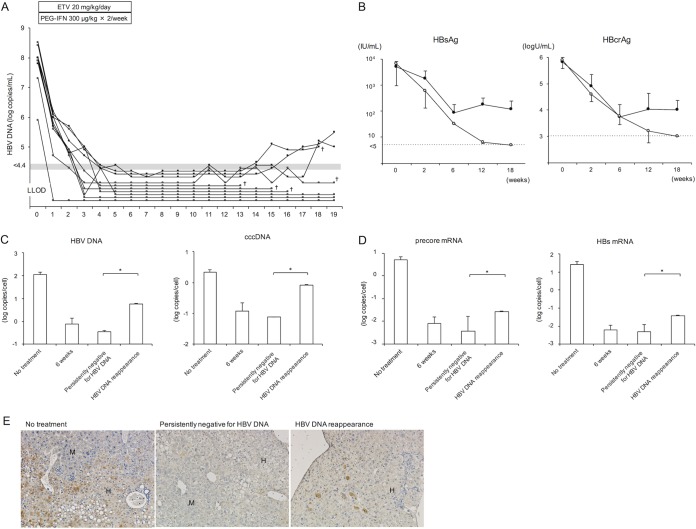
We next treated HBV-infected mice with a high dose of entecavir and PEG-IFN. We designated a dose of 20 mg/kg/day as a high dose of entecavir because no further antiviral effect was observed when more than 20 mg/kg (up to 100 mg/kg) of entecavir was administered (Fig. 2A). We also showed that 6 weeks of treatment with 300 μg/kg of PEG-IFN alone reduced mouse serum HBV DNA by approximately 3 logs (Fig. 2B). In this experiment, 13 HBV-infected mice were treated with 20 mg/kg of entecavir and 300 μg/kg of PEG-IFN for 6 weeks. Serum HBV DNA titers in these mice decreased more rapidly than those in mice treated with 2 mg/kg of entecavir and 30 μg/kg of PEG-IFN and fell below the detectable limit in all mice (Fig. 2C and 3A). Three out of the 13 mice were sacrificed at the end of the therapy. In these mice, serum HBsAg and HBcrAg values had decreased significantly, and HBeAg values had decreased below the detectable limit (Fig. 2D). Serum HBV DNA in the remaining 10 mice remained negative, but HBV DNA again became positive in three out of the 10 mice at 15, 16, and 18 weeks, respectively (Fig. 3A). Five mice were lost during examination, and the remaining five mice were sacrificed at 13 weeks after cessation of combination treatment. In mice in which serum HBV DNA reappeared, serum HBsAg and HBcrAg values also rebounded after cessation of the treatment (Fig. 3B). In contrast, the values continued to decline after cessation of the treatment in mice that remained persistently negative for serum HBV DNA. Six weeks of treatment with high-dose entecavir plus PEG-IFN resulted in a significant reduction of intrahepatic HBV DNA and cccDNA levels (Fig. 3C). By week 19 (13 weeks after cessation of the combination treatment), intrahepatic HBV DNA and cccDNA levels increased in mice in which serum HBV DNA had reappeared and decreased in mice that had remained persistently negative for serum HBV DNA. As with intrahepatic cccDNA levels, intrahepatic expression levels of the precore mRNA and HBs mRNA decreased after 6 weeks of combination treatment and remained at low levels in mice that were persistently negative for serum HBV DNA (Fig. 3D). Histological examination at 19 weeks (13 weeks after cessation of the combination treatment) showed HBcAg-positive hepatocytes in mice in which serum HBV DNA had reappeared. In contrast, no hepatocytes were positive for HBcAg in mice that had remained persistently negative for serum HBV DNA (Fig. 3E).
FIG 2.
Changes in HBV markers in mice treated with high-dose antivirals. (A to B) HBV-infected mice were treated with either 20 mg/kg or 100 mg/kg of entecavir (ETV) daily for 4 weeks (A) or 300 μg/kg of PEG-IFN-α-2a twice weekly for 6 weeks (B). (C and D) Three mice were treated with 20 mg/kg of entecavir daily and 300 μg/kg of PEG-IFN twice weekly for 6 weeks. (C) The time course of serum HBV DNA titer in each mouse. (D) Serum HBsAg, HBeAg, and HBcrAg levels in each mouse at baseline and after 6 weeks of treatment in three mice.
FIG 3.
Effect of high-dose antivirals on HBV-infected mice. Ten mice were treated with 20 mg/kg of entecavir daily and 300 μg/kg of PEG-IFN twice weekly for 6 weeks. (A) Time course of serum HBV DNA titer in each mouse. Mice in which serum HBV DNA reappeared after cessation of the treatment are shown with closed circles, and mice that remained persistently negative for serum HBV DNA after cessation of the treatment are shown with open circles. †, mice were lost at that time point, and therefore subsequent data could not be collected. Five mice were lost during examination, and the remaining five mice were sacrificed at 19 weeks. (B) Changes in serum HBsAg and HBcrAg levels. Mice in which serum HBV DNA reappeared after cessation of the treatment (closed circles) (n = 3) and mice that remained persistently negative for serum HBV DNA after cessation of the treatment (open circles) (n = 7) are shown. Data are presented as the mean ± SD. (C and D) Intrahepatic HBV DNA and HBV cccDNA (C) and precore mRNA and HBs mRNA (D) at 19 weeks in mice with serum HBV DNA reappearance after cessation of the treatment (HBV DNA reappearance; two samples of liver tissue in 2 mice) and mice persistently negative for serum HBV DNA after cessation of the treatment (persistently negative for HBV DNA; n = 3). HBV DNA, cccDNA, precore mRNA, and HBs mRNA levels were also measured in HBV-infected mice that did not receive treatment (no treatment; n = 6) or that received 6 weeks of combination treatment (6 weeks; n = 3). Data are presented as the mean ± SD. *, P < 0.05. (E) Histological analysis of liver samples. Liver samples were stained with anti-hepatitis B core (HBc) antibody. Labeled regions indicate human (H) and mouse (M) hepatocytes (original magnification, ×200). Similar histological patterns were observed in other mice in each group.
HBV hypermutation following treatment with entecavir and IFN.
IFN-α is known to induce hypermutation of the HBV genome. To analyze the mechanism of HBV regulation following extreme reduction of intrahepatic cccDNA levels via combination treatment with high-dose entecavir and PEG-IFN, HBV hypermutation was analyzed in five mice that were positive for serum HBV DNA after 2 weeks of the combination treatment (Fig. 4). In these five mice, serum HBV DNA became negative following treatment, but three of the mice again became positive for HBV DNA after cessation of the treatment. Differential DNA denaturation PCR (3D-PCR) and detection by AT-yellow agarose gel electrophoresis revealed slight increases in more heavily hypermutated HBV genomes at lower denaturation temperatures (88°C) after 2 weeks of treatment, but no significant differences were observed between mice in which serum HBV DNA reappeared and mice that remained persistently negative for serum HBV DNA.
FIG 4.
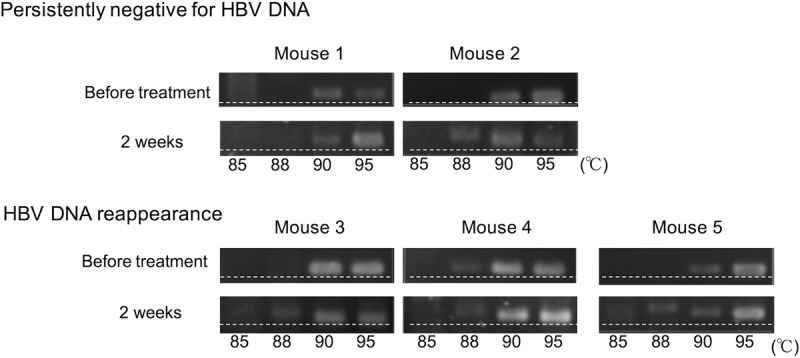
HBV hypermutation following treatment with PEG-IFN and entecavir. HBV DNA was amplified by 3D-PCR, and hypermutated genomes were detected by HA-yellow agarose gel electrophoresis in five mice that were positive for serum HBV DNA after 2 weeks of treatment with PEG-IFN plus entecavir. Serum HBV DNA remained persistently negative in two mice (persistently negative for HBV DNA; mice 1 and 2) and reappeared after cessation of the treatment in three mice (HBV DNA reappearance; mice 3, 4, and 5). More heavily hypermutated HBV DNA was observed at 88°C. The dashed lines were added to help visualize the retardation of AT-rich DNA in the HA-yellow agarose gel.
HBV methylation following IFN treatment.
Host-mediated methylation of the HBV genome at CpG II and CpG III is associated with suppression of HBV gene expression. Therefore, we analyzed HBV genome methylation in the liver following IFN treatment. Interestingly, twice weekly administration of 300 μg/kg of PEG-IFN for 6 weeks resulted in significant increases in intrahepatic HBV genome methylation at CpG III (Fig. 5).
FIG 5.
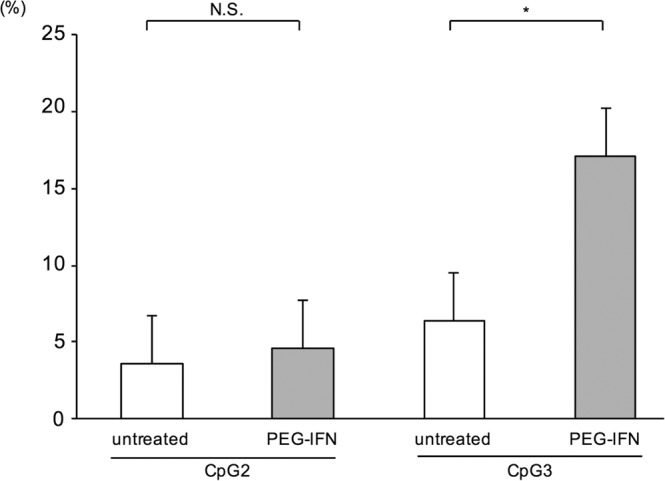
CpG methylation of the HBV genome following interferon treatment. Mice were treated with or without 300 μg/kg of PEG-IFN twice weekly. After 6 weeks of treatment, DNA was extracted from mouse livers, and the proportions of methylated sites in CpG II and CpG III islands were analyzed. Data are presented as the mean ± SD for three mice. *, P < 0.05; N.S., not significant.
DISCUSSION
Therapy is required to suppress HBV replication in patients with active chronic HBV infection to reduce the risk of progression to cirrhosis, HCC, and liver failure, but neither NA therapy nor PEG-IFN therapy is considered curative due to the inability of these drugs to directly eliminate cccDNA from hepatocytes. cccDNA is a plasmid-like minichromosome that serves as the template for the transcription of all viral mRNAs necessary for HBV protein production and viral replication (2). Transcription of pregenome RNA (pgRNA) from cccDNA is a critical step in genome amplification and HBV replication, and the viral proteins HBsAg and HBeAg are translated from HBs mRNA (pre-S/S mRNA) and precore mRNA, respectively. While NAs strongly suppress HBV replication by blocking the activity of HBV polymerase, HBV replication frequently rebounds following discontinuation of therapy or in the event of emergence of drug resistance. The antiviral activity of PEG-IFN is multifactorial and involves induction of a large class of interferon-stimulated genes (ISGs) that interfere with several aspects of the HBV life cycle, including HBV RNA transcription and translation (25). While the effects of NAs are transient, the effects of PEG-IFN may be more enduring; in addition to inhibiting the transcription of pgRNA and HBV mRNA, PEG-IFN also induces epigenetic modification and APOBEC3A cytidine deaminase-mediated editing of cccDNA (26), which is associated with a higher rate of HBsAg clearance (27). PEG-IFN administration has been shown to lead to recruitment of transcriptional corepressors and hypoacetylation of cccDNA-associated histones, as well as inhibition of binding of host transcription factors such as STAT1 and STAT2 to cccDNA (13). Although an antiviral effect of the combination of an NA and PEG-IFN-α-2a has been shown (15–18), it has not yet been shown that the synergistic activity of the two drugs in combination is more likely to be effective in durably suppressing or eliminating cccDNA.
To test this hypothesis, humanized mice were treated with entecavir and/or PEG-IFN for 12 weeks. Monotherapy with entecavir reduced serum HBV DNA levels but did not affect levels of HBsAg and HBeAg. Conversely, PEG-IFN monotherapy reduced HBsAg and HBeAg levels but had only a minimal effect on serum HBV DNA levels, suggesting that while the drugs act via different mechanisms, neither has sufficient efficacy as a monotherapy. Consistent with previous clinical reports (15–20), combination treatment with both entecavir and PEG-IFN reduced serum HBV DNA, intrahepatic HBV DNA, and cccDNA levels (Fig. 1C). While PEG-IFN treatment reduced intrahepatic precore and HBs mRNA levels, the combination of entecavir and PEG-IFN suppressed the levels of these mRNAs to a significantly greater extent than either drug alone (Fig. 1D). IFN is known to mediate cccDNA transcriptional activity by epigenetic control (13, 28), which might lead to negative serum HBV DNA levels and a strong reduction of HBsAg, reflecting attenuation of cccDNA establishment. The synergistic effect of combination therapy might also result from relief of HBV-mediated interference in the interferon signaling pathway (29).
To assess the effect of extreme reduction of intrahepatic cccDNA, HBV-infected mice were treated with a 10-fold-higher dose of entecavir plus PEG-IFN. The high-dose combination treatment resulted in a rapid decline of serum HBV DNA (Fig. 2C) and reduced HBsAg, HBeAg, and HBcrAg values (Fig. 2D). After 6 weeks of the high-dose combination treatment, intrahepatic HBV DNA and cccDNA were reduced approximately 10-fold compared to the results of treatment with the prescribed dose (Fig. 3C). Serum HBV DNA reappeared in some mice after cessation of the treatment, followed by rebound of serum HBsAg and HBcrAg values (Fig. 3B). The reappearance of viral markers after discontinuation of treatment might also be due in part to integration of HBV DNA into the host genome. In contrast, these values continued to decline after cessation of the treatment in mice that remained persistently negative for serum HBV DNA (Fig. 3B). The level of intrahepatic cccDNA is correlated with HBV activity (24), and it was reported that the levels at the end of treatment with entecavir plus PEG-IFN correlated with viral relapse after cessation of the treatment in chronic hepatitis B patients (20). Taken together, these results suggest that a high dose of entecavir plus PEG-IFN could inhibit HBV replication without an immune response through the extreme reduction of intrahepatic cccDNA levels in some mice. In these mice, serum HBV DNA remained negative until 13 weeks after cessation of the treatment. Future analysis with a longer-term follow-up is needed to verify complete HBV eradication.
Although the metabolism of entecavir and PEG-IFN is different in mice and humans, the doses of entecavir (20 mg/kg) and PEG-IFN (300 μg/kg) used in the present study are not expected to be practical for use in patients because development of severe adverse events is expected in some patients. However, our results showed conceptually that extreme reduction of intrahepatic cccDNA levels by anti-HBV therapy might make it possible to minimize HBV replication even after cessation of the treatment. Therefore, it is necessary to develop safe and effective new treatments to reduce or eliminate intrahepatic cccDNA. Lucifora et al. reported that activation of lymphotoxin-β receptor induced the degradation of cccDNA without hepatotoxicity (26). Recently, the potential for genome editing using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 to reduce cccDNA both in vitro and in vivo has been described (30). Further study is needed to develop new therapeutic approaches to eliminate cccDNA.
G-to-A hypermutation of HBV occurs as a result of deamination activity by the host APOBEC proteins and is thought to play a role in antiviral immunity. IFN treatment induces transcription of APOBEC proteins and increases hypermutation of the HBV genome (26, 31). Although combination treatment with entecavir and PEG-IFN increased hypermutation of the HBV genome in serum in the present study, no correlation was observed between HBV hypermutation and reappearance of HBV DNA (Fig. 4). Hypermutation of the HBV genome in serum was analyzed in only five mice in the present study. Further analysis using a large number of animals and HBV-infected patients to clarify the relationship between hypermutation in serum and intrahepatic HBV genomes and the HBV activity after cessation of treatment with an NA plus PEG-IFN is needed.
The 3.2-kb HBV genome contains three major CpG islands. Island I contains the start site of the S gene, island II spans a region that overlaps enhancer I/II and is proximal to the core promoter, and island III covers the start codon of the polymerase gene and the upstream region of the SP1 promoter (32, 33). It was reported that transcription of HBV cccDNA is regulated by CpG methylation during chronic infection (27, 33). In the present study, using HBV-infected mice, PEG-IFN treatment significantly increased methylation of HBV cccDNA in CpG island III (Fig. 5). This is the first report that shows that IFN induces methylation of HBV cccDNA, and the result suggests that IFN treatment inhibits HBV regulation through induction of methylation of HBV cccDNA. In a previous study, Liu et al. showed that while IFN-α durably suppresses transcription in duck hepatitis B virus (DHBV) in a chicken hepatoma cell line through epigenetic modification of cccDNA, the effect was due to reduced acetylation of DHBV histone H3 and not due to DNA methylation (34). The reason for this difference is unclear, but it may result from differences in the experimental systems or between HBV and DHBV or be related the expanded repertoire of ISGs activated by PEG-IFN relative to IFN (35), and changes in both acetylation and methylation are likely involved. Nonetheless, it was difficult to analyze methylation of the HBV genome in the livers of mice in which undetectable viremia was maintained due to lower cccDNA levels. Further analysis is needed to clarify the correlation between CpG methylation and IFN treatment response in HBV-infected patients.
To our knowledge, this is the first report describing elimination of HBV without involvement of the cellular immune response in an animal model. Persistent loss of serum HBV DNA and loss of HBV markers using a high dose of entecavir plus PEG-IFN might facilitate control of HBV in part through IFN-induced methylation of HBV cccDNA. Although it is not clear whether HBV was completely eliminated from hepatocytes in this study, the absence of viremia for more than 12 weeks suggests the elimination of replication-competent HBV, which might result in a practical cure.
MATERIALS AND METHODS
Generation of HBV-infected human hepatocyte chimeric mice.
Generation of the uPA+/+ SCID+/+ mice and transplantation of human hepatocytes were performed as described previously (22). Frozen human hepatocytes obtained from the same donor were transplanted into all mice. All animal protocols described in this study were performed in accordance with the guidelines of the local committee for animal experiments, and all animals received humane care. Human hepatocyte chimeric mice were injected with 5 × 105 copies of HBV-positive human serum intravenously at 8 weeks after hepatocyte transplantation. Human serum samples were obtained from a patient with chronic hepatitis B infection who had high titer of HBeAg-positive HBV DNA genotype C (1.0 × 107 copies/ml).
Measurement of HBV DNA and HBV markers in serum.
Serum HBV DNA levels were measured quantitatively using a TaqMan-based assay according to the protocol provided by the manufacturer (Cobas AmpliPrep/Cobas TaqMan HBV test, v2.0; Roche Diagnostics, Tokyo, Japan). The lower detection limit of real-time PCR (RT-PCR) for HBV DNA was 4.4 log copies/ml (36). Quantification of HBsAg and HBeAg was performed using Abbott Architect platforms (Abbott, Diagnostic Division, Ireland) as recommended by the manufacturer. Serum HBcrAg levels were measured using a Cleia HBcrAg assay kit with a fully automated analyzer system (Lumipulse system; Fujirebio Inc., Tokyo, Japan).
Treatment of mice with entecavir and/or PEG-IFN.
Mice developed stable viremia (approximately 8 log IU/ml) by 8 weeks after HBV infection. HBV-infected mice were treated with 2 mg/kg of entecavir (Baraclude solution; Bristol-Myers-Squibb, Munich, Germany) and/or 30 μg/kg PEG-IFN-α-2a (Chugai Pharmaceutical Co. Ltd., Tokyo, Japan) for 12 weeks. Entecavir was administered orally once per day, and PEG-IFN-α-2a was administered subcutaneously twice per week.
DNA extraction and quantification of intrahepatic HBV DNA and cccDNA.
DNA was extracted from 20 mg of mouse liver using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Quantification of intrahepatic HBV DNA and cccDNA was performed using an ABI Prism 7300 (Applied Biosystems, Carlsbad, CA, USA). Before measuring intrahepatic cccDNA expression, selective degradation of single-stranded DNA was performed in 20 μl of solution with 10 U of S1 nuclease (TakaraBio, Shiga, Japan) for 1 μg of DNA with 2 μl of 10× S1 nuclease buffer and incubation at room temperature for 15 min. The reaction was terminated by the addition of 1 μl of 12.5 mM EDTA. Quantification of cccDNA expression was performed with TaqMan gene expression master mix (Applied Biosystems) using forward primer (nucleotides [nt] 1521 to 1545) 5′-GGGGCGCACCTCTCTTTACGCGGTC-3′, reverse primer (nt 1862 to 1886) 5′-CAAGGCACAGCTTGGAGGCTTGAAC-3′, and TaqMan MGB probe (nt 1685 to 1704) 5-FAM-AACGACCGACCTTGAGGCAT-MGB-3. The concentration of forward and reverse primers was 300 nM, and the probe concentration was 100 nM. After 2 min of incubation at 50°C and 10 min of polymerase activation at 95°C, PCR was performed for 45 cycles (95°C for 10 s and 65°C for 30 s). Results were normalized by the cell numbers of the liver samples. Cell number was estimated by measuring the β-actin gene level using the TaqMan gene expression assay (Applied Biosystems).
RNA extraction and quantification of intrahepatic mRNA expression level.
RNA was extracted from liver samples using NucleoSpin RNA II (TaKaRa Bio). RT-PCR was performed using random primers and ReverTra Ace (Toyobo Co., Ltd., Osaka, Japan). Quantification of mRNA expression levels was performed using Power SYBR green master mix (Applied Biosystems). The primers used for quantification of the expression level of precore mRNA were 5′-GGTCTGTTCACCAGCACCAT-3′ and 5′-GGAAAGAAGTCAGAAGGCAA-3′, those for detection of core promoter transcripts were 5′-CCGGAAAGCTTGAGCTCTTCTT-3′ and 5′-CACAGAATAGCTTGCCTGAGTG-3′, and those for measurement of total HBV RNA transcription levels were 5′-CCAGGAACATCAACCACCAG-3′ and 5′-CGAACCACTGAACAAATGGC-3′. All amplifications were performed as recommended. The levels of pgRNA transcripts were calculated by subtracting precore mRNA levels from levels of total core protein transcripts. HBs transcripts were calculated by subtracting total core promoter transcripts from total HBV RNA transcripts.
Histochemical analysis of mouse liver.
Immunohistochemical staining using antibodies against hepatitis B core antigen (HBcAg) (Dako Diagnostika, Hamburg, Germany) were performed as described previously (23). Immunoreactive materials were visualized using a streptavidin-biotin staining kit (Vectastain ABC kit; Vector Laboratories Inc., Burlingame, CA, USA) and diaminobenzidine.
Amplification and analysis of hypermutated HBV genomes by 3D-PCR.
Detection of hypermutated genomes was performed using 3D-PCR as described previously (37). As AT-rich DNA melts at lower temperatures than GC-rich DNA, use of lower denaturation temperatures (85°C and 88°C) during PCR allows differential amplification of AT-rich genomes or variants within a quasispecies. The amplification conditions included an initial denaturation step at 83 to 95°C for 5 min, followed by 45 cycles of denaturation at 83 to 95°C for 1 min, annealing at 50°C for 30 s, and extension at 72°C for 30 s and then 10 min of final extension. The PCR products were analyzed on a 2% agarose gel containing AT-yellow (Resolve-It kit; Vector Laboratories, Inc., Burlingame, CA, USA). Gel preparation and electrophoresis were performed according to the instructions provided by the manufacturer. AT-yellow binds to the DNA minor groove at regions of four consecutive A/T base pairs. Using agarose gels containing AT-yellow, sequence-specific DNA ligands bind to DNA in a sequence-dependent manner and retard the electrophoretic migration of the DNA. This ability allows for the detection of G-to-A hypermutation of the HBV genome by separation of DNAs of similar size but containing different sequences in an agarose gel.
Analysis of HBV genome methylation using ultradeep sequencing.
We used Methyl Primer Express software v1.0 (Applied Biosystems) for detection of CpG islands and design of primers for bisulfite sequencing PCR. To analyze methylation within the HBV genome, the location of the islands was mapped within the HBV isolate used to infect the humanized mice. The CpG islands were defined based on the following criteria: a GC content of ≥50%, an observed-to-expected CpG dinucleotide ratio of ≥0.60, and a sequence window longer than 100 bp. Bisulfite sequencing primers were designed separately for both CpG islands II (nt 1228 to 1663) and III (nt 2295 to 2446) to avoid overlap between CpG sites (Table 1) (32). DNA samples were modified using the Cells-to-CpG bisulfite conversion kit (Applied Biosystems) according to the manufacturer's specifications. The bisulfite-modified DNA was amplified by nested PCR with AmpliTaq Gold 360 master mix (Life Technologies, Carlsbad, CA, USA) using the following thermal profile: initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 30 s, followed by 7 min of final extension at 72°C. The PCR products were purified, and an adaptor-ligated library was prepared as described previously (38). Sequence analysis was performed using an Ion PGM sequencer (Life Technologies). Sequence data quality was examined using FastQC before and after trimming with Trimmomatic version 36 with default settings. Reads were mapped to the HBV reference sequence (accession number NC_003977.2) using bwa-meth version 0.2.0 with default settings, and methylation data were tabulated using PileOMeth version 0.1.13 with the mergeContext option. Mean methylation percentages were compared between interferon-treated and untreated mice using Student's t test (R version 3.3.1).
TABLE 1.
Primers for bisulfite sequencing PCR
| Target and primer direction and sequencing round | Primer sequence, 5′→3′ |
|---|---|
| HBV CpG island II | |
| Forward, first | TTAGGGTGTGTTGTTAATTGGA |
| Reverse, first | TAAAAACCCAAACRACCC |
| Forward, second | TGGTTGTTAGGGTGTGTTG |
| Reverse, second | ACCRACAAATAAAAAAACACAA |
| HBV CpG island III | |
| Forward, first | GGAGTGTGGATTYGTATTT |
| Reverse, first | TAAAATTTCCCACCTTATAAATC |
| Forward, second | AGGTAGGTTTTTTAGAAGAAGAAT |
| Reverse, second | AAAAAATACTAACATTAAAATTCCC |
Statistical analysis.
Levels of intrahepatic HBV DNA, cccDNA, and mRNA expression were compared by Mann-Whitney U tests. P values less than 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank Rie Akiyama and Yoko Inoue for their expert technical help and Emi Nishio and Akemi Sata for technical assistance. Part of this work was carried out at the Analysis Center of Life Science, Natural Science Center for Basic Research and Development, Hiroshima University.
This research is partially supported by research funding from the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (AMED) (grant number 16fk0310017h0004). There was no additional external funding received for this study.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Kazuaki Chayama received honoraria from MSD K.K. and Bristol-Myers Squibb and research funding from AbbVie, Dainippon Sumitomo Pharma, and The Institute of Physical and Chemical Research (RIKEN). Michio Imamura received honoraria and research funding from Bristol-Myers Squibb. Masataka Tsuge received research funding from Bristol-Myers Squibb.
M.I., N.H., and K.C. conceived and designed the study. T.U., H.K., C.N.H., N.H., M.T., H.A.-C., Y.Z., G.N.M., D.M., and H.O. performed experiments and procedures. H.A. and K.C. administered the project. Y.I. and C.T. provided resources. T.U., M.I., C.N.H., N.H., and K.C. wrote the article.
REFERENCES
- 1.Trépo C, Chan HLY, Lok A. 2014. Hepatitis B virus infection. Lancet 384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Nassal M. 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 3.Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Schall RA, Bornstein JD, Kitrinos KM, Subramanian GM, McHutchison JG, Heathcote EJ. 2013. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet 381:468–475. doi: 10.1016/S0140-6736(12)61425-1. [DOI] [PubMed] [Google Scholar]
- 4.Chang TT, Liaw YF, Wu SS, Schiff E, Han KH, Lai CL, Safadi R, Lee SS, Halota W, Goodman Z, Chi YC, Zhang H, Hindes R, Iloeje U, Beebe S, Kreter B. 2010. Long-term entecavir therapy results in the reversal of fibrosis/cirrhosis and continued histological improvement in patients with chronic hepatitis B. Hepatology 52:886–893. doi: 10.1002/hep.23785. [DOI] [PubMed] [Google Scholar]
- 5.Dienstag JL, Goldin RD, Heathcote EJ, Hann HWL, Woessner M, Stephenson SL, Gardner S, Gray DF, Schiff ER. 2003. Histological outcome during long-term lamivudine therapy. Gastroenterology 124:105–117. doi: 10.1053/gast.2003.50013. [DOI] [PubMed] [Google Scholar]
- 6.Hosaka T, Suzuki F, Kobayashi M, Seko Y, Kawamura Y, Sezaki H, Akuta N, Suzuki Y, Saitoh S, Arase Y, Ikeda K, Kobayashi M, Kumada H. 2013. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology 58:98–107. doi: 10.1002/hep.26180. [DOI] [PubMed] [Google Scholar]
- 7.Wong GL-H, Chan HL-YH-Y, Mak CW-H, Lee SK-Y, Ip ZM-Y, Lam AT-H, Iu HW-H, Leung JM-S, Lai JW-Y, Lo AO, Chan HY, Wong VW-S. 2013. Entecavir treatment reduces hepatic events and deaths in chronic hepatitis B patients with liver cirrhosis. Hepatology 58:1537–1547. doi: 10.1002/hep.26301. [DOI] [PubMed] [Google Scholar]
- 8.Allweiss L, Volz T, Lütgehetmann M, Giersch K, Bornscheuer T, Lohse AW, Petersen J, Ma H, Klumpp K, Fletcher SP, Dandri M. 2014. Immune cell responses are not required to induce substantial hepatitis B virus antigen decline during pegylated interferon-alpha administration. J Hepatol 60:500–507. doi: 10.1016/j.jhep.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Volz T, Allweiss L, Ben MBarek M, Warlich M, Lohse AW, Pollok JM, Alexandrov A, Urban S, Petersen J, Lütgehetmann M, Dandri M. 2013. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol 58:861–867. doi: 10.1016/j.jhep.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Chan HLY, Thompson A. 2011. Hepatitis B surface antigen quantification: why and how to use it in 2011—a core group report. J Hepatol 55:1121–1131. doi: 10.1016/j.jhep.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Lau GKK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, Chang WY, Berg T, Flisiak R, McCloud P, Pluck N. 2005. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med 352:2682–2695. doi: 10.1056/NEJMoa043470. [DOI] [PubMed] [Google Scholar]
- 12.Tangkijvanich P, Chittmittraprap S, Poovorawan K, Limothai U, Khlaiphuengsin A, Chuaypen N, Wisedopas N, Poovorawan Y. 2016. A randomized clinical trial of peginterferon alpha-2b with or without entecavir in patients with HBeAg-negative chronic hepatitis B: role of host and viral factors associated with treatment response. J Viral Hepat 23:417–438. doi: 10.1111/jvh.12467. [DOI] [PubMed] [Google Scholar]
- 13.Belloni L, Allweiss L. 2012. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest 122:529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen HLA, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TMK, Gerken G, de Man RA, Niesters HGM, Zondervan P, Hansen B, Schalm SW. 2005. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 365:123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- 15.Brouwer WP, Xie Q, Sonneveld MJ, Zhang N, Zhang Q, Tabak F, Streinu-Cercel A, Wang JY, Idilman R, Reesink HW, Diculescu M, Simon K, Voiculescu M, Akdogan M, Mazur W, Reijnders JGP, Verhey E, Hansen BE, Janssen HLA. 2015. Adding pegylated interferon to entecavir for hepatitis B e antigen-positive chronic hepatitis B: a multicenter randomized trial (ARES study). Hepatology 61:1512–1522. doi: 10.1002/hep.27586. [DOI] [PubMed] [Google Scholar]
- 16.Marcellin P, Ahn SH, Ma X, Caruntu FA, Tak WY, Elkashab M, Chuang WL, Lim SG, Tabak F, Mehta R, Petersen J, Foster GR, Lou L, Martins EB, Dinh P, Lin L, Corsa A, Charuworn P, Subramanian GM, Reiser H, Reesink HW, Fung S, Strasser SI, Trinh H, Buti M, Gaeta GB, Hui AJ, Papatheodoridis G, Flisiak R, Chan HLY. 2016. Combination of tenofovir disoproxil fumarate and peginterferon α-2a increases loss of hepatitis B surface antigen in patients with chronic hepatitis B. Gastroenterology 150:134–144.e10. doi: 10.1053/j.gastro.2015.09.043. [DOI] [PubMed] [Google Scholar]
- 17.Chi H, Hansen BE, Guo S, Zhang NP, Qi X, Chen L, Guo Q, Arends P, Wang J-Y, Verhey E, de Knegt RJ, Xie Q, Janssen HLA. 2017. Pegylated interferon alfa-2b add-on treatment in hepatitis B virus envelope antigen-positive chronic hepatitis B patients treated with nucleos(t) ide analogue: a randomized, controlled trial (PEGON). J Infect Dis 215:1085–1093. doi: 10.1093/infdis/jix024. [DOI] [PubMed] [Google Scholar]
- 18.Cao Z, Liu Y, Ma L, Lu J, Jin Y, Ren S, He Z, Shen C, Chen X. 13 April 2017. A potent HBsAg response in subjects with inactive HBsAg carrier treated with pegylated-interferon alpha. Hepatology doi: 10.1002/hep.29213. [DOI] [PubMed] [Google Scholar]
- 19.Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M, Fischer C, Currie G, Brosgart C, Petersen J. 2006. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology 44:675–684. doi: 10.1002/hep.21282. [DOI] [PubMed] [Google Scholar]
- 20.Hagiwara S, Kudo M, Osaki Y, Matsuo H, Inuzuka T, Matsumoto A, Tanaka E, Sakurai T, Ueshima K, Inoue T, Yada N, Nishida N. 2013. Impact of peginterferon alpha-2b and entecavir hydrate combination therapy on persistent viral suppression in patients with chronic hepatitis B. J Med Virol 995:987–995. doi: 10.1002/jmv.23564. [DOI] [PubMed] [Google Scholar]
- 21.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat Med 7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 22.Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, Soeno Y, Asahina K, Hino H, Asahara T, Yokoi T, Furukawa T, Yoshizato K. 2004. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol 165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuge M, Hiraga N, Takaishi H, Noguchi C, Oga H, Imamura M, Takahashi S, Iwao E, Fujimoto Y, Ochi H, Chayama K, Tateno C, Yoshizato K. 2005. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis B virus. Hepatology 42:1046–1054. doi: 10.1002/hep.20892. [DOI] [PubMed] [Google Scholar]
- 24.Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. 2006. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology 44:694–702. [DOI] [PubMed] [Google Scholar]
- 25.Konerman MA, Lok AS. 2016. Interferon treatment for hepatitis B. Clin Liver Dis 20:645–665. doi: 10.1016/j.cld.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou W-M, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W-C, Wang M-R, Kong L-B, Ren W-G, Zhang Y-G, Nan Y-M. 2011. Peginterferon alpha-based therapy for chronic hepatitis B focusing on HBsAg clearance or seroconversion: a meta-analysis of controlled clinical trials. BMC Infect Dis 11:165. doi: 10.1186/1471-2334-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Christen V, Duong F, Bernsmeier C, Sun D, Nassal M, Heim MH. 2007. Inhibition of alpha interferon signaling by hepatitis B virus. J Virol 81:159–165. doi: 10.1128/JVI.01292-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy EM, Kornepati AVR, Cullen BR. 2015. Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antiviral Res 123:188–192. doi: 10.1016/j.antiviral.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J. 2014. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 9:e110442. doi: 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivekanandan P, Thomas D, Torbenson M. 2008. Hepatitis B viral DNA is methylated in liver tissues. J Viral Hepat 15:103–107. doi: 10.1111/j.1365-2893.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, Guo JT. 2013. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog 9:e1003613. doi: 10.1371/journal.ppat.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dill MT, Makowska Z, Trincucci G, Gruber AJ, Vogt JE, Filipowicz M, Calabrese D, Krol I, Lau DT, Terracciano L, Van Nimwegen E, Roth V, Heim MH. 2014. Pegylated IFN-α regulates hepatic gene expression through transient Jak/STAT activation. J Clin Invest 124:1568–1581. doi: 10.1172/JCI70408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goedel S, Rullkoetter M, Weisshaar S, Mietag C, Leying H, Boehl F. 2009. Hepatitis B virus (HBV) genotype determination by the COBAS AmpliPrep/COBAS TaqMan HBV Test, v2.0 in serum and plasma matrices. J Clin Virol 45:232–236. doi: 10.1016/j.jcv.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Fujimoto Y, Ochi H, Abe H, Maekawa T, Yatsuji H, Shirakawa K, Takaori-Kondo A, Chayama K. 2007. Dual effect of APOBEC3G on hepatitis B virus. J Gen Virol 88:432–440. doi: 10.1099/vir.0.82319-0. [DOI] [PubMed] [Google Scholar]
- 38.Uchida T, Hiraga N, Imamura M, Yoshimi S. 2016. Elimination of HCV via a non-ISG-mediated mechanism by vaniprevir and BMS-788329 combination therapy in human hepatocyte chimeric mice. Virus Res 213:62–68. doi: 10.1016/j.virusres.2015.11.010. [DOI] [PubMed] [Google Scholar]