ABSTRACT
Acinetobacter baumannii is emerging as a multidrug-resistant nosocomial pathogen of increasing threat to human health worldwide. Pili are important bacterial virulence factors, playing a role in attachment to host cells and biofilm formation. The Csu pilus, which is assembled via the chaperone-usher secretion system, has been studied in A. baumannii ATCC 19606. Here we show that, in opposition to previous reports, the common laboratory strain ATCC 17978 produces Csu pili. We found that, although ATCC 17978 was resistant to sulfamethoxazole (Smx) and trimethoprim (Tmp), subinhibitory concentrations of these antibiotics abolished the expression of Csu and consequently produced a dramatic reduction in biofilm formation by ATCC 17978. Smx and Tmp acted synergistically to inhibit the enzymatic systems involved in the bacterial synthesis of tetrahydrofolate (THF), which is required for the synthesis of nucleotides. The effects of these antibiotics were partially relieved by exogenous THF addition, indicating that Smx and Tmp turn off Csu assembly by inducing folate stress. We propose that, for Acinetobacter, nanomolar concentrations of Smx and Tmp represent a “danger signal.” In response to this signal, Csu expression is repressed, allowing biofilm dispersal and escape from potentially inhibitory concentrations of antibiotics. The roles of antibiotics as signaling molecules are being increasingly acknowledged, with clear implications for both the treatment of bacterial diseases and the understanding of complex microbial interactions in the environment.
KEYWORDS: Acinetobacter, folate biosynthesis, folate stress, pilus assembly
INTRODUCTION
Pili formed by Gram-negative bacteria play an important role in the ability of these organisms to adhere to surfaces, to form biofilms, and to invade host cells and can affect motility (1). The chaperone-usher (CU) family of pili has been the focus of many studies, and the P-pili and type I pili of uropathogenic Escherichia coli play particularly important roles in establishing infection (2–4). A CU pathway for pilus biogenesis is present in most medically relevant Acinetobacter species (5). The genes for this CU pathway are encoded in a single six-gene operon, termed csu, and include csuA/B, csuA, csuB, csuC, csuD, and csuE (6). In the presence of the CsuC chaperone, CsuA/B polymerizes to form the major pilus subunit (7). Additional experimental and bioinformatic analyses have provided evidence that CsuD functions as the usher and CsuE as a tip adhesin, while CsuA and CsuB may play roles as minor pilin subunits (5, 6, 8). Despite its being conserved in many sequenced Acinetobacter baumannii strains, the role of Csu has been characterized only for A. baumannii ATCC 19606, an isolate used in many laboratories (5, 6). Genetic studies in this strain have shown that mutation of csuC or csuE completely abolishes expression of Csu pili and prevents biofilm formation (5). Additionally, the two-component regulatory system BfmRS is involved in Csu expression, as a bfmR mutant did not express CsuA/B and was unable to form biofilms (6). Interestingly, previous genome sequence analyses suggested that other A. baumannii strains, including the commonly used ATCC 17978 (17978) strain, encode a nonfunctional Csu operon due to inactivating single-nucleotide polymorphisms (9, 10). However, those observations have not been phenotypically confirmed.
We recently resequenced A. baumannii ATCC 17978 (GenBank accession no. CP012004), which led to the identification of a previously unreported plasmid, pAB3. We found that, when cells were cultured under standard laboratory conditions, pAB3 was readily lost by a subset of 17978 cells, leading to activation of the type VI secretion system (T6SS) (11), and we suggested that this may be a differentiation process that yields two distinct bacterial types. Originally, these two cell types were termed Ab17978T6+ (containing no pAB3 plasmid [pAB3−]) and Ab17978T6− (containing the pAB3 plasmid [pAB3+]), in reference to their T6SS-related phenotypes. Here, we update our nomenclature and refer to these cells as 17978pAB3− and 17978pAB3+, respectively, to indicate cells that either do not or do harbor the plasmid. Importantly, the chromosomal sequences of 17978pAB3− and 17978pAB3+ are identical. In addition to playing a regulatory role for T6SS, we sought to determine whether pAB3 regulated additional chromosomal genes, such as Csu.
Here, we present data showing that Csu pili are expressed and are essential for biofilm formation in A. baumannii ATCC 17978. We found that subinhibitory concentrations of the antifolate antibiotics sulfamethoxazole (Smx) and trimethoprim (Tmp), which are used to maintain the pAB3 plasmid, are able to repress CsuA/B expression. This result indicates that pilus expression is tightly linked to folate metabolism.
RESULTS
A. baumannii ATCC 17978 produces Csu pili, which are required for biofilm formation.
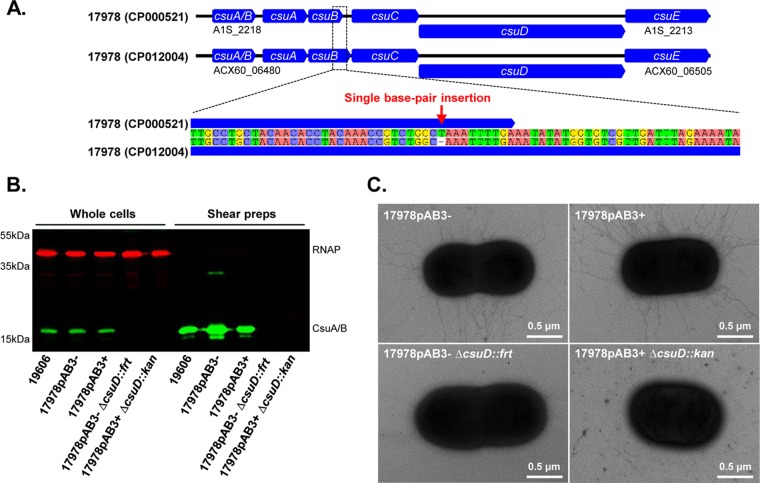
To investigate the effects of carrying the pAB3 plasmid on gene expression, we carried out transcriptome sequencing (RNA-seq) experiments to compare the transcriptomes of 17978pAB3− and 17978pAB3+. This analysis indicated that T6SS was regulated by the plasmid (12) but, unexpectedly, we found that the Csu pili were also induced ∼5-fold in 17978pAB3− cells. Previous comparative genomic analyses using the original 17978 sequence (GenBank accession no. CP000521) reported that the csuB gene contained a single-base-pair insertion, resulting in a truncated coding sequence, which led to the hypothesis that 17978 cells were unable to produce functional Csu pili (9, 10). However, inspection of the recently updated genome sequence (GenBank accession no. CP012004) revealed that the entire csuB gene is actually intact and does not contain an insertion (Fig. 1A) (11). Therefore, we decided to investigate whether Csu pili were assembled and to determine whether their expression was plasmid regulated in 17978, as suggested by the RNA-seq experiments.
FIG 1.
Expression of Csu pili in A. baumannii ATCC 17978. (A) Schematic diagram of the csu gene clusters in the previous A. baumannii ATCC 17978 genome sequence (GenBank accession no. CP000521) and the recently corrected A. baumannii ATCC 17978 genome sequence (GenBank accession no. CP012004). The previous csuB gene (A1S_2216) contained a thymine base pair insertion, while the updated genome sequence of the csuB gene (ACX60_06490) does not contain this insertion. (B) Confirmation of Csu pilus expression by Western blotting. Total cell lysates and shear preparation samples of A. baumannii ATCC 19606 and ATCC 17978 wild-type strains, with and without the pAB3 plasmid (17978pAB3+ and 17978pAB3−, respectively), and csuD mutant strains (17978pAB3− ΔcsuD::frt and 17978pAB3+ ΔcsuD::kan) were subjected to SDS-PAGE, followed by immunoblotting. Immunoblotting was performed using the CsuA/B-specific antibody (green bands), and RNAP was used as the loading control (red bands). CsuA/B antiserum reacted with an ∼19-kDa protein in 19606, 17978pAB3−, and 17978pAB3+ samples that was absent in the csu mutant samples, indicating that functional Csu pili are present not only in the 19606 strain but also in the 17978 strain and Csu pili are not expressed in the 17978pAB3− ΔcsuD::frt and 17978pAB3+ ΔcsuD::kan strains. (C) Transmission electron microscopic images showing the presence of Csu pili in 17978pAB3− and 17978pAB3+ wild-type cells. Csu pili were not detectable on the surface of csuD mutant strains (17978pAB3− ΔcsuD::frt and 17978pAB3+ ΔcsuD::kan).
We detected CsuA/B, the proposed major pilin protein, by Western blotting using an anti-CsuA/B antibody in whole cells and shear surface preparations of both 17978pAB3− and 17978pAB3+ cells (Fig. 1B). Importantly, CsuA/B was also detected in A. baumannii ATCC 19606 (Fig. 1B), a strain that was previously shown to express functional Csu pili (5). Unmarked deletion of csuD in 17978pAB3− (csuD::frt) and marked deletion in 17978pAB3+ (csuD::kan) resulted in complete loss of CsuA/B expression in whole cells and shear preparations (Fig. 1B). Both 17978pAB3− and 17978pAB3+ cells had visible surface pili when imaged by electron microscopy (EM), but pili were absent in their respective csuD mutant strains (Fig. 1C). Loss of CsuA/B expression in the csuD mutant is consistent with previous reports showing that mutation of csu genes causes inhibitory feedback of the entire system (5, 13). The entire csu cluster, with its native promoter, was cloned to generate the complementation vector pCsuFull (Fig. 2A). We then introduced pCsuFull into the 17978pAB3− csuD mutant, which restored CsuA/B expression and export to the cell surface to wild-type levels (Fig. 2B). This result was confirmed by the presence of visible pilus structures on the cell surface of the complemented strain, as observed by EM (Fig. 2C).
FIG 2.
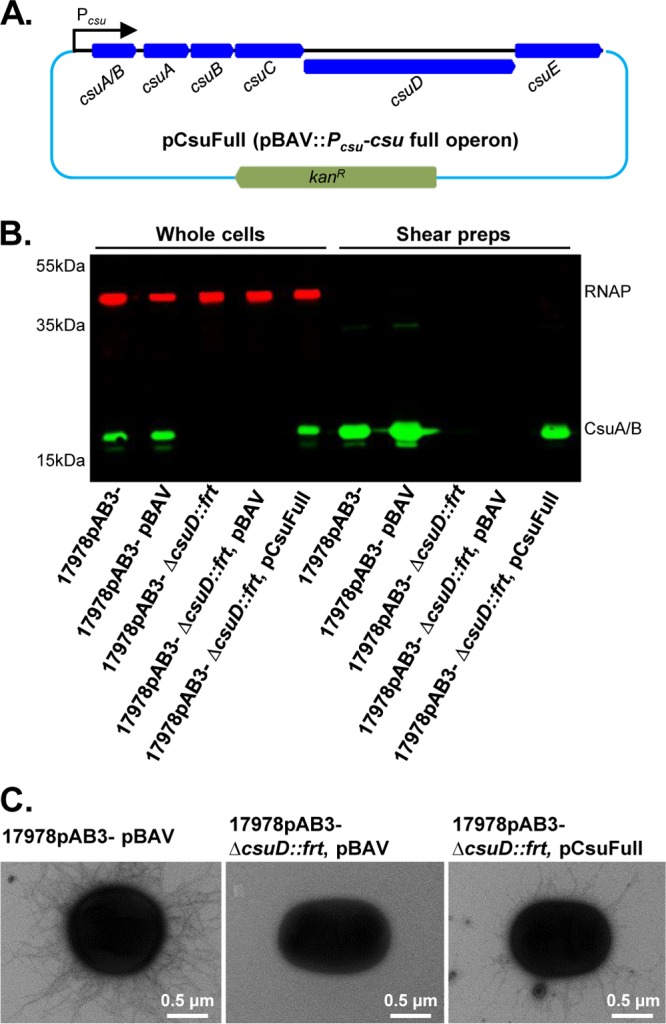
Complementation of Csu pili in 17978pAB3− ΔcsuD::frt. (A) Schematic diagram of the complementation plasmid pCsuFull (pBAV::csu full operon). The entire csu gene cluster, including its own promoter region, was cloned into the pBAV shuttle vector, which confers resistance to kanamycin. The pCsuFull plasmid was then electroporated in 17978pAB3− ΔcsuD::frt competent cells to complement the mutant in trans. (B) Confirmation of the expression of Csu pili in complemented cells by Western blotting. Total cell lysates and shear preparation samples were subjected to SDS-PAGE and immunoblotting with the anti-CsuA/B antibody (green bands); RNAP was used as a loading control (red bands). CsuA/B was detected in the 17978pAB3− wild-type strain and its vector control strain (17978pAB3− pBAV) but not in the 17978pAB3− ΔcsuD::frt strain and its vector control strain (17978pAB3− ΔcsuD::frt pBAV). The complemented strain (17978pAB3− ΔcsuD::frt pCsuFull) restored the CsuA/B expression. (C) Detection of Csu pili on the surface of the 17978pAB3− ΔcsuD::frt pCsuFull complemented strain. Transmission electron microscopic analysis confirmed the presence of Csu pili on wild-type cells (17978pAB3− pBAV) and complemented strain cells (17978pAB3− ΔcsuD::frt pCsuFull), whereas mutant cells (17978pAB3− ΔcsuD::frt pBAV) did not show Csu pili.
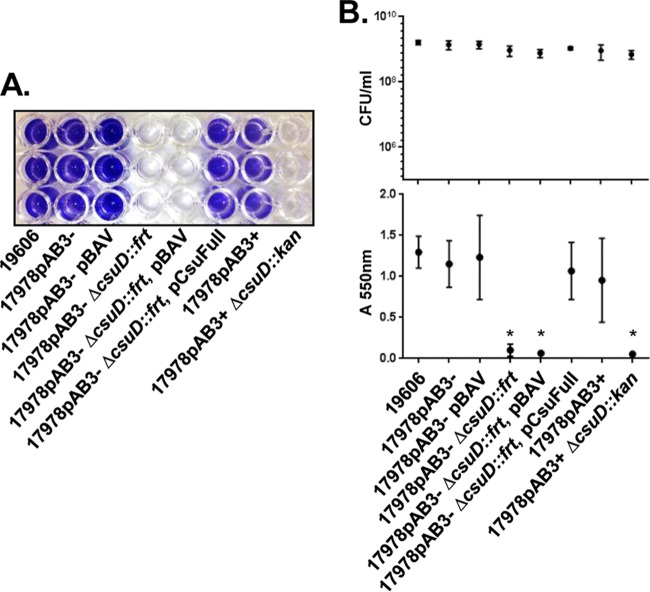
Because Csu pili have been implicated in biofilm formation in A. baumannii ATCC 19606, we next tested the ability of 17978 and the csuD mutant strains to form biofilms on a polystyrene surface by using a crystal violet (CV) staining method. The 17978pAB3− and 17978pAB3+ cells formed equally robust biofilms on both polystyrene test tubes and 96-well plates, whereas the mutants lacking csuD were completely unable to form any visible biofilm structures (Fig. 3A; also see Fig. S1, top, in the supplemental material). It is interesting to note that the csuD mutant cells seemed to grow more slowly than wild-type cells and primarily as pellets at the bottom of the wells, especially when the plates were incubated without shaking (Fig. S1, bottom). To exclude the possibility that the lack of biofilm production was linked to different growth rates, biofilm assays were repeated with cultures normalized on the basis of the optical density at 600 nm (OD600). Equivalent bacterial loads were subsequently verified by serial dilution and CFU counts (Fig. 3B, top). Under these OD600-normalized conditions, the csuD mutants of both 17978pAB3− and 17978pAB3+ remained unable to form biofilms, as assessed by the CV staining assay. Importantly, complementation of the 17978pAB3− csuD::frt mutant restored the biofilm formation phenotype, in agreement with our experiments showing a return of CsuA/B expression (Fig. 3B, bottom). Together, these data suggest that Csu pili are essential for biofilm formation in 17978. Furthermore, these results indicate that 17978 cells, regardless of the presence or absence of pAB3, produce Csu pili that are present on the cell surface at comparable levels.
FIG 3.
Requirement for Csu pili for biofilm formation in A. baumannii ATCC 17978. (A) Representative crystal violet assay image of A. baumannii strains on polystyrene surfaces. The biofilms were stained with crystal violet and dissolved in 10% acetic acid solution. A. baumannii ATCC 17978 wild-type and complemented strains, but not the csuD mutant strain, formed biofilms. The endogenous pAB3 plasmid and the pBAV shuttle vector did not affect biofilm formation. The A. baumannii ATCC 19606 strain was used as a control. (B) Quantitative determination of biofilm formation. To normalize cell growth rates, A. baumannii strains were grown in LB medium and collected at OD600 values of 1.0 to 1.2. To count CFU, collected cells were plated on LB medium (top); to quantify the biofilms, absorbance was measured at OD550 (bottom). Points indicate mean values, and error bars indicate standard deviations of triplicate samples. *, P ≤ 0.002, significant reduction of biofilm production in csuD mutant strains, compared to 17978pAB3−.
Subinhibitory concentrations of Smx and Tmp repress Csu expression and prevent biofilm formation.
The observations that 17978pAB3− and 17978pAB3+ express equivalent levels of Csu pili and have the same capacity to form biofilms contradicted the RNA-seq data, which indicated that Csu pilus expression was repressed by the plasmid. This apparent contradiction prompted us to revisit our RNA-seq protocol. The pAB3 plasmid in 17978pAB3+ imparts resistance to the antibiotic Smx. Smx kills 17978pAB3− cells at concentrations of >200 μg/ml and, since pAB3 can be lost in 1 to 5% of cells during laboratory growth (11), Smx can be used to maintain an isogenic 17978pAB3+ population. Although 17978pAB3− and 17978pAB3+ cells exhibit similar resistance to Tmp, the addition of both Smx and Tmp, at 30 and 6 μg/ml, respectively, is sufficient to inhibit the growth of 17978pAB3− cells. As both antibiotics target the folate biosynthesis pathway, selection can be maintained at a lower antibiotic concentration. Therefore, we often grow 17978pAB3+ cells in the presence of both Smx and Tmp. For our RNA-seq experiments, 17978pAB3+ was grown overnight with 30 μg/ml Smx and 6 μg/ml Tmp; 70 μl of this overnight culture was then diluted into 10 ml of fresh medium without the addition of antibiotics, corresponding to an initial OD600 of 0.03, and grown at 37°C until the cultures reached an OD600 of 0.55. These cultures were then used for RNA extraction and sequencing.
We hypothesized that, despite the >140-fold dilution of Smx and Tmp in the cultures used for RNA extraction, the low levels of these antibiotics might affect Csu expression. To test this, we first cultured 17978pAB3+ cells in the presence of 30 μg/ml Smx and 6 μg/ml Tmp at and determined that, under these conditions, expression of CsuA/B was completely inhibited, resulting in total loss of biofilm formation (Fig. 4A to C). Dimethyl sulfoxide (DMSO), the solvent used for preparation of Smx and Tmp solutions, did not affect biofilm formation; therefore, the antibiotics were responsible for the observed phenotype (data not shown). EM imaging of cells confirmed that, in the presence of Smx and Tmp, pili were not visible on the cell surface of 17978pAB3+ cells (Fig. 4D). In addition, we observed that the csuA/B transcription levels in 17978pAB3+ cells were dramatically reduced (>103-fold) in the presence of 30 μg/ml Smx and 6 μg/ml Tmp (Fig. S2). These results corresponded to our Western blotting and RNA-seq results. Then, we tested whether treating the cultures with low concentrations of Smx and Tmp, mimicking the conditions under which we prepared the RNA samples, would result in Csu repression. Western blot analysis showed that, when 17978pAB3+ bacteria were grown overnight in the presence of 30 μg/ml Smx and 6 μg/ml Tmp, resuspended in fresh medium containing 60 ng/ml Smx and 12 ng/ml Tmp (500-fold dilution), and cultured for 4 h, production of CsuA/B was dramatically reduced (Fig. S3). These results demonstrate that subinhibitory concentrations of Smx and Tmp repress Csu expression.
FIG 4.
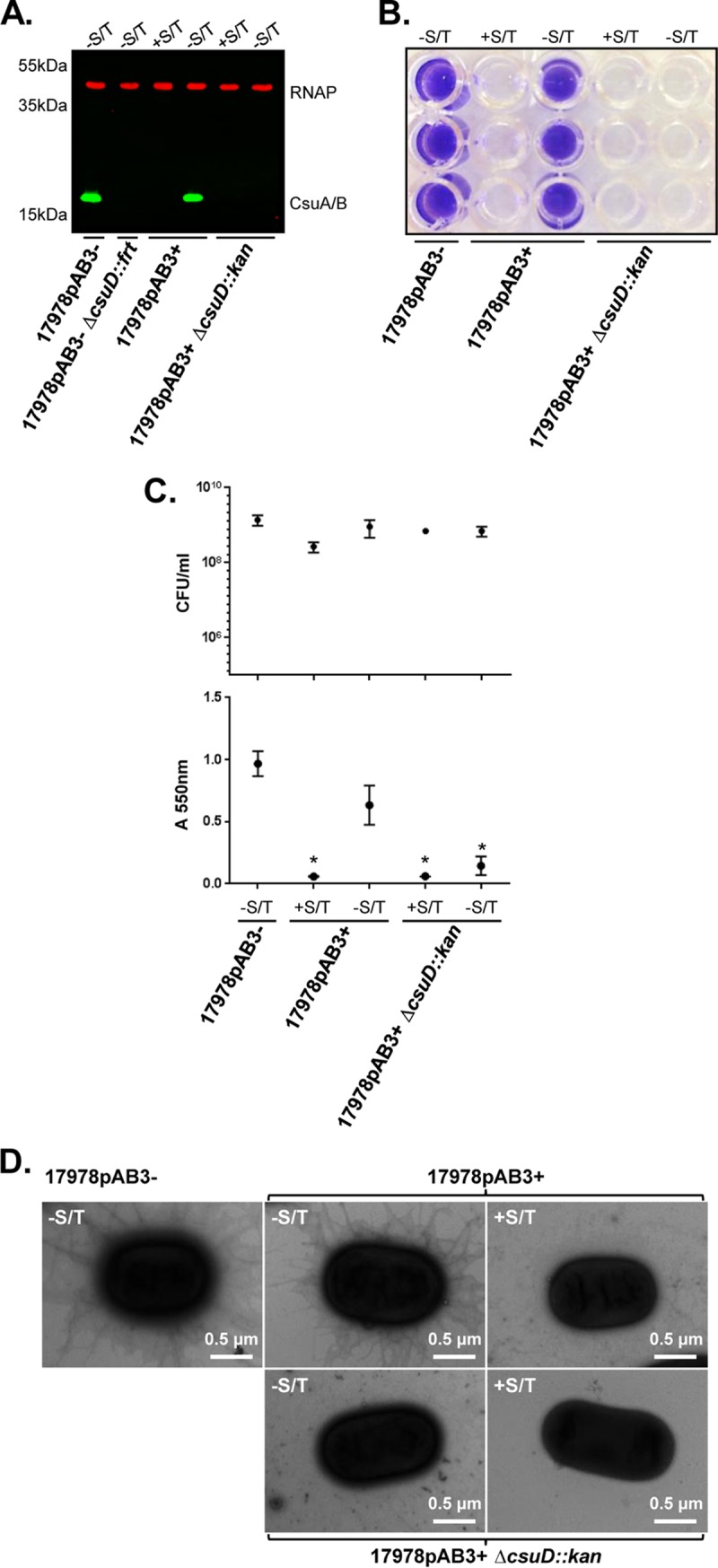
Abolishment of both Csu pilus expression and biofilm formation by Smx and Tmp. (A) Inhibition of CsuA/B expression by Smx and Tmp. The 17978pAB3+ cells were grown in LB medium with or without 30 μg/ml Smx and 6 μg/ml Tmp (S/T); 17978pAB3− cells were sensitive to the combination of Smx and Tmp. Under Smx/Tmp-treated conditions, CsuA/B was not detected in 17978pAB3+ lysates. Immunoblot was performed using CsuA/B-specific antibody (green bands); RNAP was used as a loading control (red bands). (B) Representative crystal violet assay image of A. baumannii strains with and without Smx and Tmp. The 17978pAB3+ strain treated with Smx and Tmp was not able to form biofilms. The csuD mutant was also unable to form biofilms, with or without treatment with Smx and Tmp, which corresponded to CsuA/B immunoblot results. (C) Quantitative measurements of biofilm formation by A. baumannii strains with and without Smx and Tmp, including CFU counts of A. baumannii biofilms (top) and absorbance at 550 nm following crystal violet staining (bottom). Points indicate mean values, and error bars indicate standard deviations of triplicate samples. *, P ≤ 0.005, significant reduction of biofilm production in 17978pAB3+ cells with Smx and Tmp and csuD mutant strains with and without Smx and Tmp, compared to 17978pAB3− cells without Smx and Tmp. (D) Transmission electron microscopic images confirming Csu pilus repression on A. baumannii surfaces by Smx and Tmp. This result corresponded to CsuA/B immunoblot and biofilm assay results.
Folate stress results in repression of Csu expression.
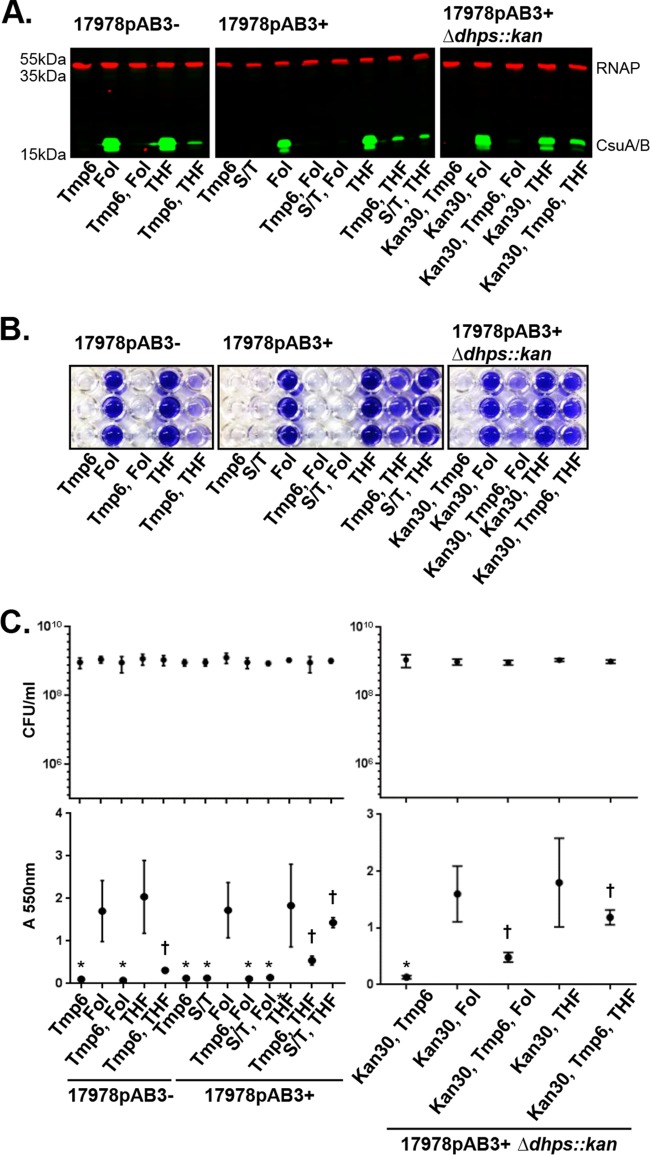
We next sought to investigate the mechanisms by which Smx and Tmp inhibit CsuA/B expression and biofilm formation. The addition of subinhibitory concentrations of Tmp (6 μg/ml) or Smx (30 or 150 μg/ml) to 17978pAB3− cells completely inhibited expression of CsuA/B, as assessed by Western blotting (Fig. 5A), without affecting cell growth (Fig. S4); 17978pAB3− cells treated with either antibiotic were unable to form biofilms, consistent with the lack of CsuA/B expression (Fig. 5B and C). Subinhibitory concentrations of Tmp alone inhibited CsuA/B expression in 17978pAB3+ cells and blocked the formation of biofilms (Fig. 5A to C). Interestingly, Smx did not affect CsuA/B expression or biofilm formation in 17978pAB3+ cells, whereas the combination of Smx and Tmp completely abolished CsuA/B expression and biofilm formation (Fig. 5A to C).
FIG 5.
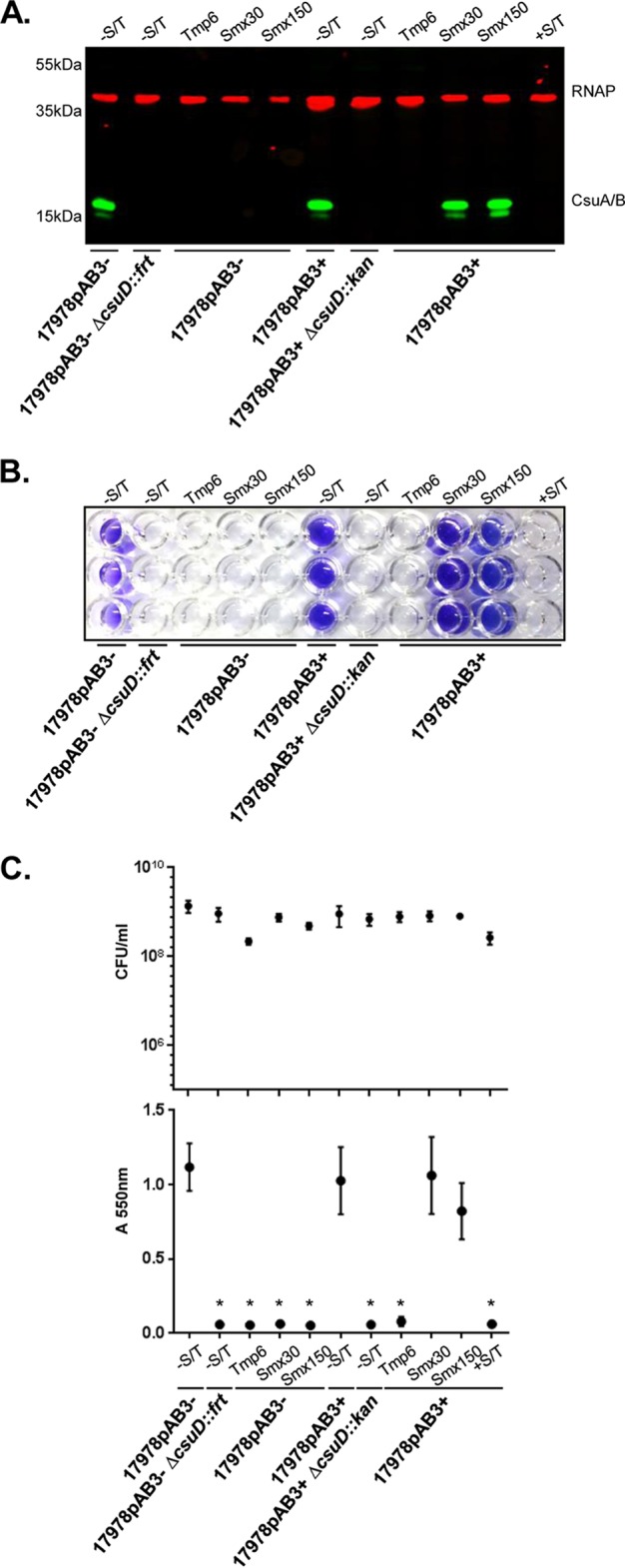
Effects of Smx and Tmp, alone and in combination, on Csu pilus expression and biofilm formation. (A) Verification by Western blotting of CsuA/B expression with Smx and Tmp (S/T), alone and in combination. Smx or Tmp single treatment was able to inhibit CsuA/B expression in 17978pAB3− cells, whereas Smx was unable to inhibit CsuA/B expression in 17978pAB3+ cells. Immunoblot was performed using CsuA/B-specific antibody (green bands); RNAP was used as loading control (red bands). (B) Representative crystal violet assay image of A. baumannii strains treated with Smx and Tmp, alone or in combination. (C) CFU counts (top) and biofilm quantification through the crystal violet assay (bottom). Points indicate mean values, and error bars indicate standard deviations of triplicate samples. *, P ≤ 0.005, significant reduction of biofilm production, compared to 17978pAB3− cells without Smx and Tmp.
A. baumannii cells utilize folate-derived one-carbon metabolism, which is essential for generating purines, dTMP, methionine, and glycine (14). Smx inhibits dihydropteroate synthesis via the dihydropteroate synthase (DHPS) enzyme by competing with p-aminobenzoic acid for DHPS binding, whereas Tmp prevents tetrahydrofolate (THF) synthesis by binding to the dihydrofolate reductase (DHFR) enzyme. Ultimately, both antibiotics functionally and synergistically inhibit the synthesis of folate pathway intermediates, including the essential cofactor THF (Fig. 6), which is why they are used together clinically to treat a wide variety of bacterial infections (15, 16). Our results suggest that antibiotics targeting folate biosynthesis affect the expression of Csu pili and that 17978pAB3+ resistance to Smx provides protection from the effects of this antibiotic. The resistance of 17978pAB3+ to Smx is provided by a pAB3-encoded dhps gene. The susceptibility of 17978pAB3+ cells to Tmp-induced CsuA/B repression and the resistance to Smx-induced repression suggested that dhps was mediating resistance to the antibiotic and its effects on CsuA/B expression. To test this, we replaced dhps in 17978pAB3+ with a kanamycin resistance cassette, which sensitized these cells to Smx, similar to 17978pAB3− cells. Growth of 17978pAB3+ dhps::kan cells in the presence of subinhibitory concentrations of Tmp or Smx completely inhibited CsuA/B expression and biofilm formation, with a phenotype identical to that of 17978pAB3− (Fig. 7). These results show that DHPS is responsible for the resistance of 17978pAB3+ to the effects of Smx on CsuA/B expression.
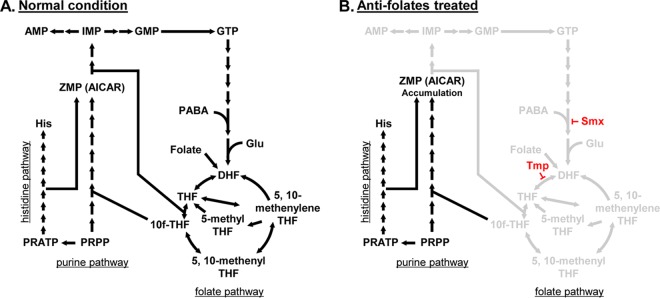
FIG 6.
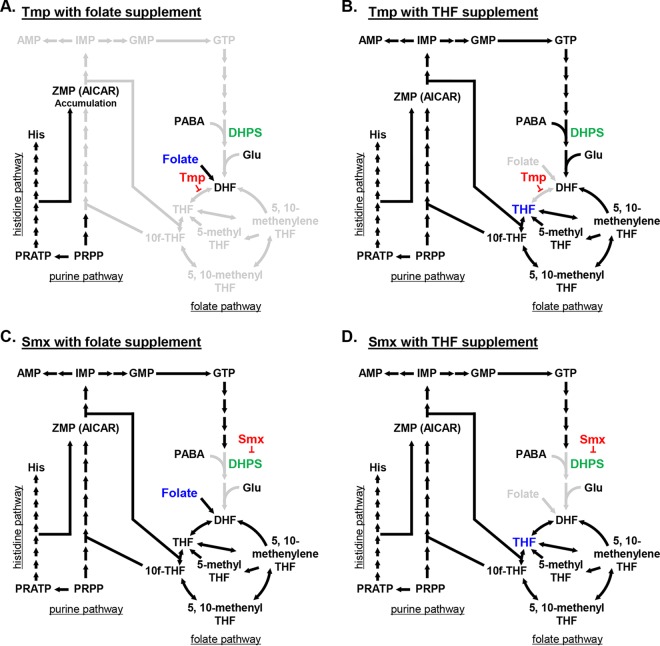
Schematic diagrams of the de novo purine biosynthesis pathway and one-carbon metabolism. (A) Schematic diagram of the folate, purine, and histidine pathways under normal conditions. (B) Schematic diagram of the folate, purine, and histidine pathways under antifolate treatment conditions. Antifolate antibiotics disrupt folate-derived one-carbon metabolism, inhibiting THF synthesis and accumulating ZMP (AICAR). PRPP, 5-phosphoribosyl-1-pyrophosphate; PRATP, phosphoribosyl-ATP; PABA, para-aminobenzoic acid.
FIG 7.
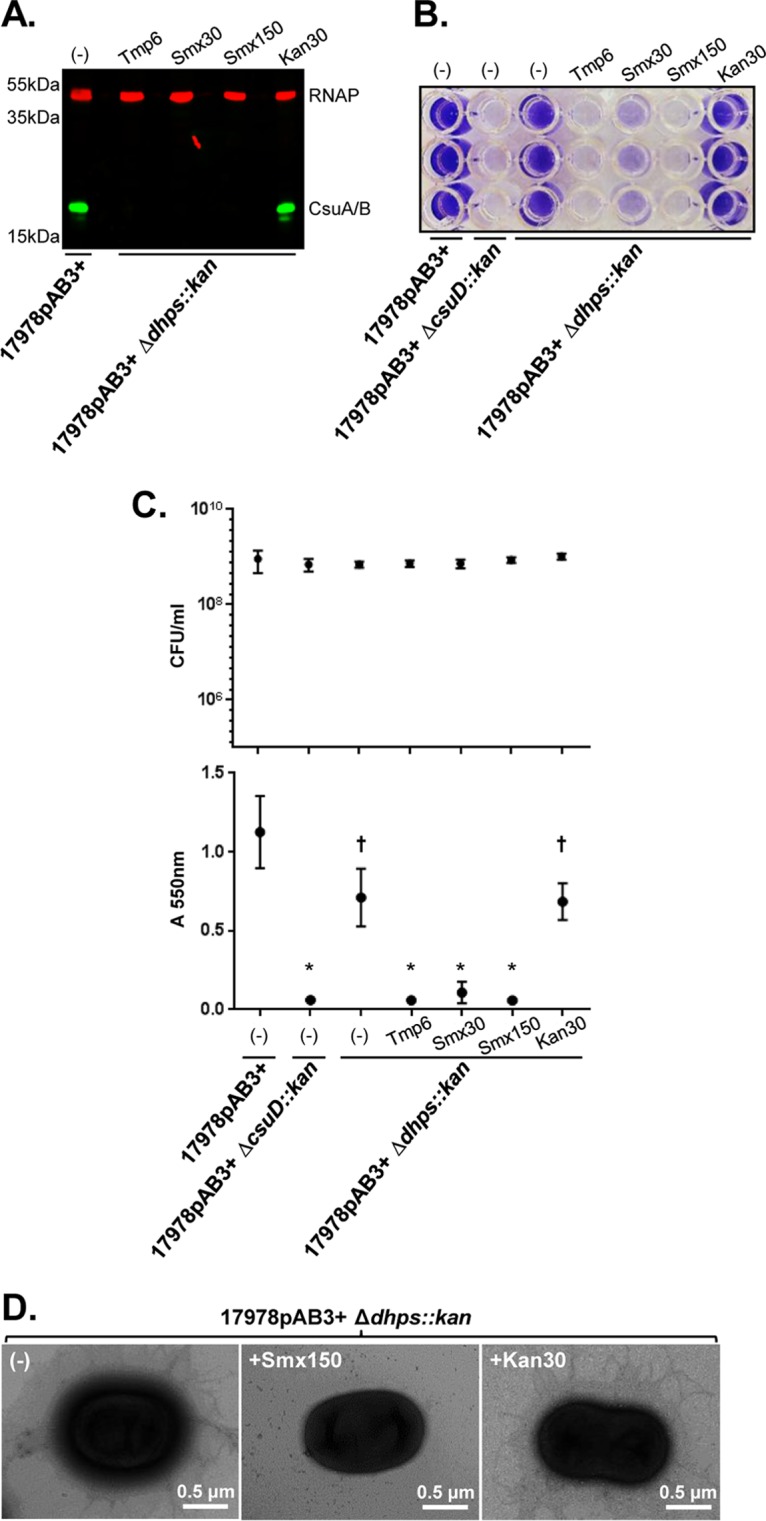
Dihydropteroate synthase (DHPS) responsibility for pilus expression and biofilm formation. (A) CsuA/B expression in 17978pAB3+ Δdhps::kan cells with Tmp or Smx treatment, as determined by Western blotting with the anti-CsuA/B antibody (green bands); RNAP was used as loading control (red bands). CsuA/B is expressed in the presence of kanamycin but not in the presence of both antifolate antibiotics. (B) Representative crystal violet assay image of A. baumannii strains with Tmp or Smx treatment. The 17978pAB3+ Δdhps::kan cells were unable to form biofilms under antifolate antibiotic treatment conditions. (C) CFU counts (top) and biofilm crystal violet quantification (bottom). Points indicate mean values, and error bars indicate standard deviations of triplicate samples. *, P ≤ 0.003; †, P ≤ 0.05, significant reduction of biofilm production, compared to 17978pAB3+ cells. (D) Transmission electron microscopic images confirming Csu pilus repression on 17978pAB3+ Δdhps::kan surfaces by Smx but not kanamycin (Kan).
Additionally, we tested whether these antibiotics were able to disperse preformed biofilms. Either individually or together, Smx and Tmp could not disperse preformed biofilms of A. baumannii 17978pAB3− and pAB3+ cells, even at 5- or 10-fold higher concentrations (Fig. S5).
Antibiotic-induced pilus repression is rescued by exogenous folate biosynthesis intermediates.
We reasoned that, if Csu expression and biofilm formation in 17978 were downregulated by the folate stress caused by Tmp and Smx, relieving the stress by supplementing the cultures with exogenous THF and folate should rescue Csu expression. To test this hypothesis, we added exogenous THF or folate to cells grown in the presence of subinhibitory concentrations of Tmp and/or Smx. THF partially restored Csu pilus expression and biofilm formation in both 17978pAB3− and 17978pAB3+ cells treated with Tmp (Fig. 8). However, folate supplementation could not restore these phenotypes. The phenotype of Csu expression and biofilm formation was also recovered with THF, but not folate, in 17978pAB3+ dhps::kan cells treated with Tmp (Fig. 8). Interestingly, with subinhibitory concentrations of Smx, both THF and folate supplementation could partially restore Csu pilus expression and biofilm formation in both 17978pAB3− and 17978pAB3+ dhps::kan cells (Fig. 9). While exogenous THF was able to partially relieve folate stress in the presence of both antibiotics, folate was effective only in the Smx-treated cells. An explanation for these results could be that Tmp blocks the activity of DHFR, which is involved in a later step in the pathway (Fig. 10A and B). To confirm that biofilm formation is not mediated by factors independent of Csu pili, which may be induced by THF supplementation, we tested biofilm formation by csuD mutants treated with folate or THF. The csuD mutants of both 17978pAB3− and 17978pAB3+ were unable to form biofilms when folate or THF was supplemented (Fig. S6), suggesting that the biofilm restoration with THF under folate stress was due to restoration of Csu expression and not other factors. These results support the hypothesis that Csu expression is downregulated in response to folate stress (Fig. 10).
FIG 8.
Recovery of Csu pilus and biofilm repression with THF but not folate supplements under Tmp-mediated folate stress conditions. (A) CsuA/B expression with folate (Fol) or THF supplements, as determined by Western blotting with the anti-CsuA/B antibody (green bands); RNAP was used as loading control (red bands). Folate stress was generated by Tmp. S/T, Smx and Tmp; Kan, kanamycin. (B) Representative crystal violet assay images of A. baumannii strains with folate or THF supplements. THF but not folate was able to relieve folate stress generated by Tmp, resulting in partially restored biofilm formation. (C) CFU counts (top) and biofilm crystal violet assay quantification (bottom). Points indicate mean values, and error bars indicate standard deviations of triplicate samples. *, P ≤ 0.005; †, P ≤ 0.05, significant reduction of biofilm production, compared to 17978pAB3− cells with folate.
FIG 9.
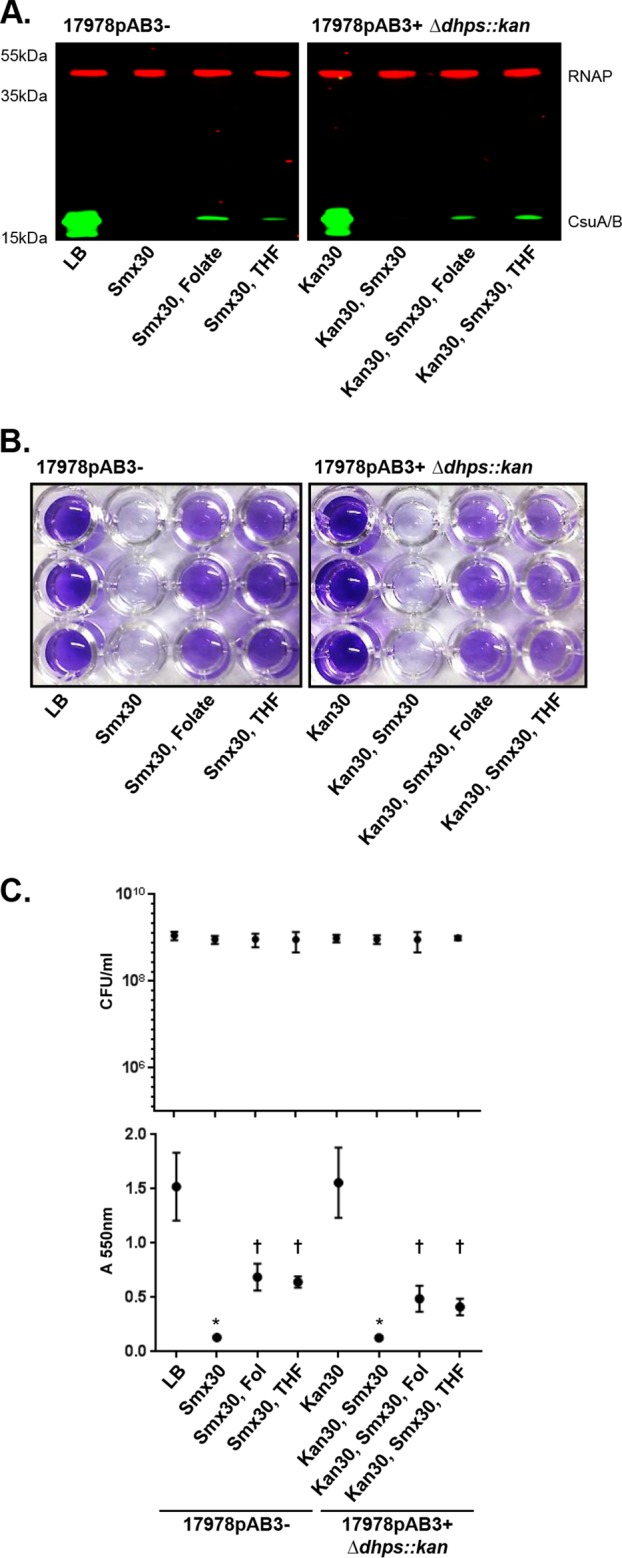
Recovery of Csu pilus and biofilm repression with both THF and folate supplements under Smx-mediated folate stress conditions. (A) CsuA/B expression with folate or THF supplements, as determined by Western blotting with the anti-CsuA/B antibody (green bands); RNAP was used as loading control (red bands). Folate stress was generated by Smx. Kan, kanamycin. (B) Representative crystal violet assay images of 17978pAB3− and 17978pAB3+ Δdhps::kan cells with folate or THF supplements. Both folate and THF supplements were able to relieve folate stress generated by Smx, resulting in partially restored biofilm formation. (C) CFU counts (top) and biofilm crystal violet assay quantification (bottom). Points indicate mean values, and error bars indicate standard deviations of triplicate samples. *, P ≤ 0.002; †, P ≤ 0.05, significant reduction of biofilm production, compared to 17978pAB3− cells with LB.
FIG 10.
Simplified models of the folate, purine, and histidine pathways with different antifolate treatments. (A) Although imported folate is converted to dihydrofolate (DHF), DHF is unable to be converted to the other intermediates and is unable to relieve folate stress. (B) Supplemented THF is converted to other intermediates, relieving folate stress generated by Tmp. (C and D) Although Smx interferes with dihydropteroate synthesis, both folate and THF can be converted to the downstream intermediates of the folate pathway, resulting in relief of the folate stress generated by Smx. PRPP, 5-phosphoribosyl-1-pyrophosphate; PRATP, phosphoribosyl-ATP; PABA, para-aminobenzoic acid.
Folate stress downregulates Csu expression in multiple Acinetobacter strains.
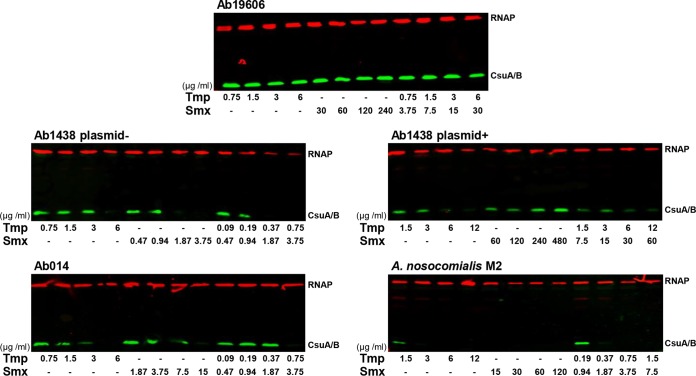
To investigate whether the effects of subinhibitory concentrations of Tmp and Smx were strain specific or represented a widespread phenomenon, we performed CsuA/B immunoblotting and biofilm assays with several medically relevant Acinetobacter species. We found that the clinical isolates A. baumannii Ab1438 and Ab014 (11, 17) and Acinetobacter nosocomialis M2 (18) repressed their Csu pilus expression under folate stress conditions (Fig. 11). However, biofilms were not affected by folate stress in any of those strains, suggesting that biofilm formation is not Csu dependent and is determined by other factors in Ab1438, Ab014, and M2 (Fig. S7). Interestingly, in the commonly used A. baumannii ATCC 19606 strain, Csu pilus expression and biofilm formation were not affected by folate stress (Fig. 11; also see Fig. S7). A recent study revealed that ATCC 19606 contains sulfonamide resistance genes in its genome (19), which could confer insensitivity to the antibiotics.
FIG 11.
Effects of antifolates on Csu piliation in other Acinetobacter strains. The expression of Csu pili was confirmed by Western blotting with anti-RNAP (red bands) and anti-CsuA/B (green bands) antibodies. Concentrations of Tmp and Smx are shown below each lane.
DISCUSSION
Model bacterial strains are essential for standardized experimental investigations into the genetic mechanisms used by pathogens to cause disease. A. baumannii ATCC 17978 has served as such a model system for research on pathogenic Acinetobacter spp. and was the first strain of this species to have its whole genome sequenced (20). The development of genetic tools to study the biology of A. baumannii in the ATCC 17978 background has greatly increased our understanding of this species (21). However, the recent genome resequencing of A. baumannii ATCC 17978 suggests that some prior assumptions regarding chromosome composition in this strain are likely incorrect (11, 20); this includes the finding of a novel plasmid, pAB3, which in the original genome sequence was assembled as part of the chromosome. The work presented here further demonstrates that Csu pili are in fact expressed by this strain, in which they play a major role in biofilm formation.
Our prior RNA-seq experiments suggested that the pAB3 plasmid regulates not only T6SS but also pilus expression. However, 17978pAB3+ and 17978pAB3− expressed pili at comparable levels, indicating that the presence or absence of this plasmid did not have an appreciable effect on Csu expression. These pili were detected by EM on the cell surface of 17978 cells of the wild-type strains but were absent in strains lacking csuD. Mutation of csuD resulted in the loss of expression of the major CsuA/B pilin. These results are in agreement with previous reports investigating the role of Csu pili in A. baumannii ATCC 19606, which showed that mutation of other csu genes, including csuC and csuE, prevented CsuA/B expression (5). We also found that Csu pili constituted the major factor influencing biofilm formation by 17978 cells on polystyrene surfaces, as the csuD mutants of both 17978pAB3+ and 17978pAB3− showed a complete lack of CV staining in our biofilm assay.
The apparent contradiction between the RNA-seq data and the levels of Csu pilins is explained by the finding that subinhibitory concentrations of antibiotics influence Csu expression and function. Maintenance of a homogeneous population of 17978pAB3+ cells requires antibiotic selection. Smx can be used at high concentrations to select for the plasmid, but we routinely use the combination of Smx and Tmp to decrease the antibiotic levels while maintaining selective pressure. When cells are cultured overnight with high concentrations of these antibiotics and diluted >140-fold prior to RNA or protein extraction, Csu pilins are repressed by folate stress and not by the plasmid, as we initially thought.
Tmp and Smx target the folate biosynthesis pathway (22). Folate-dependent one-carbon metabolism is critical in most living organisms (e.g., bacteria, plants, and mammals). Bacterial and plant cells are able to synthesize folates through a de novo biosynthesis pathway, while animals cannot synthesize these essential cofactors for one-carbon metabolism and must acquire them from dietary sources (23–25). Not only folate but also THF and its various derivatives (the active form of folate) are vital cofactors for enzymes involved in one-carbon transfer reactions (26). This folate-mediated metabolic pathway is involved in key metabolic functions of cell replication, including the biosynthesis of nucleic acids, amino acids, and pantothenate (14, 27). The findings that subinhibitory concentrations of antibiotics targeting folate synthesis negatively affect pilus production in A. baumannii 17978 and that exogenous THF and folate restore piliation demonstrate that the signaling activity of Tmp and Smx is linked to folate stress. Folate stress and its transcriptional effects have been reported previously for E. coli (28–30). Here we showed, for the first time, that this phenomenon is prevalent in Acinetobacter strains.
Concentrations of antibiotics below therapeutic levels are now recognized as having wide-ranging effects on bacterial physiology (31). This has led to the hypothesis that antibiotics may act as signaling molecules rather than outright antibacterial agents, especially in the natural environment (31). Numerous studies have provided support for this hypothesis, finding a wide variety of phenotypic changes caused by subinhibitory antibiotic concentrations (28, 32–35). In certain species, up to 5% of the promoters appear to be affected (36). In general, most of these responses tend to increase biofilm production, with concomitant increases in resistance to the signaling antibiotic (28, 32–35, 37). In A. baumannii, subinhibitory concentrations of imipenem or levofloxacin-meropenem have been shown to increase biofilm, type IV pili, and surface motility, which are associated with overexpression of the AdeFGH efflux pump (33, 35, 37). In agreement with our results, previous reports have shown that subinhibitory concentrations of Tmp inhibit P fimbria expression on uropathogenic E. coli (28), quinolones inhibit type 1 fimbria expression on E. coli (32), and azithromycin inhibits piliation of Neisseria gonorrhoeae (34). It is tempting to speculate that Smx and Tmp at low concentrations act as a “danger signal.” In such a scenario, Csu expression is repressed upon mild folate stress, allowing biofilm dispersal and escape from potentially inhibitory concentrations of antibiotics.
Defects in folate metabolism can manifest biochemically as an accumulation of a late intermediate in purine biosynthesis, namely, 5-aminoimidazole-4-carboxamide ribonucleotide (ZMP). A recent study showed that ZMP accumulated significantly under folate stress induced by antifolate antibiotics in Clostridium beijerinckii (38). Those authors showed that increases in the ZMP and 5-aminoimidazole-4-carboxamide riboside 5′-triphosphate (ZTP) pool resulting from folate stress could trigger changes in gene expression (38). It was proposed that this might occur through a riboswitch-mediated mechanism in diverse Proteobacteria species (38–40). It is tempting to speculate that a similar mechanism operates in Acinetobacter strains. Using riboswitch prediction software (RibEx), we detected a putative riboswitch in the csu promoter region (37). In addition, we found possible riboswitches in regions upstream from bfmR and bfmS, a two-component system that regulates the transcription of Csu pili in A. baumannii (Fig. S8) (6). In future work, we will investigate whether these putative riboswitches are involved in the folate stress response in Acinetobacter strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. Strains were routinely grown in lysogeny broth (LB) liquid medium at 37°C, with shaking (200 rpm). Ampicillin (100 μg/ml), kanamycin (7.5 or 30 μg/ml), tetracycline (5 μg/ml), carbenicillin (400 μg/ml), Smx (30 or 150 μg/ml), Tmp (6 μg/ml), folate (final concentration, 0.5 mM), and tetrahydrofolate (final concentration, 0.5 mM) were added when necessary.
Construction of the csuD mutant, Csu pilus complemented strains, and the dhps mutant.
Primers used in this study are listed in Table S2. Construction of the csuD deletion mutant in A. baumannii ATCC 17978 was described previously (21). Briefly, a csuD-deleted kanamycin cassette flanked by Flp recombinase target (FRT) sites and recombineering DNA was amplified from plasmid pKD04 using primers CsuD_mutant_F and Csu_mutant_R. A. baumannii ATCC 17978 pAB3− or pAB3+ cells carrying pAT04 were inoculated into LB medium with 5 μg/ml tetracycline to maintain the plasmid. Isopropyl-β-d-thiogalactopyranoside (IPTG) (2 mM) was added, and the bacteria were grown for 2 to 3 h, to mid-log phase, at 37°C. After three washes in ice-cold 10% glycerol and 1,000-fold concentration, 100 μl of bacterial suspension was mixed with 5 μg of PCR product and electroporated in a 2-mm cuvette at 1.8 kV. After outgrowth in 4 ml of rich medium containing 2 mM IPTG, the bacteria were pelleted, plated on agar plates containing 7.5 μg/ml kanamycin, and incubated overnight at 37°C. The kanamycin-resistant transformants were confirmed for inactivation of the csuD gene with the kanamycin cassette by PCR. To delete the kanamycin cassette from the mutant, marked mutant cells were passed two or three times in LB medium without tetracycline (with only 7.5 μg/ml kanamycin) for curing of the pAT04 plasmid. To confirm the pAT04 curing, cells were cultured with 5 μg/ml tetracycline. Then, these tetracycline-sensitive marked mutant cells were cultured, and electrocompetent cells were prepared as described above. The mutant strains were transformed with pAT03 expressing Flp recombinase and were plated on LB agar with 2 mM IPTG and 400 μg/ml carbenicillin. The resistant transformants were analyzed by PCR to confirm the loss of the kanamycin cassette. Just as for pAT04 plasmid curing, clean mutant cells were passaged two or three times in LB medium without carbenicillin, and then the pAT03 curing was confirmed by PCR.
To complement the csuD mutant, we constructed two different shuttle vectors. First, the DNA sequences of the csuD gene and its native promoter (Pcsu) were PCR amplified from genomic DNA of A. baumannii ATCC 17978 using the primer pairs pBAV_Pcsu_F/Pcsu_CsuD_R and Pcsu_CsuD_F/CsuD_pBAV_R, respectively. The amplified DNA fragments were ligated by overlapping PCR, yielding the Pcsu-csuD DNA fragment. E. coli-A. baumannii shuttle vector pBAV DNA fragments were PCR amplified using primers pBAV_F and pBAV_R. These two DNA fragments (pBAV and Pcsu-csuD) were then fused using the In-Fusion HD EcoDry cloning kit (Clontech Laboratories, Inc.), yielding plasmid pCsuD (pBAV::Pcsu-csuD). A second complementation vector contained the entire csu cluster (csuA/B-csuA-csuB-csuC-csuD-csuE) with its native promoter (Pcsu) instead only the csuD gene with Pcsu. The Pcsu-csuA/B-csuA-csuB-csuC-csuD-csuE (Pcsu-csu full operon) DNA sequence was PCR amplified from genomic DNA of A. baumannii ATCC 17978 using the primers CsuFull_comp_F and CsuFull_comp_R. The amplified Pcsu-csu full operon DNA fragments and the pBAV shuttle vector were digested using the restriction enzyme PstI and cloned together, yielding plasmid pCsuFull (pBAV::Pcsu-csu full operon). These plasmids were then electroporated into 17978pAB3− ΔcsuD mutant cells, followed by selection with kanamycin. The resistant transformants were analyzed by PCR, to confirm the presence of the plasmid in the transformants (17978pAB3− ΔcsuD pCsuD or 17978pAB3− ΔcsuD pCsuFull) (11).
To construct the dhps mutant, dhps-deleted recombineering DNA was amplified from plasmid pBAV1k-t5-gfp using the primers DHPS_mutant_F and DHPS_mutant_R. Subsequent steps to obtain a dhps mutant were the same as those for construction of the csuD mutant, as described above.
Pilus shear preparations.
Each A. baumannii strain was streaked onto agar plates and incubated overnight at 37°C. Bacteria were collected from the agar plates and resuspended in 5 ml of phosphate-buffered saline (PBS), yielding a concentrated cell suspension. Appropriate dilutions were made, and cell suspensions were normalized to an OD600 of 50. Cells were vortex-mixed at high speed for 1 min, followed by incubation on ice for 1 min. After centrifugation at 16,000 × g for 1 min at 4°C, the resultant supernatants were transferred to new tubes. The pili in the cell-free supernatants were precipitated for 30 min with cold trichloroacetic acid (TCA) (final concentration, 20%), on ice. The precipitated pilus proteins were collected by centrifugation at 16,000 × g for 1 min at 4°C. The supernatant was removed, and the pellet was washed twice with cold acetone to remove residual TCA. The tube was then dried completely, and the proteins bound to the tube wall were dissolved with 1× Laemmli buffer.
Immunoblot analysis.
To test the expression of Csu pili, whole-cell pellets and pilus shear preparation samples were obtained from different A. baumannii strains under different conditions. The harvested cell pellets were solubilized by boiling in 1× Laemmli buffer. The method for shear sample preparation is described above. Samples were loaded on 12% SDS-PAGE gels for separation. Following separation, proteins were transferred to nitrocellulose membranes, and CsuA/B was visualized using a mouse anti-CsuA/B specific antibody, followed by IRDye 680-labeled goat anti-mouse IgG antibodies (LI-COR Biosciences, Lincoln, NE). For loading control, RNA polymerase (RNAP) was targeted; RNAP was visualized using a rabbit anti-RNAP specific antibody, followed by IRDye 800-labeled goat anti-rabbit IgG antibodies (LI-COR Biosciences). Images were obtained using a LI-COR Odyssey imaging system (LI-COR Biosciences).
Transmission electron microscopic analysis.
For negative staining and analysis by transmission electron microscopy, bacterial suspensions were allowed to absorb for 10 min onto freshly glow-discharged Formvar/carbon-coated copper grids. The grids were washed in distilled water and stained for 1 min with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA). Excess liquid was gently wicked off, and the grids were allowed to air dry. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA) equipped with an AMT 8-megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA).
Biofilm formation analysis.
Bacterial cells were inoculated into polystyrene culture tubes (500 μl of medium per tube) and incubated under aerobic conditions (37°C, 200 rpm) to an OD600 of 1.0. Cells were serially diluted in PBS (pH 7.0) and plated on LB agar plates to measure CFU.
Bacterial cell culture broths were discarded from the culture tubes. The cell culture tubes were washed five times with water and then stained with CV. Six hundred microliters of 0.1% CV (dissolved in water) was added. The culture tubes were incubated for 10 min at room temperature before removal of the CV staining solution and then were washed five times with water. After washing, 600 μl of 10% acetic acid solution was added to dissolve the biofilm-bound CV by gently shaking the culture tubes. To quantify the biofilm formation ability, CV dissolved in acetic acid solution was dispensed onto sterile, clear, round-bottomed, 96-well, polystyrene plates (Corning Inc.) (200 μl per well), and absorbance was measured at 550 nm.
Reverse transcription-PCR.
Early exponentially growing A. baumannii cells (OD600 of 0.5 to 0.6) were treated with RNAprotect, followed by total RNA isolation using the RNeasy minikit (Qiagen Inc.). Contaminating DNA in the RNA samples was removed by digestion for 2 h at 37°C with RNase-free DNase I (Thermo Scientific), followed by RNeasy mini purification. For reverse transcription (RT)-PCR, cDNA was prepared from 1 μg RNA using a high capacity RNA-to-cDNA kit (Applied Biosystems), according to the manufacturer's protocol. Real-time quantitative PCR (qPCR) was performed using Power SYBR green PCR master mix reagents (Applied Biosystems) on a ViiA7 real-time PCR system (Applied Biosystems), following the manufacturer's suggested protocol. The A. baumannii rpoB gene was used as a reference gene (41). The gene (rpoB and csuA/B)-specific primers used for qPCR are listed in Table S2.
Statistical analysis.
The significance of the differences between the mean values of the groups was analyzed as follows. Unless otherwise stated, normality was checked for all data using the Shapiro-Wilk normality test. If the data passed the normality test, then a multiple-comparison analysis was performed by using one-way analysis of variance (ANOVA) followed by the Brown-Forsythe test. If the data did not pass the normality test, then a multiple-comparison analysis was performed by using the Kruskal-Wallis test followed by the Dunn test. P values of ≤0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the imaging laboratory of molecular microbiology at Washington University, especially W. Beatty, and B. Anthony for EM analysis. We thank Luis Actis for sharing the anti-CsuA/B antibody. We thank Florencia Haurat, Gisela Di Venanzio, and Eleonora Garcia Vescovi for critical reading of the manuscript.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (grant R01AI125363). B.S.W. is the recipient of Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship-Doctoral and Alberta Innovates-Technology Future awards and a Stephen I. Morse fellowship.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00778-17.
REFERENCES
- 1.Waksman G, Hultgren SJ. 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melican K, Sandoval RM, Kader A, Josefsson L, Tanner GA, Molitoris BA, Richter-Dahlfors A. 2011. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog 7:e1001298. doi: 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busch A, Waksman G. 2012. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci 367:1112–1122. doi: 10.1098/rstb.2011.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soto GE, Hultgren SJ. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol 181:1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 6.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 7.Pakharukova N, Garnett JA, Tuittila M, Paavilainen S, Diallo M, Xu Y, Matthews SJ, Zavialov AV. 2015. Structural insight into archaic and alternative chaperone-usher pathways reveals a novel mechanism of pilus biogenesis. PLoS Pathog 11:e1005269. doi: 10.1371/journal.ppat.1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pakharukova N, Tuittila M, Paavilainen S, Zavialov A. 2015. Crystallization and preliminary X-ray diffraction analysis of the Csu pili CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Acinetobacter baumannii. Acta Crystallogr F Struct Biol Commun 71:770–774. doi: 10.1107/S2053230X15007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. 2011. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett 323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 10.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. 2014. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber BS, Hennon SW, Wright MS, Scott NE, de Berardinis V, Foster LJ, Ayala JA, Adams MD, Feldman MF. 2016. Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 7:e01253-16. doi: 10.1128/mBio.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Breij A, Gaddy J, van der Meer J, Koning R, Koster A, van den Broek P, Actis L, Nibbering P, Dijkshoorn L. 2009. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC19606T to human airway epithelial cells and their inflammatory response. Res Microbiol 160:213–218. doi: 10.1016/j.resmic.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Suh JR, Herbig AK, Stover PJ. 2001. New perspectives on folate catabolism. Annu Rev Nutr 21:255–282. doi: 10.1146/annurev.nutr.21.1.255. [DOI] [PubMed] [Google Scholar]
- 15.Falagas ME, Vardakas KZ, Roussos NS. 2015. Trimethoprim/sulfamethoxazole for Acinetobacter spp.: a review of current microbiological and clinical evidence. Int J Antimicrob Agents 46:231–241. doi: 10.1016/j.ijantimicag.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Palomino JC, Martin A. 2016. The potential role of trimethoprim-sulfamethoxazole in the treatment of drug-resistant tuberculosis. Future Microbiol 11:539–547. doi: 10.2217/fmb.16.2. [DOI] [PubMed] [Google Scholar]
- 17.Tilley D, Law R, Warren S, Samis JA, Kumar A. 2014. CpaA a novel protease from Acinetobacter baumannii clinical isolates deregulates blood coagulation. FEMS Microbiol Lett 356:53–61. doi: 10.1111/1574-6968.12496. [DOI] [PubMed] [Google Scholar]
- 18.Niu C, Clemmer KM, Bonomo RA, Rather PN. 2008. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol 190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamidian M, Hall RM. 2017. Acinetobacter baumannii ATCC 19606 carries GIsul2 in a genomic island located in the chromosome. Antimicrob Agents Chemother 61:e01991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burchall JJ. 1973. Mechanism of action of trimethoprim-sulfamethoxazole. II. J Infect Dis 128(Suppl):437–441. [DOI] [PubMed] [Google Scholar]
- 23.Fox JT, Stover PJ. 2008. Folate-mediated one-carbon metabolism. Vitam Horm 79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 24.Cossins EA, Chen L. 1997. Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45:437–452. doi: 10.1016/S0031-9422(96)00833-3. [DOI] [PubMed] [Google Scholar]
- 25.Bermingham A, Derrick JP. 2002. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- 26.Schirch V. 1998. Mechanism of folate-requiring enzymes in one-carbon metabolism, p 211–252. In Sinnott M. (ed), Comprehensive biological catalysis, vol 1 Academic Press, New York, NY. [Google Scholar]
- 27.Basset GJC, Quinlivan EP, Gregory JF, Hanson AD. 2005. Folate synthesis and metabolism in plants and prospects for biofortification. Crop Sci 45:449–453. doi: 10.2135/cropsci2005.0449. [DOI] [Google Scholar]
- 28.Dean EA, Kessler RE. 1988. Quantitation of effects of subinhibitory concentrations of trimethoprim on P fimbria expression and in vitro adhesiveness of uropathogenic Escherichia coli. J Clin Microbiol 26:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang EY, Mohler AM, Rohlman CE. 1997. Protein expression in response to folate stress in Escherichia coli. J Bacteriol 179:5648–5653. doi: 10.1128/jb.179.17.5648-5653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowska E, Kuczynska-Wisnik D, Bak M, Lipinska B. 2003. Trimethoprim induces heat shock proteins and protein aggregation in E. coli cells. Curr Microbiol 47:286–289. doi: 10.1007/s00284-002-4007-z. [DOI] [PubMed] [Google Scholar]
- 31.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breines DM, Burnham JC. 1994. Modulation of Escherichia coli type 1 fimbrial expression and adherence to uroepithelial cells following exposure of logarithmic phase cells to quinolones at subinhibitory concentrations. J Antimicrob Chemother 34:205–221. doi: 10.1093/jac/34.2.205. [DOI] [PubMed] [Google Scholar]
- 33.Dhabaan GN, AbuBakar S, Cerqueira GM, Al-Haroni M, Pang SP, Hassan H. 2015. Imipenem treatment induces expression of important genes and phenotypes in a resistant Acinetobacter baumannii isolate. Antimicrob Agents Chemother 60:1370–1376. doi: 10.1128/AAC.01696-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorby GL, McGee ZA. 1990. Antimicrobial interference with bacterial mechanisms of pathogenicity: effect of sub-MIC azithromycin on gonococcal piliation and attachment to human epithelial cells. Antimicrob Agents Chemother 34:2445–2448. doi: 10.1128/AAC.34.12.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He X, Lu F, Yuan F, Jiang D, Zhao P, Zhu J, Cheng H, Cao J, Lu G. 2015. Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob Agents Chemother 59:4817–4825. doi: 10.1128/AAC.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A 99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abreu-Goodger C, Merino E. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res 33:W690–W692. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim PB, Nelson JW, Breaker RR. 2015. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell 57:317–328. doi: 10.1016/j.molcel.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jungermann KA, Schmidt W, Kirchniawy FH, Rupprecht EH, Thauer RK. 1970. Glycine formation via threonine and serine aldolase: its interrelation with the pyruvate formate lyase pathway of one-carbon unit synthesis in Clostridium kluyveri. Eur J Biochem 16:424–429. doi: 10.1111/j.1432-1033.1970.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 40.Locasale JW. 2013. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeld N, Bouchier C, Courvalin P, Perichon B. 2012. Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob Agents Chemother 56:2504–2510. doi: 10.1128/AAC.06422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.